Abstract
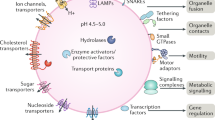
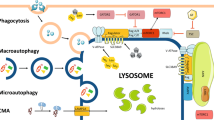
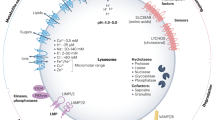
Lysosomes are catabolic organelles that govern numerous cellular processes, including macromolecule degradation, nutrient signalling and ion homeostasis. Aberrant changes in lysosome abundance are implicated in human diseases. Here we outline the mechanisms of lysosome biogenesis and turnover, and discuss how changes in the lysosome pool impact physiological and pathophysiological processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Liu, G. Y. & Sabatini, D. M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020).
Lawrence, R. E. & Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019).
Kim, J., Kundu, M., Viollet, B. & Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Yim, W. W.-Y. & Mizushima, N. Lysosome biology in autophagy. Cell Discov. 6, 6 (2020).
Kim, J. & Guan, K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019).
de Araujo, M. E. G., Liebscher, G., Hess, M. W. & Huber, L. A. Lysosomal size matters. Traffic 21, 60–75 (2020).
Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2020).
Napolitano, G. & Ballabio, A. TFEB at a glance. J. Cell Sci. 129, 2475–2481 (2016).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009).
Settembre, C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Martina, J. A., Chen, Y., Gucek, M. & Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012).
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012).
Vega-Rubin-de-Celis, S., Peña-Llopis, S., Konda, M. & Brugarolas, J. Multistep regulation of TFEB by MTORC1. Autophagy 13, 464–472 (2017).
Napolitano, G. et al. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat. Commun. 9, 3312 (2018).
Martínez-Fábregas, J. et al. Lysosomal protease deficiency or substrate overload induces an oxidative-stress mediated STAT3-dependent pathway of lysosomal homeostasis. Nat. Commun. 9, 5343 (2018).
Sakamaki, J.-I. et al. Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell 66, 517–532 (2017).
Annunziata, I. et al. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat. Commun. 10, 3623 (2019).
Ravichandran, M. et al. Coordinated transcriptional and catabolic programs support iron-dependent adaptation to RAS–MAPK pathway inhibition in pancreatic cancer. Cancer Discov. 12, 2198–2219 (2022).
Chauhan, S. et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell 50, 16–28 (2013).
Shingu, T. et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat. Genet. 49, 75–86 (2017).
Yang, C. & Wang, X. Lysosome biogenesis: regulation and functions. J. Cell Biol. 220, e202102001 (2021).
Coutinho, M. F., Prata, M. J. & Alves, S. Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol. Genet. Metab. 105, 542–550 (2012).
Saftig, P. & Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 (2009).
Johnson, D. E., Ostrowski, P., Jaumouillé, V. & Grinstein, S. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212, 677–692 (2016).
Medina, D. L. et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430 (2011).
Mutvei, A. P. et al. Rap1-GTPases control mTORC1 activity by coordinating lysosome organization with amino acid availability. Nat. Commun. 11, 1416 (2020).
Rabanal-Ruiz, Y. et al. mTORC1 activity is supported by spatial association with focal adhesions. J. Cell Biol. 220, e202004010 (2021).
Pu, J., Guardia, C. M., Keren-Kaplan, T. & Bonifacino, J. S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129, 4329–4339 (2016).
Yin, Q. et al. CDK4/6 regulate lysosome biogenesis through TFEB/TFE3. J. Cell Biol. 219, e201911036 (2020).
Hung, Y.-H., Chen, L. M.-W., Yang, J.-Y. & Yang, W. Y. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 4, 2111 (2013).
Wang, L., Klionsky, D. J. & Shen, H.-M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 24, 186–203 (2022).
Thurston, T. L. M., Wandel, M. P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012).
Maejima, I. et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 (2013).
Aits, S. & Jäättelä, M. Lysosomal cell death at a glance. J. Cell Sci. 126, 1905–1912 (2013).
Bussi, C. et al. Lysosomal damage drives mitochondrial proteome remodelling and reprograms macrophage immunometabolism. Nat. Commun. 13, 7338 (2022).
Yoshida, Y. et al. Ubiquitination of exposed glycoproteins by SCFFBXO27 directs damaged lysosomes for autophagy. Proc. Natl Acad. Sci. USA 114, 8574–8579 (2017).
Chauhan, S. et al. TRIMs and galectins globally cooperate and TRIM16 and Galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell 39, 13–27 (2016).
Papadopoulos, C., Kravic, B. & Meyer, H. Repair or lysophagy: dealing with damaged lysosomes. J. Mol. Biol. 432, 231–239 (2020).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Luzio, J. P., Hackmann, Y., Dieckmann, N. M. G. & Griffiths, G. M. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6, a016840 (2014).
Bright, N. A., Gratian, M. J. & Luzio, J. P. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15, 360–365 (2005).
Jahreiss, L., Menzies, F. M. & Rubinsztein, D. C. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 9, 574–587 (2008).
McNally, E. K., Karim, M. A. & Brett, C. L. Selective lysosomal transporter degradation by organelle membrane fusion. Dev. Cell 40, 151–167 (2017).
Golden, C. K. et al. A two-tiered system for selective receptor and transporter protein degradation. PLoS Genet. 18, e1010446 (2022).
Yang, X. et al. ESCRT, not intralumenal fragments, sorts ubiquitinated vacuole membrane proteins for degradation. J. Cell Biol. 220, e202012104 (2021).
Zhang, W. et al. A conserved ubiquitin- and ESCRT-dependent pathway internalizes human lysosomal membrane proteins for degradation. PLoS Biol. 19, e3001361 (2021).
Oku, M. et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216, 3263–3274 (2017).
Zhu, L., Jorgensen, J. R., Li, M., Chuang, Y.-S. & Emr, S. D. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife 6, e26403 (2017).
He, C.-W. et al. Membrane recruitment of Atg8 by Hfl1 facilitates turnover of vacuolar membrane proteins in yeast cells approaching stationary phase. BMC Biol. 19, 117 (2021).
Lee, C., Lamech, L., Johns, E. & Overholtzer, M. Selective lysosome membrane turnover is induced by nutrient starvation. Dev. Cell 55, 289–297 (2020).
Andrews, N. W. & Corrotte, M. Plasma membrane repair. Curr. Biol. 28, R392–R397 (2018).
Glunde, K. et al. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia 5, 533–545 (2003).
Davidson, S. M. & Vander Heiden, M. G. Critical functions of the lysosome in cancer biology. Annu. Rev. Pharmacol. Toxicol. 57, 481–507 (2017).
Xu, J. et al. Chloroquine treatment induces secretion of autophagy-related proteins and inclusion of Atg8-family proteins in distinct extracellular vesicle populations. Autophagy 18, 2547–2560 (2022).
Minakaki, G. et al. Autophagy inhibition promotes SNCA/α-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 14, 98–119 (2018).
Solvik, T. A. et al. Secretory autophagy maintains proteostasis upon lysosome inhibition. J. Cell Biol. 221, e202110151 (2022).
Radulovic, M. et al. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, e99753 (2018).
Skowyra, M. L., Schlesinger, P. H., Naismith, T. V. & Hanson, P. I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018).
Eriksson, I., Wäster, P. & Öllinger, K. Restoration of lysosomal function after damage is accompanied by recycling of lysosomal membrane proteins. Cell Death Dis. 11, 370 (2020).
Tan, J. X. & Finkel, T. A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature 609, 815–821 (2022).
Radulovic, M. et al. Cholesterol transfer via endoplasmic reticulum contacts mediates lysosome damage repair. EMBO J. 41, e112677 (2022).
Bhattacharya, A. et al. A lysosome membrane regeneration pathway depends on TBC1D15 and autophagic lysosomal reformation proteins. Nat. Cell Biol. 25, 685–698 (2023).
Nakamura, S. et al. LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat. Cell Biol. 22, 1252–1263 (2020).
Jia, J. et al. Galectins control mTOR in response to endomembrane damage. Mol. Cell 70, 120–135 (2018).
Cui, Z. et al. Structure of the lysosomal mTORC1–TFEB–Rag–Ragulator megacomplex. Nature 614, 572–579 (2023).
Napolitano, G., Di Malta, C. & Ballabio, A. Non-canonical mTORC1 signaling at the lysosome. Trends Cell Biol. 32, 920–931 (2022).
Napolitano, G. et al. A substrate-specific mTORC1 pathway underlies Birt–Hogg–Dubé syndrome. Nature 585, 597–602 (2020).
Goodwin, J. M. et al. GABARAP sequesters the FLCN–FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis. Sci. Adv. 7, eabj2485 (2021).
Lawrence, R. E. et al. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 366, 971–977 (2019).
Tsun, Z.-Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013).
SA, N. et al. Induction of lysosomal and mitochondrial biogenesis by AMPK phosphorylation of FNIP1. Science 380, eabj5559 (2023).
Wang, S. et al. Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis. Autophagy 15, 1954–1969 (2019).
Settembre, C. et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647–658 (2013).
McGrath, M. J. et al. Defective lysosome reformation during autophagy causes skeletal muscle disease. J. Clin. Invest. 131, e135124 (2021).
Martina, J. A., Diab, H. I., Brady, O. A. & Puertollano, R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35, 479–495 (2016).
Yorimitsu, T., Nair, U., Yang, Z. & Klionsky, D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 (2006).
Korolchuk, V. I. et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 (2011).
Li, X. et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016).
Mejlvang, J. et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 217, 3640–3655 (2018).
Medina, D. L. et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 (2015).
Wang, W. et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl Acad. Sci. USA 112, E1373–E1381 (2015).
Bandyopadhyay, U. et al. Leucine retention in lysosomes is regulated by starvation. Proc. Natl Acad. Sci. USA 119, e2114912119 (2022).
Abu-Remaileh, M. et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017).
Buel, G. R., Dang, H., Asara, J. M., Blenis, J. & Mutvei, A. P. Prolonged deprivation of arginine or leucine induces PI3K/Akt-dependent reactivation of mTORC1. J. Biol. Chem. 298, 102030 (2022).
Germic, N., Frangez, Z., Yousefi, S. & Simon, H.-U. Regulation of the innate immune system by autophagy: neutrophils, eosinophils, mast cells, NK cells. Cell Death Differ. 26, 703–714 (2019).
Clarke, A. J. & Simon, A. K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 19, 170–183 (2019).
Levine, B., Mizushima, N. & Virgin, H. W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Koo, I. C. et al. Role for lysosomal enzyme beta-hexosaminidase in the control of mycobacteria infection. Proc. Natl Acad. Sci. USA 105, 710–715 (2008).
Watts, C. Lysosomes and lysosome-related organelles in immune responses. FEBS Open Bio. 12, 678–693 (2022).
Gray, M. A. et al. Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr. Biol. 26, 1955–1964 (2016).
Bretou, M. et al. Lysosome signaling controls the migration of dendritic cells. Sci. Immunol. 2, eaak9573 (2017).
Samie, M. & Cresswell, P. The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat. Immunol. 16, 729–736 (2015).
Pastore, N. et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12, 1240–1258 (2016).
Tattoli, I. et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012).
Visvikis, O. et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896–909 (2014).
Mohamud, Y. et al. Coxsackievirus B3 targets TFEB to disrupt lysosomal function. Autophagy 17, 3924–3938 (2021).
Campbell, G. R., Rawat, P., Bruckman, R. S. & Spector, S. A. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog. 11, e1005018 (2015).
Villegas, F. et al. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell 24, 257–270 (2019).
Young, N. P. et al. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 30, 535–552 (2016).
Marks, H. et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604 (2012).
Xu, Y. & Yang, X. Autophagy and pluripotency: self-eating your way to eternal youth. Trends Cell Biol. 32, 868–882 (2022).
Gong, J. et al. Phosphorylation of ULK1 by AMPK is essential for mouse embryonic stem cell self-renewal and pluripotency. Cell Death Dis. 9, 38 (2018).
Tan, A., Prasad, R. & Jho, E.-H. TFEB regulates pluripotency transcriptional network in mouse embryonic stem cells independent of autophagy-lysosomal biogenesis. Cell Death Dis. 12, 343 (2021).
Betschinger, J. et al. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153, 335–347 (2013).
Yuizumi, N. et al. Maintenance of neural stem-progenitor cells by the lysosomal biosynthesis regulators TFEB and TFE3 in the embryonic mouse telencephalon. Stem Cells 39, 929–944 (2021).
Kobayashi, T. et al. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat. Commun. 10, 5446 (2019).
García-Prat, L. et al. TFEB-mediated endolysosomal activity controls human hematopoietic stem cell fate. Cell Stem Cell 28, 1838–1850 (2021).
Mortensen, M. et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208, 455–467 (2011).
Ho, T. T. et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017).
García-Prat, L. et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016).
Bohl, B., Jabali, A., Ladewig, J. & Koch, P. Asymmetric Notch activity by differential inheritance of lysosomes in human neural stem cells. Sci. Adv. 8, eabl5792 (2022).
Loeffler, D. et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 573, 426–429 (2019).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Aman, Y. et al. Autophagy in healthy aging and disease. Nat. Aging 1, 634–650 (2021).
Liang, R. et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell 26, 359–376 (2020).
Perera, R. M., Di Malta, C. & Ballabio, A. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu. Rev. Cancer Biol. 3, 203–222 (2019).
Kawakami, A. & Fisher, D. E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Invest. 97, 649–656 (2017).
Kauffman, E. C. et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat. Rev. Urol. 11, 465–475 (2014).
Argani, P. et al. Primary renal neoplasms with the ASPL–TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 159, 179–192 (2001).
Perera, R. M. et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365 (2015).
Alesi, N. et al. TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat. Commun. 12, 4245 (2021).
Asrani, K. et al. An mTORC1-mediated negative feedback loop constrains amino acid-induced FLCN–Rag activation in renal cells with TSC2 loss. Nat. Commun. 13, 6808 (2022).
Shin, H. R. & Zoncu, R. The lysosome at the intersection of cellular growth and destruction. Dev. Cell 54, 226–238 (2020).
Kimmelman, A. C. & White, E. Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 (2017).
Di Malta, C. et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science 356, 1188–1192 (2017).
Ploper, D. et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl Acad. Sci. USA 112, E420–E429 (2015).
Calcagnì, A. et al. Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. eLife 5, e17047 (2016).
Zhitomirsky, B. & Assaraf, Y. G. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget 6, 1143–1156 (2015).
Fassl, A. et al. Increased lysosomal biomass is responsible for the resistance of triple-negative breast cancers to CDK4/6 inhibition. Sci. Adv. 6, eabb2210 (2020).
Zhitomirsky, B. & Assaraf, Y. G. Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget 8, 45117–45132 (2017).
Kundu, S. T. et al. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat. Commun. 9, 2731 (2018).
Yun, S. et al. TFEB links MYC signaling to epigenetic control of myeloid differentiation and acute myeloid leukemia. Blood Cancer Discov. 2, 162–185 (2021).
Bonam, S. R., Wang, F. & Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 18, 923–948 (2019).
Parenti, G., Medina, D. L. & Ballabio, A. The rapidly evolving view of lysosomal storage diseases. EMBO Mol. Med. 13, e12836 (2021).
Settembre, C. et al. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 17, 119–129 (2008).
Lapierre, L. R. et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013).
Hughes, A. L. & Gottschling, D. E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 (2012).
Dobrowolski, R. et al. Presenilin deficiency or lysosomal inhibition enhances Wnt signaling through relocalization of GSK3 to the late-endosomal compartment. Cell Rep. 2, 1316–1328 (2012).
Yap, C. C., Digilio, L., McMahon, L. P., Garcia, A. D. R. & Winckler, B. Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J. Cell Biol. 217, 3141–3159 (2018).
Cheng, X.-T. et al. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217, 3127–3139 (2018).
Overly, C. C. & Hollenbeck, P. J. Dynamic organization of endocytic pathways in axons of cultured sympathetic neurons. J. Neurosci. 16, 6056–6064 (1996).
Thottacherry, J. J., Sathe, M., Prabhakara, C. & Mayor, S. Spoiled for choice: diverse endocytic pathways function at the cell surface. Annu. Rev. Cell Dev. Biol. 35, 55–84 (2019).
Hu, Y.-B., Dammer, E. B., Ren, R.-J. & Wang, G. The endosomal–lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 4, 18 (2015).
Demetriades, C., Plescher, M. & Teleman, A. A. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat. Commun. 7, 10662 (2016).
Menon, S. et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014).
Meng, D. et al. ArfGAP1 inhibits mTORC1 lysosomal localization and activation. EMBO J. 40, e106412 (2021).
Acknowledgements
We thank F. R. Maxfield and T. E. McGraw for helpful discussions. This work was supported by grants from the Swedish Research Council (grant no. 2020-01126), Swedish Cancer Society (grant nos 21 0437 JIA and 20 1188 Pj), Åke Wibergs Foundation, Lars Hierta’s Memory Foundation and Magnus Bergvall Foundation to A.P.M.; and National Institutes of Health grant no. R01 GM051405 to J.B. We apologize to those whose work was not included in this review due to limitations in scope and space.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Andrea Ballabio and Xiaochen Wang for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mutvei, A.P., Nagiec, M.J. & Blenis, J. Balancing lysosome abundance in health and disease. Nat Cell Biol 25, 1254–1264 (2023). https://doi.org/10.1038/s41556-023-01197-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-023-01197-7