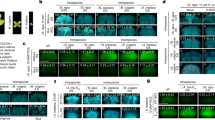
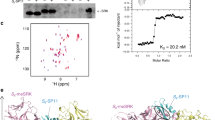
Abstract
Self-incompatibility and recurrent transitions to self-compatibility have shaped the extant mating systems underlying the nonrandom mating critical for speciation in angiosperms. Linkage between self-incompatibility and speciation is illustrated by the shared pollen rejection pathway between self-incompatibility and interspecific unilateral incompatibility (UI) in the Brassicaceae. However, the pollen discrimination system that activates this shared pathway for heterospecific pollen rejection remains unknown. Here we show that Stigma UI3.1, the genetically identified stigma determinant of UI in Arabidopsis lyrata × Arabidopsis arenosa crosses, encodes the S-locus-related glycoprotein 1 (SLR1). Heterologous expression of A. lyrata or Capsella grandiflora SLR1 confers on some Arabidopsis thaliana accessions the ability to discriminate against heterospecific pollen. Acquisition of this ability also requires a functional S-locus receptor kinase (SRK), whose ligand-induced dimerization activates the self-pollen rejection pathway in the stigma. SLR1 interacts with SRK and interferes with SRK homomer formation. We propose a pollen discrimination system based on competition between basal or ligand-induced SLR1–SRK and SRK–SRK complex formation. The resulting SRK homomer levels would be sensed by the common pollen rejection pathway, allowing discrimination among conspecific self- and cross-pollen as well as heterospecific pollen. Our results establish a mechanistic link at the pollen recognition phase between self-incompatibility and interspecific incompatibility.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the Article and the Supplementary Information. Source data are provided with this paper.
References
Cutter, A. D. Reproductive transitions in plants and animals: selfing syndrome, sexual selection and speciation. New Phytol. 224, 1080–1094 (2019).
Pickup, M. et al. Mating system variation in hybrid zones: facilitation, barriers and asymmetries to gene flow. New Phytol. 224, 1035–1047 (2019).
Bedinger, P. A., Broz, A. K., Tovar-Mendez, A. & McClure, B. Pollen–pistil interactions and their role in mate selection. Plant Physiol. 173, 79–90 (2017).
Nasrallah, J. B. Stop and go signals at the stigma–pollen interface of the Brassicaceae. Plant Physiol. 193, 927–948 (2023).
Charlesworth, D., Vekemans, X., Castric, V. & Glemin, S. Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytol. 168, 61–69 (2005).
Nasrallah, J. B. Self-incompatibility in the Brassicaceae: regulation and mechanism of self-recognition. Curr. Top. Dev. Biol. 131, 435–452 (2019).
Zhao, H. et al. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell 34, 579–596 (2022).
Goldberg, E. E. et al. Species selection maintains self-incompatibility. Science 330, 493–495 (2010).
Foxe, J. P. et al. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl Acad. Sci. USA 106, 5241–5245 (2009).
Kitashiba, H. & Nasrallah, J. B. Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed. Sci. 64, 23–37 (2014).
Hiscock, S. J. & Dickinson, H. G. Unilateral incompatibility within the Brassicaceae: further evidence for the involvement of the self-incompatibility (S)-locus. Theor. Appl. Genet. 86, 744–753 (1993).
Lewis, D. & Crowe, L. K. Unilateral interspecific incompatiility in flowering plants. Heredity 12, 233–256 (1958).
Murfett, J. et al. S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell 8, 943–958 (1996).
Li, W. & Chetelat, R. T. A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330, 1827–1830 (2010).
Li, L. et al. Evolution of interspecies unilateral incompatibility in the relatives of Arabidopsis thaliana. Mol. Ecol. 27, 2742–2753 (2018).
Huang, J. et al. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature 614, 303–308 (2023).
Pfennig, K. S. Reinforcement as an initiator of population divergence and speciation. Curr. Zool. 62, 145–154 (2016).
Li, W. & Chetelat, R. T. Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl Acad. Sci. USA 112, 4417–4422 (2015).
Martin, F. W. The genetic control of unilateral incompatibility between two tomato species. Genetics 56, 391–398 (1967).
Qin, X. et al. A farnesyl pyrophosphate synthase gene expressed in pollen functions in S-RNase-independent unilateral incompatibility. Plant J. 93, 417–430 (2018).
Udagawa, H. et al. Genetic analysis of interspecific incompatibility in Brassica rapa. Theor. Appl. Genet. 121, 689–696 (2010).
Broz, A. K. & Bedinger, P. A. Pollen-pistil interactions as reproductive barriers. Annu. Rev. Plant Biol. 72, 615–639 (2021).
Umbach, A. L., Lalonde, B. A., Kandasamy, M. K., Nasrallah, J. B. & Nasrallah, M. E. Immunodetection of protein glycoforms encoded by two independent genes of the self-incompatibility multigene family of Brassica. Plant Physiol. 93, 739–747 (1990).
Naithani, S., Chookajorn, T., Ripoll, D. R. & Nasrallah, J. B. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl Acad. Sci. USA 104, 12211–12216 (2007).
Lalonde, B. A. et al. A highly conserved Brassica gene with homology to the S-locus-specific glycoprotein structural gene. Plant Cell 1, 249–258 (1989).
Inaba, R. & Nishio, T. Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105, 1159–1165 (2002).
Franklin, T. M., Oldknow, J. & Trick, M. SLR1 function is dispensable for both self-incompatible rejection and self-compatible pollination processes in Brassica. Sex. Plant Reprod. 9, 203–208 (1996).
Luu, D. T., Marty-Mazars, D., Trick, M., Dumas, C. & Heizmann, P. Pollen-stigma adhesion in Brassica spp involves SLG and SLR1 glycoproteins. Plant Cell 11, 251–262 (1999).
Nasrallah, M. E., Liu, P., Sherman-Broyles, S., Boggs, N. A. & Nasrallah, J. B. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl Acad. Sci. USA 101, 16070–16074 (2004).
Tsuchimatsu, T. et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464, 1342–1346 (2010).
Liu, P., Sherman-Broyles, S., Nasrallah, M. E. & Nasrallah, J. B. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17, 734–740 (2007).
Sherman-Broyles, S. et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19, 94–106 (2007).
Rea, A. C., Liu, P. & Nasrallah, J. B. A transgenic self-incompatible Arabidopsis thaliana model for evolutionary and mechanistic studies of crucifer self-incompatibility. J. Exp. Bot. 61, 1897–1906 (2010).
Nasrallah, M. E., Liu, P. & Nasrallah, J. B. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297, 247–249 (2002).
Fujii, S. et al. A stigmatic gene confers interspecies incompatibility in the Brassicaceae. Nat. Plants 5, 731–741 (2019).
Murase, K. et al. Mechanism of self/nonself-discrimination in Brassica self-incompatibility. Nat. Commun. 11, 4916 (2020).
Ma, R. et al. Structural basis for specific self-incompatibility response in Brassica. Cell Res. 12, 1320–1329 (2016).
Giranton, J. L., Dumas, C., Cock, J. M. & Gaude, T. The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl Acad. Sci. USA 97, 3759–3764 (2000).
Shimosato, H. et al. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 19, 107–117 (2007).
Zhang, L. et al. FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol. 31, 3004–3016.e3004 (2021).
Fujii, S., Kubo, K. & Takayama, S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2, 16130 (2016).
Turelli, M. & Moyle, L. C. Asymmetric postmating isolation: Darwina’s corollary to Haldanea’s rule. Genetics 176, 1059–1088 (2007).
Bechtold, N., Ellis, J. & Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199 (1993).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Liu, H. et al. Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res. 18, 422–432 (2008).
Waadt, R. et al. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516 (2008).
Kubiasova, K. et al. Cytokinin fluoroprobe reveals multiple sites of cytokinin perception at plasma membrane and endoplasmic reticulum. Nat. Commun. 11, 4285 (2020).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Guo, Y. L., Zhao, X., Lanz, C. & Weigel, D. Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 157, 937–946 (2011).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Yang, Z., Wong, W. S. & Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 (2005).
Boggs, N. A., Nasrallah, J. B. & Nasrallah, M. E. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5, e1000426 (2009).
Tang, C. et al. The evolution of selfing in Arabidopsis thaliana. Science 317, 1070–1072 (2007).
Acknowledgements
We thank NASC and ABRC for providing A. thaliana, B. oleracea and B. rapa seeds; L. Comai (University of California Davis) for providing A. arenosa seeds; and J. Kudla (Universität Münster) for providing vectors for BIFC assays. This work was supported by the Natural Science Foundation of China (NSFC) (numbers 32170353, 32370234, 31970310 and 32100269), Major Research Plan from the Ministry of Science and Technology of China (number 2013CB945100) and Program for New Century Excellent Talents in University (NECT-08-0529) to P.L.
Author information
Authors and Affiliations
Contributions
B.L., M.L., W.L., Q.C. and H.Z. performed the construction of plasmids, transformation of A. thaliana, analyses of the progenies of the transformants and pollination assays. B.L. performed Co-IP. M.L. and J.Q. performed BIFC experiments. J.X. performed bioinformatic analyses. P.L. designed the study and wrote the paper. J.B.N., M.E.N. and Y.X. were involved in designing the experiments and writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Alignment of the protein sequences and domain architecture of SLR1 and eSRK from eight Arabidopsis, Capsella, and Brassica species.
The alignments of representative SLR1 sequences (upper panel) and eSRK + transmembrane (TM) sequences (lower panel) from A. lyrata (Al), A. halleri (Ah), A. thaliana (At), A. arenosa (Aa), C. grandiflora (Cg), B. rapa (Br), and B. oleracea (Bo) are shown. The domain architecture of SLR1 and eSRK proteins and their 12 conserved cysteine residues, which are characteristic of the so-called S-domain (SD) protein family, are shown above the alignments. Also shown for eSRKs are the hypervariable (hv) regions which contain most residues responsible for the binding of SRK with SCR (purple dots) and residues involved in SRK homodimerization (blue dots) as determined from the high-resolution crystal structure of the eSRK-SCR complex41.
Extended Data Fig. 2 The gene tree of SLR1, but not that of SRK proteins, is concordant with the species tree in representative Brassicaceae species.
The gene tree includes SLR1 and SRK proteins from representative Brassicaceae species. The blue filled triangles mark A. thaliana and B. rapa for which nineteen and four SLR1 alleles have been reported, respectively. To the right of the gene tree, the colored circles represent A. lyrata (Al; blue), A. halleri (Ah; azure), A. arenosa (Aa; yellow), A. thaliana (At; grey), C. grandiflora (Cg; pink), C. rubella (Cr; black), B. rapa (Br; red), and B. oleracea (Bo; purple), and the self-incompatible (SI) or self-compatible (SC) state of each species is indicated in parentheses. The species tree (in blue) was constructed by the coalesced sequences (3,778) of genome-wide single-copy orthogroups. The bootstrap values and genetic distance are shown.
Extended Data Fig. 3 The AlSLR1 transgene confers the ability to discriminate against A. arenosa pollen on the stigmas of the A. thaliana Old-1 accession.
Microscopic images of pollinated stigmas (a) and the number of pollen tubes per stigma (b) observed in a representative T1 Old-1:AlSR1 plant. The germination and tube growth of A. arenosa pollen observed on wild-type Old-1 stigmas were largely abolished in Old-1:AlSLR1 plants on stage-13 but not stage-14 stigmas. Thus, the interspecific incompatibility acquired by Old-1 was transient as for Wei-1 (Extended Data Table 1). The incompatibility response of Old-1:AlSLR1 was weaker than that of Wei-1:AlSLR1 (Figs. 2b and 2c), as Old-1:AlSLR1 stigmas supported the growth of a somewhat larger number pollen tubes on the stigma papilla cells. Scale bars, 50 μm. The dots indicate individual data points. Error bars are ± SEM, n (in cyan) indicates the number of stigmas. *P < 0.05 (two-tailed Students t-test). Each experiment was repeated at least three times with consistent results.
Extended Data Fig. 4 The AlSLR1 transgene does not confer the ability to discriminate against A. arenosa pollen on the stigmas of the A. thaliana Col-0, Ws, and Mt-0 accessions.
a,b,c The microscopic images to the left show that the germination and tube growth of A. arenosa pollen on A. thaliana stigmas were not significantly altered by expression of the AlSLR1 transgene in Col-0 (a), Ws (b), and Mt-0 (c). This conclusion was confirmed by the number of pollen tubes observed per stigma in 12, 13 and 14 stage flowers as shown in the graphs to the right. The results of manual pollinations are shown for a representative T1 transgenic plant of each of the three accessions. Similar results were also observed in AlSLR1 transformants of the RLD, No, Nd-0, and C24 accessions (see Extended Data Table 1, Figs. 3b and 3c). Scale bars, 50 μm. The dots indicate individual data points. Error bars are ± SEM, n (in cyan) indicates the number of stigmas. Each experiment was repeated at least three times with consistent results.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 4
Unprocessed gels for Fig. 4a.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, B., Li, M., Qiu, J. et al. A pollen selection system links self and interspecific incompatibility in the Brassicaceae. Nat Ecol Evol (2024). https://doi.org/10.1038/s41559-024-02399-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-024-02399-4