Abstract
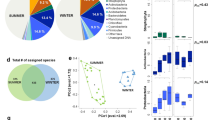
Grass pollen is the world’s most harmful outdoor aeroallergen. However, it is unknown how airborne pollen assemblages change across time and space. Human sensitivity varies between different species of grass that flower at different times, but it is not known whether temporal turnover in species composition match terrestrial flowering or whether species richness steadily accumulates over the grass pollen season. Here, using targeted, high-throughput sequencing, we demonstrate that all grass genera displayed discrete, temporally restricted peaks of incidence, which varied with latitude and longitude throughout Great Britain, revealing that the taxonomic composition of grass pollen exposure changes substantially across the grass pollen season.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All sequence data are available at the Sequence Read Archive (SRA) using the project accession number SUB4136142. Archived sequence data were used to generate Fig. 2 and Supplementary Figs. 1–6, 8–10). Data on first flowering dates used in Supplementary Fig. 5 were obtained from Nature’s Calendar, Woodland Trust and are available upon request. The sequence analysis pipeline is available at https://github.com/colford/nbgw-plant-illumina-pipeline.
References
Blackley, C. H. Experimental Researches on the Causes and Nature of Catarrhus Æstivus (hay-fever Or Hay-asthma) (Bailliere, 1873).
Marks, G., Pearce, N., Strachan, D. & Asher, I. The Global Asthma Report 2014 16–21 (Global Asthma Network, 2014).
Bauchau, V. Eur. Respir. J. 24, 758–764 (2004).
Bousquet, P.-J. et al. Allergy 62, 301–309 (2007).
García‐Mozo, H. Allergy 72, 1849–1858 (2017).
Emberlin, J. et al. Aerobiologia 16, 373–379 (2000).
Emberlin, J. et al. Grana 33, 94–99 (1994).
Mander, L., Li, M., Mio, W., Fowlkes, C. C. & Punyasena, S. W. Proc. R. Soc. Lond. B 280, 20131905 (2013).
Kmenta, M. et al. World Allergy Organ. J. 10, 31 (2017).
Cope, C. & Gray, A. Grasses of the British Isles (Botanical Society of the British Isles, 2009)..
Estrella, N., Menzel, A., Krämer, U. & Behrendt, H. Int. J. Biometeorol. 51, 49–59 (2006).
Skjøth, C. A., Sommer, J., Stach, A., Smith, M. & Brandt, J. Clin. Exp. Allergy 37, 1204–1212 (2008).
McInnes, R. N. et al. Sci. Total Environ. 599–600, 483–499 (2017).
van Ree, R., van Leeuwen, W. A. & Aalberse, R. C. J. Allergy Clin. Immunol. 102, 184–190 (1998).
Petersen, A. et al. J. Allergy Clin. Immunol. 107, 856–862 (2001).
Jung, S. et al. PLoS ONE 13, 1–12 (2018).
Moingeon, P., Peltre, G. & Bergmann, K.-C. Clin. Exp. Allergy Rev. 8, 12–14 (2008).
de Weger, L. A. et al. Clin. Transl. Allergy 1, 18 (2011).
Kraaijeveld, K. et al. Mol. Ecol. Resour. 15, 8–16 (2015).
Korpelainen, H. & Pietiläinen, M. Nord. J. Bot. 35, 602–608 (2017).
Deiner, K. et al. Mol. Ecol. 26, 5872–5895 (2017).
Creer, S. et al. Methods Ecol. Evol. 7, 1008–1018 (2016).
de Vere, N. et al. PLoS ONE 7, e37945 (2012).
England, P. H. GP In-Hours Consultations Bulletin: 25 August 2016 Week 33 (PHE Real-time Syndromic Surveillance Team, 2016).
Frame, J. & Laidlaw, A. S. Improved Grassland Management (The Crowood Press, 2011).
RGCL. Recommended Grass and Clover Lists (British Grassland Society, 2017).
Rousseau, D.-D. et al. Geophys. Res. Lett. 30, 1765 (2003).
D'Amato, G. et al. Allergy 62, 976–990 (2007).
West, J. S. & Kimber, R. B. E. Ann. Appl. Biol. 166, 4–17 (2015).
Hirst, J. M. Ann. App. Biol. 39, 257–265 (1952).
Adams-Groom, B., Emberlin, J., Corden, J., Millington, W. & Mullins, J. Aerobiologia 18, 117–123 (2002).
Skjøth, C. A., Baker, P., Sadyś, M. & Adams-Groom, B. Urban Climate 14, 414–428 (2015).
Galán, C. et al. Aerobiologia 33, 293–295 (2017).
Hawkins, J. et al. PLoS ONE 10, e0134735 (2015).
Kress, W. J. & Erickson, D. L. PLoS ONE 2, e508 (2007).
Miya, M. et al. R. Soc. Open Sci. 2, 150088 (2015).
de Vere, N. et al. Sci. Rep. 7, 42838 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Bioinformatics 30, 2114–2120 (2014).
Magoč, T. & Salzberg, S. L. Bioinformatics 27, 2957–2963 (2011).
Stace, C. New Flora of the British Isles (Cambridge Univ. Press, 2010).
Camacho, C. et al. BMC Bioinformatics 10, 421 (2008).
Wang, Y., Naumann, U., Wright, S. T. & Warton, D. I. Methods Ecol. Evol. 3, 471–474 (2012).
McMurdie, P. J. & Holmes, S. PLoS Comput. Biol. 10, e1003531 (2014).
Dixon, P. J. Veg. Sci. 14, 927–930 (2003).
Acknowledgements
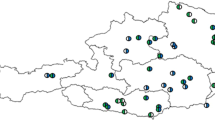
We thank J. Kenny, P. Koldkjær, R. Gregory and A. Lucaci for sequencing support; J. Winn for ArcGIS assistance with Fig. 1; W. Grail and the technical support staff at Bangor University; the Botanic Gardens Conservation International (BGCI) for access to the list of plant collections in the National Gardens in the United Kingdom and Ireland; the Met Office network for providing additional observational grass pollen count data; the Woodland Trust and the Centre for Ecology & Hydrology for supplying the UK Phenology Network data and the citizen scientists who have contributed to the latter scheme. We acknowledge the computational services and support of the Supercomputing Wales project, which is part-funded by the European Regional Development Fund (ERDF) via Welsh Government. This work was supported by the Natural Environment Research Council (https://nerc.ukri.org/), awarded to S.C. (NE/N003756/1), C.A.S. (NE/N002431/1), N.J.O. (NE/N002105/1), and G.W.G., N.d.V. and M.H. (NE/N001710/1). IBERS Aberystwyth receives strategic funding from the BBSRC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
S.C., N.d.V., G.W.G., R.N.M., N.J.O., C.A.S., Y.C., B.W.W. and G.L.B. conceived and designed the study; B.A.-G., G.L.B., G.M.P., A.E., R.N., S.P., K.S. and N.S. collected samples and counted pollen; G.L.B. performed laboratory work, supported by S.C.; N.d.V., C.P., C.R.F., L.J., G.L.B and S.C. contributed methods; C.A. and D.B.R. contributed materials; C.P. and G.L.B. analysed the data; and G.L.B., C.P. and S.C. produced the first draft of the manuscript. All authors contributed substantially to the final submitted manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary Figures 1–10, Supplementary Tables 1–6 and Supplementary References
Rights and permissions
About this article
Cite this article
Brennan, G.L., Potter, C., de Vere, N. et al. Temperate airborne grass pollen defined by spatio-temporal shifts in community composition. Nat Ecol Evol 3, 750–754 (2019). https://doi.org/10.1038/s41559-019-0849-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0849-7
This article is cited by
-
The role of automatic pollen and fungal spore monitoring across major end-user domains
Aerobiologia (2024)
-
Using qPCR and microscopy to assess the impact of harvesting and weather conditions on the relationship between Alternaria alternata and Alternaria spp. spores in rural and urban atmospheres
International Journal of Biometeorology (2023)
-
Summer pollen flora in rural and urban central England dominated by nettle, ryegrass and other pollen missed by the national aerobiological network
Aerobiologia (2022)
-
DNA metabarcoding identifies urban foraging patterns of oligolectic and polylectic cavity-nesting bees
Oecologia (2022)
-
Environmental DNA reveals diversity and abundance of Alternaria species in neighbouring heterogeneous landscapes in Worcester, UK
Aerobiologia (2022)