Abstract
Macrophages present a spectrum of phenotypes that mediate both the pathogenesis and resolution of atherosclerotic lesions. Inflammatory macrophage phenotypes are pro-atherogenic, but the stimulatory factors that promote these phenotypes remain incompletely defined. Here we demonstrate that microbial small RNAs (msRNA) are enriched on low-density lipoprotein (LDL) and drive pro-inflammatory macrophage polarization and cytokine secretion via activation of the RNA sensor toll-like receptor 8 (TLR8). Removal of msRNA cargo during LDL re-constitution yields particles that readily promote sterol loading but fail to stimulate inflammatory activation. Competitive antagonism of TLR8 with non-targeting locked nucleic acids was found to prevent native LDL-induced macrophage polarization in vitro, and re-organize lesion macrophage phenotypes in vivo, as determined by single-cell RNA sequencing. Critically, this was associated with reduced disease burden in distinct mouse models of atherosclerosis. These results identify LDL-msRNA as instigators of atherosclerosis-associated inflammation and support alternative functions of LDL beyond cholesterol transport.
Similar content being viewed by others
Main
Atherosclerosis is a pathophysiological process driven by dyslipidaemia and unchecked chronic inflammation1,2. Pharmacological lowering of low-density lipoprotein (LDL)-C levels is a primary clinical target to reduce the risk of cardiovascular disease (CVD); yet, residual risk remains for many with properly managed plasma lipid levels3. Results of the CANTOS trial support that targeting inflammation—monoclonal antibody (canakinumab) against interleukin 1β (IL1β)—in the absence of reducing plasma lipids is a viable approach to mitigating cardiovascular events4. Nonetheless, inhibiting facilitators of systemic immune responses (that is, cytokines) may be problematic for patients at risk of infection. A more desirable approach may be to target upstream, inciting stimuli that promote atherosclerosis-associated inflammation; however, identification of such stimulating factors has proven elusive.
Macrophages have long been the focus of atherosclerosis research owing to their ability to enhance inflammation and recruit immune cells, as well as their propensity to become engorged with cholesterol to yield foam cells5. Macrophage scavenger receptor-mediated uptake of modified (for example, oxidized) LDL is widely regarded as a crucial event in foam cell formation within atheromas6. The importance of scavenger receptors and the impact of LDL modifications have been demonstrated by genetic studies in mice7,8,9,10 and coordinated signalling between scavenger receptors and pro-inflammatory cell signalling in macrophages has been reported in atherosclerosis11,12. Nevertheless, the inability of anti-oxidation therapies to improve clinical outcomes in CVD has introduced questions as to whether LDL oxidation is a cause or consequence of atherosclerosis13,14. Furthermore, single-cell RNA-sequencing (scRNA-seq) studies have found that transcriptomes of lipid-loaded foam cells within the atheroma are not associated with inflammatory phenotypes15,16,17, supporting earlier studies that activation of sterol-sensing machinery suppresses inflammatory gene expression18. On the contrary, other groups have reported that native LDL (nLDL) promote pro-inflammatory macrophage phenotypes and cytokine secretion from macrophages19,20. Moreover, small dense LDL particles, which carry reduced sterol cargo, are associated with increased risk of CVD21. Collectively, these studies indicate that a cholesterol-independent mechanism may link nLDL to pro-inflammatory macrophage phenotypes in the lesion and the chronic inflammation that underlies atherosclerotic CVD (ASCVD). In this Article, we provide evidence that nLDL transports microbial small RNA (msRNA) that propagate atherosclerosis through activation of toll-like receptor 8 (TLR8) and subsequent inflammatory macrophage polarization.
Results
nLDL induces inflammatory activation in macrophages
nLDL particles can accumulate in human atherosclerotic lesions to concentrations >2 mg ml−1, prompting foam cell formation through fluid-phase pinocytosis22,23,24. Nevertheless, the impact of nLDL on the macrophage phenotype is incompletely defined. We exposed phorbol 12-myristate 13-acetate (PMA)-differentiated human THP-1 macrophages to human nLDL (0.5 mg ml−1) and observed 365 upregulated genes and 141 downregulated genes using high-throughput RNA sequencing (RNA-seq; Fig. 1a and Supplementary Table 1). Using a network analysis, many of these genes were linked to transcription factors nuclear factor kappa B (NF-κB) and CCAAT/enhancer binding protein beta (C/EBPβ), which are established regulators of inflammation and atherosclerosis (Fig. 1b,c and Supplementary Tables 2 and 3). We validated dose- and time-dependent nLDL-induced changes to NF-κB and C/EBPβ transcriptional networks by quantitative polymerase chain reaction (qPCR) for messenger RNA levels and multi-plex enzyme-linked immunosorbent assay (ELISA) for secreted cytokines (Fig. 1d,e and Extended Data Fig. 1a,b). nLDL treatments resulted in accumulation of apolipoprotein B (APOB), the essential structural protein of LDL, within macrophages and the formation of lipid droplets, as evidenced by lipid-droplet protein perilipin 2 (PLIN2) (Fig. 1f and Extended Data Fig. 1c), observations that are supported by previous studies24,25. Intracellular increases of IL-1β protein and inflammatory cell signalling factors, inhibitor of NF-κB kinase subunit beta (IKKβ) and NF-κB inhibitor alpha (IκBα)—transcriptional targets of NF-κB—further validated RNA-seq results (Fig. 1f). Although circulating monocytes have been found to internalize LDL26, we found that nLDL treatment did not stimulate expression of IL1B, IL6 (encoding interleukin-6) or TNF (encoding tumour necrosis factor α) from freshly isolated human CD14+ monocytes (Fig. 1g). However, when monocytes were differentiated with granulocyte–macrophage colony-stimulating factor (GM-CSF) and interferon gamma (IFNγ) towards a pro-inflammatory macrophage phenotype, nLDL induced both IL1B and IL6, comparable to results observed in THP-1 macrophages. Intriguingly, primary monocytes differentiated to alternatively activated macrophages (macrophage colony-stimulating factor (M-CSF), IL-4 and IL-13) did not induce IL1B or IL6 expression in response to nLDL, but repressed TNF (Fig. 1g). Subsequent experiments with independent donors confirmed GM-CSF/IFNγ-differentiated primary macrophages were activated by both time- and dose-dependent nLDL treatments (Fig. 1h–k). Similar results were obtained by treating primary mouse bone-marrow-derived macrophages (BMDMs) with nLDL (Extended Data Fig. 1d–f). Collectively, these data support that nLDL activates macrophage cytokine expression and secretion, probably through NF-κB.
a, Differentially expressed transcripts (mRNA) of THP-1 macrophages treated with 0.5 mg ml−1 nLDL for 24 h identified by RNA-seq (n = 3 biological replicates). FDR (α = 0.05)-adjusted P values after Wald test. log2FC >1.0, adjusted P < 0.05 (P adj). b, Transcription factor network enrichment analysis of nLDL-altered genes. Benjamini–Hochberg multiple testing correction adjusted P values generated based on hypergeometric distribution. Adjusted P < 0.05. c, Differentially expressed transcripts of a within the RelA (NF-κB p65) cluster. d,e, mRNA expression by qPCR (relative quantitative value (RQV)) (d) and secreted cytokine levels by ELISA of selected targets of NF-κB and C/EBPβ following exposure to 0.5 mg ml−1 nLDL for 3, 6 and 24 h relative to cells receiving no treatment for 24 h (control (Ctr)) (n = 3 biological replicates) (e). f, Immunoblots of selected proteins of THP-1 cells exposed to varied doses of nLDL for 24 h (n = 3 biological replicates). g, mRNA expression of primary human CD14+ cells from peripheral blood either freshly isolated or differentiated with GM-CSF and IFNγ, or M-CSF, IL-4 and IL-13, and treated with 0.5 mg mL−1 nLDL for 24 h (n = 3 healthy human donors). h–j, mRNA expression (h and j) and cytokine secretion (i and k) of primary human CD14+ PBMC differentiated with GM-CSF and IFNγ following treatment with nLDL for indicated time (h and i) or dose (j and k) (n = 4 biological replicates). Data are mean ± s.e.m. In d, e, h and j, a one-way ANOVA, Benjamini, Krieger and Yekutieli FDR (q = 0.05) for experimental groups relative to Ctr. *q < 0.05, **q < 0.01. In g, a two-way ANOVA or Sidak’s multiple comparison test was performed, *P < 0.05, **P < 0.01. In i and k, one-way ANOVA and Dunnett’s multiple comparison test on experimental groups relative to Ctr group were carried out, *P < 0.05, **P < 0.01. Numerical source data, statistics, exact P values and q values are provided.
nLDL activates the RNA sensor TLR8 in macrophages
Macrophage toll-like receptors (TLR) facilitate innate immunity by recognizing pathogen-associated molecular patterns (PAMPs), which have been extensively studied in atherosclerosis27. Extracellular PAMPs are detected by surface TLRs (TLR1, -2, -4, -5 and -6), whereas intracellular PAMPs (that is, nucleic acids) are recognized by endosomal TLRs (TLR3, -7, -8 and -9). Surface TLR2, TLR4 and TLR6 have been directly implicated in the pathogenesis of atherosclerosis through their recognition of modified LDL particles (for example, oxidized LDL) and lipoprotein (a) (Lp(a)), which has similar density to nLDL and is a natural source of oxidized phospholipids11,28. Although LDL preparations from healthy donors were consistent with nLDL (Extended Data Fig. 2a–c), it is possible that our preparations could contain Lp(a) and/or oxidized phospholipid cargo capable of signalling through TLR2; however, chemical inhibition of TLR2 with C29 (ref. 29) failed to blunt nLDL-induced cytokine transcription (Fig. 2a) or secretion of IL-6 (Fig. 2b). Furthermore, nLDL treatments of primary macrophages failed to upregulate HMOX1 (Fig. 2a), a key gene of the Mox macrophage phenotype driven by exposure to oxidized phospholipids30. Conversely, cytokine and HMOX1 expression were robustly induced in response to the TLR2 ligand PAM3CSK4, which was abrogated by C29 (Extended Data Fig. 2d). The potential involvement of TLRs in nLDL-induced polarization was confirmed by chelation of intracellular calcium31, which abrogated secretion of TNF-α in response to nLDL, lipopolysaccharide (LPS; TLR4 ligand) and single-stranded RNA oligonucleotide (ssRNA40; TLR8 ligand; Fig. 2c). However, disruption of endo-lysosome maturation with bafilomycin A1 reduced TNF-α secretion in response to only nLDL− and ssRNA40, supporting the involvement of an endosomal receptor in nLDL-induced cytokine secretion. To further resolve this, individual TLRs were co-expressed in HEK293T cells with an NF-κB promoter-driven luciferase reporter and validated by treatment with TLR-specific ligands, or TNF-α for cells expressing no TLR (Fig. 2d). We observed activation of nLDL-driven NF-κB activity only in the presence of TLR8, an endosomal receptor that recognizes ssRNA and products of ssRNA catabolism32,33. Although TLR7 and TLR8 share some ligands, only TLR8 was required for nLDL-induced NF-κB activity (Extended Data Fig. 2e). The levels of TLR8 expression in HEK293T cells were found to be comparable to endogenous TLR8 expression levels observed in primary human and THP-1 macrophages, although it is suspected that high TLR8 turnover is present in THP-1 cells (Fig. 2e). Nevertheless, partial silencing of TLR8 with small interfering RNAs (siRNAs) blunted nLDL-induced expression and secretion of IL-6 and TNF-α in THP-1 macrophages (Fig. 2f,g). Intriguingly, siRNA against TLR7 blunted nLDL-induced cytokine expression in these cells nearly as effective as siRNA against TLR8, without effecting TLR8 expression (Extended Data Fig. 2f). However, THP-1 macrophages failed to induce a robust response to the TLR7 agonist R848, and partial silencing of TLR7 did not impede modest induction of IL1B (Extended Data Fig. 2g), bringing into question the functionality of TLR7 in these conditions. Conversely, silencing TLR8 expression strongly impaired the macrophage response to ssRNA40 (Extended Data Fig. 2h). We speculate that our siRNA results may be partially linked to off-target effects of siRNA on TLR8 activity34,35. To address this issue, we also selectively inhibited TLR8 activity with CU-CPT9a36, a selective chemical inhibitor of TLR8 activity, and we observed a significant reduction in cytokine expression and secretion in response to nLDL and ssRNA40 treatments (Fig. 2h,i). Thus, we conclude that TLR8 is necessary and sufficient for nLDL-induced activation of NF-κB in human macrophages.
a,b, mRNA expression by qPCR (a) and IL-6 secretion (b) of primary human CD14+ macrophages differentiated with GM-CSF and IFNγ that were pre-treated with vehicle (Veh; DMSO) or C29 for 30 min and then incubated with nLDL or without (Ctr) for 24 h (n = 4 biological replicates). c, TNF-α secretion of THP-1 macrophages pre-treated with vehicle (Veh; DMSO), bafilomycin A1 (Baf-A1; 500 µM) or EGTA-AM for 45 min, followed by exposure to LPS (100 ng ml−1), ssRNA40 (1 μg ml−1), nLDL (0.5 mg ml−1) or untreated (Ctr) for 6 h (n = 3 biological replicates). d, Normalized NF-κB-driven luciferase activity of HEK293T cells over expressing an empty vector, individual or tandem human TLRs following treatments with vehicle (Ctr), cognate TLR ligand(s) or nLDL (0.5 mg ml−1) (n = 3 biological replicates). e, Immunoblots of HEK293T cells transfected with the indicated plasmids relative to endogenous expression in primary human macrophages or THP-1 macrophages (representative blot of three independent experiments). TLR8-O denotes full-length protein. TLR8-X represents proteolytically processed (active) peptides. f,g, mRNA expression (f) (n = 4 biological replicates) and cytokine secretion (g) (n = 3 biological replicates) of THP-1 macrophages electroporated with scrambled (siScr) or TLR8-targeted (siTLR8) siRNAs or without siRNA (Ctr) following 24 h of nLDL exposure (0.5 mg ml−1). h,i, mRNA expression (h) and cytokine secretion (i) of THP-1 macrophages pre-treated with CU-CPT9a microparticles (30 min) followed by 24 h of nLDL exposure (n = 3 biological replicates). Data are mean ± s.e.m. In a, b and i, two-way ANOVA and Sidak’s multiple comparisons test were performed relative to Ctr, *P < 0.05, **P < 0.01. In c and f–h, two-way ANOVA, Benjamini, Krieger and Yekutieli FDR were carried out (q = 0.05), *q < 0.05, **q < 0.01, ***q < 0.001. Numerical source data, statistics, exact P values and q values are provided.
The functionality of TLR8 in mice has been controversial, as murine TLR7 recognizes ligands of both human TLR7 and TLR8 and is required for detection of synthetic and viral ssRNA37,38. Additionally, mouse TLR8 lacks a five-amino-acid motif that is necessary for proteolytic processing, folding and activity of human TLR8 (refs. 39,40). Nonetheless, canonical signalling domains required for activation of NF-κB remain conserved and are activated in the presence of synthetic thymidine homopolymer oligonucleotides41, suggesting the possibility of undiscovered natural ligands. We found that over-expression of mouse TLR8 permitted nLDL activation of NF-κB, despite a lack of responsiveness to synthetic ssRNA and nucleoside analogues (Extended Data Fig. 2i). Moreover, BMDMs isolated from wild-type and Tlr7-deficient mice showed similar sensitivity to nLDL treatments, indicating TLR7 is not necessary for nLDL-induced macrophage activation in mice (Extended Data Fig. 2j). Although nLDL was capable of activating both mouse and human TLR8 in luciferase-reporter assays, the chemical inhibitor CU-CPT9a was effective at blocking only human TLR8 activity (Extended Data Fig. 2k). Collectively, these data support a conserved sensitivity of TLR8 to nLDL in both mice and humans.
nLDL-msRNAs mediate inflammatory signalling in macrophages
We previously reported that lipoproteins of mice are enriched with microbial small RNA (msRNA) that are probably derived from microbes of our internal and external environment42, which could plausibly be responsible for TLR8 activation. We therefore quantified the sRNAs of human LDL by high-throughput small RNA (sRNA) sequencing (Fig. 3a). LDL-sRNA reads that aligned to the human reference genome were prioritized, which revealed human sRNAs derived from parent ribosomal RNA (rRNAs; rDR) and transfer RNA (tRNAs; tDR) to be most abundant, followed by microRNAs (miRNA; Fig. 3b). Reads that failed to align to the human genome were then aligned concurrently to structural RNA databases (that is, rRNA (SILVA) and tRNA (GtRNA-db)), as well as to curated genome databases for bacteria and other microbes (Fig. 3c,d and Extended Data Fig. 3a–d). On average, non-host sRNA were approximately 16 times more abundant than host sRNAs, with bacterial rDRs explaining approximately 30% of all reads (Fig. 3a,c). Alignment to bacterial genome databases found enrichment for msRNAs derived from Proteobacteria, although one individual showed enrichment for Firmicutes msRNAs, and several showed high abundances of Actinobacteria msRNAs (Fig. 3d and Extended Data Fig. 3b). In addition, alignment to fungi and algae genome databases also found substantial mapping relative to the host genome (Extended Data Fig. 3c,d). To distinguish the inflammatory potential of host and non-host sRNAs on LDL, sRNAs with comparable size to those found on LDL were isolated from human cells (representing host sRNAs) and from Escherichia coli (representing non-host msRNA) (Fig. 3e) and transfected in equal concentrations to THP-1 macrophages. Remarkably, host-derived sRNAs were unable to activate inflammatory signalling in human macrophages, whereas foreign msRNAs promoted the expression of IL1B, IL6 and IL12B, comparable to the synthetic ssRNA ORN06 (Fig. 3f). Lastly, to determine if sRNA cargo confer nLDL’s inflammatory capacity in macrophages, reconstituted LDL (rLDL) particles lacking sRNAs were generated by selectively removing the neutral lipid core of nLDL and replacing it with cholesteryl linoleate spiked with a fluorescent cholesteryl ester43. Phospholipid-to-protein ratios were similar for rLDL and nLDL, and exogenous lipid loading was confirmed by fluorescence (Extended Data Fig. 3e,f). Although APOB levels were similar between rLDL and nLDL (Extended Data Fig. 3g), rLDL particles lacked sRNA cargo, as determined by bioanalyzer analysis (Fig. 3g) and qPCR (Fig. 3h and Extended Data Fig. 3h). Although macrophages showed robust uptake of both rLDL and nLDL, as determined by increased cellular cholesterol levels and lipid staining (Fig. 3i and Extended Data Fig. 3i), only nLDL was found to increase inflammatory gene expression and cytokine secretion in treated macrophages (Fig. 3j,k). These results support that the msRNA cargo confer nLDL’s pro-inflammatory effects in macrophages.
a, Total RNA were isolated from DGUC-LDL obtained from 20 healthy subjects and sRNA were quantified by sRNA-seq. Relative proportion of sRNA-seq reads identified as host, non-host, unknown or too short for conservative mapping with TIGER. b, Host sRNA abundance quantified as RPM (small nucleolar RNA-derived RNA (snoDR); y-RNA-derived RNA (yDR); small nuclear RNA-derived RNA (snDR)). c, Alignment of non-host sRNAs to the SILVA rRNA database and grouped by taxa and quantified as RPM. d, Voronoi plot representing relative abundance of sRNAs (segment size; each segment represents a single subject) linked to bacterial phyla (colour) identified by mapping to a database of bacterial genomes linked to the human microbiome. e, total RNA and sRNA isolated from human macrophages (THP-1) and bacteria (E. coli), as well as total RNA isolated from selected nLDL subjects of a–d and ORN06 (20-mer ORN) were assessed by a Bioanalyzer (PicoChip) (representative image of three independent experiments). f, mRNA expression of THP-1 macrophages transfected with 1 μg ml−1 of isolated human sRNA, bacterial sRNA or ORN06 for 24 h (n = 3 biological replicates). g, total RNA of nLDL and rLDL assessed by a Bioanalyzer (PicoChip) (representative image of four independent preparations). h, Quantification of representative miRNA across independent preparations of rLDL and matched nLDL, or buffer negative control (n = 4 independent preparations). i, Cellular cholesterol levels in THP-1 macrophages following treatment with nLDL and rLDL or without treatment (UNT) for 24 h (n = 4 biological replicates). j,k, mRNA expression (j) and cytokine secretion (k) of THP-1 cells after treatment with 0.5 mg ml−1 nLDL or rLDL for 24 h (n = 4 biological replicates). Data are mean ± s.e.m. In f, h and j–k, one-way ANOVA and Benjamini, Krieger and Yekutieli FDR were performed (q = 0.05), *q < 0.05, **q < 0.01, ***q < 0.001, ****q < 0.0001. In i, one-way ANOVA with Dunnett’s multiple comparison test were carried out, *P < 0.05. Numerical source data, statistics, exact P values and q values are provided.
msRNA are present on lipoproteins of GF mice
Based on the findings of Fig. 3, reduced LDL-msRNA levels in vivo may confer protection against ASCVD. Commensal flora are abundant in both mice and humans and have been extensively associated with ASCVD44. We hypothesized that mice lacking commensal microbiota (that is, germ-free (GF) mice), would have reduced levels of msRNA on their lipoproteins. Therefore, plasma from two cohorts of specific-pathogen-free (SPF) and facility-matched GF mice were collected for fractionation and isolation of high-density lipoprotein (HDL), the primary lipoprotein of mice (Extended Data Fig. 4a,b). Based on sRNA-seq, the levels of host sRNAs were similar between SPF and GF mice, including the number of reads mapped to individual host miRNA, tRNA or rRNA transcripts (Extended Data Fig. 4c,d). Moreover, the levels of non-host sRNAs were also similar between GF and SPF control mice. For example, we failed to identify any changes to overall depth of non-host msRNA based on total counts for fungi, algae or bacteria (Extended Data Fig. 4c, e–i). Assessing differential abundance of sRNA alignment counts for 374 bacterial genomes in the HMB (human microbiome) and ENV (environmental) databases revealed modest changes in individual species between GF and SPF mice (Extended Data Fig. 4j and Supplementary Table 4). Consequently, the relationship between lipoprotein sRNA and the commensal microbiome is unclear.
nt-LNA inhibits nLDL activation of macrophage TLR8
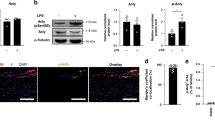
Chemically modified (for example, 2′-O-methyl and locked nucleic acid (LNA)) anti-sense oligonucleotides have been reported to impair ssRNA activation of TLR7/8, independent of transcriptional silencing34,45,46. We hypothesized that we could exploit these off-target effects to antagonize nLDL-induced TLR8 signalling by using a non-targeting (that is, scrambled) LNA. Hence, we screened commercially available control non-targeting LNAs (nt-LNA) for TLR8 antagonism using NF-κB (luciferase) reporter assays (Fig. 4a and Supplementary Table 5). nt-LNAs proved to be powerful inhibitors of TLR8-mediated responses to ssRNA ligands (ssRNA40/ORN06) but provided poor-to-ineffective inhibition of CL075, which stimulates TLR8 at an active site separate from that of ssRNA33. Replacing LNA nucleotides of nt-LNA-A with DNA nucleotides (nt-DNA-A) abolished antagonism of TLR8’s ORN detection and potentiated the response to CL075 (Extended Data Fig. 5a). Conversely, replacing RNA nucleotides of the canonical TLR8-stimulating ORN ssRNA40 with LNA nucleotides (ssLNA40) abrogated TLR8’s response to ORN, but not CL075 (Fig. 4a and Extended Data Fig. 5a). As little as 15% LNA nucleotides within nt-LNA-A was found to be effective at antagonizing TLR8’s ORN-driven activity. Together with results from dose–response assays (Fig. 4b), these data support that nt-LNA function as competitive antagonists of TLR8’s ORN recognition site through an LNA chemistry-driven effect. Although both sequences were effective, the nt-LNA-A oligonucleotide, referred to herein as nt-LNA, was selected for further experiments (Fig. 4a). nt-LNA pre-treatments in THP-1 macrophages dose dependently blocked activation of cells by ssRNA40, but not CL075, LPS (TLR4 ligand) or poly I:C (TLR3 ligand) (Extended Data Fig. 5b). Similarly, nt-LNA was also effective at blocking the induction of cytokine expression and secretion of IL-6 in response to ssRNA40 in primary human macrophages (Extended Data Fig. 5c,d). Pre-treatment of HEK293T cells over expressing mouse or human TLR8 with nt-LNA reduced nLDL-stimulated NF-κB-driven luciferase activity by approximately 50% (Fig. 4c). Likewise, nt-LNA pre-treatments blocked nLDL-induced gene expression (Fig. 4d) and IL-6 secretion (Fig. 4e) in primary human macrophages. Similar results were obtained from THP-1 human macrophages, which tended to have more robust responses for CEBP/β targets CCL3, CXCL2 and CLEC4E that were also blocked by nt-LNA (Fig. 4f). Concordantly, nt-LNA treatments also blunted nLDL-induced IL-6 and TNF-α secretion (Fig. 4g) and intracellular IL-1β and IKKβ protein levels in THP-1 macrophages (Fig. 4h). To further illustrate nt-LNA’s mechanism of TLR8 antagonism, primary BMDMs were pre-treated with IFNγ, which has previously been demonstrated to promote proteosome-based turnover of IκBα, removing a negative feedback loop on NF-κB activity47, in the presence or absence of nt-LNA. nLDL-mediated inductions of lL1b and IL6 were significantly reduced by nt-LNA treatments in BMDMs (Extended Data Fig. 5e) and modest impairment of IL-6 secretion were observed (Extended Data Fig. 5f). nLDL were found to be rapidly internalized by BMDMs, as evidenced by increased intracellular APOB protein levels and promoted lipid-droplet formation, as assessed by PLIN2 protein levels, both of which were not impaired by nt-LNA treatments (Extended Data Fig. 5g). nLDL treatments were associated with reduced IKKβ protein, probably due to enhanced proteosome activity, which was blocked by nt-LNA pre-treatments. As expected, IFNγ pre-treatment resulted in minimally detectable levels of IkBα, which were unexpectedly restored by nt-LNA pre-treatment, and further stabilized in the presence of nLDL. Collectively, these results support that nt-LNA is an effective competitive antagonist for nLDL-msRNA-mediated activation of TLR8.
a, Normalized NF-κB-driven luciferase activity of HEK293T cells over expressing human TLR8 and treated with agonists CL075 (2.5 μg ml−1), ssRNA40 (2 μg ml−1) and ORN06 (2 μg ml−1), in the presence or absence (Veh = DOTAP) of 2 μg ml−1 indicated LNA-oligonucleotide (n = 4 biological replicates). b, Dose–response curves for ORN06-induced luciferase activity in HEK293T cells over expressing human TLR8 co-treated with vehicle (white), nt-LNA-A 0.5 μg ml−1 (blue) or nt-LNA-A 2.5 μg ml−1 (purple) (n = 4 biological replicates). c, NF-κB-driven luciferase activity of HEK293T cells over expressing mouse or human TLR8 treated with nLDL (2.5 mg ml−1) with (purple) or without (white) nt-LNA-A (2.5 μg ml−1) (n = 4 biological replicates). d,e, mRNA expression (d) and cytokine secretion (e) of primary human CD14+ PBMC differentiated with GM-CSF and IFNγ following 24 h treatment with or without 0.5 mg ml−1 nLDL in the presence or absence of 2.5 μg ml−1 nt-LNA (n = 4 biological replicates). f–h, mRNA expression (f) (n = 4 biological replicates), cytokine secretion (g) (n = 4 biological replicates) and immunoblotting (h) of THP-1 macrophages following 24 h treatment with or without 0.5 mg ml−1 nLDL in the presence or absence of 2.5 μg ml−1 nt-LNA (n = 3 biological replicates). Data are mean ± s.e.m. Two-way ANOVA (a and f) or one-way ANOVA and Dunnett’s multiple comparison test (d, e and g), *P < 0.05, **P < 0.01, ***P < 0.001. Non-linear regression with extra sum-of-squares F test (two-sided) (b). Two-way ANOVA with Sidak’s multiple comparison test (c), ***P < 0.001. Numerical source data, statistics, exact P values and q values are provided.
nt-LNA inhibits atherosclerosis in mice
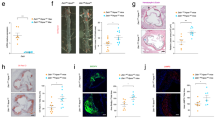
We next hypothesized that TLR8 antagonism with nt-LNA could reduce atherosclerosis-associated inflammation and disease burden in vivo. We first performed a small study in male and female hypercholesterolemic (Apoe−/−) mice fed a Western diet for 4 weeks receiving weekly doses of either saline control (Ctr), nt-LNA-A or nt-LNA-B (Fig. 5a). We observed that female mice developed larger legions in the aortic sinus in this period relative to males (Extended Data Fig. 6a,b). When normalized by sex, both nt-LNA-A and nt-LNA-B had similar reductions in the Oil-Red O (ORO)-positive lesion area, of nearly 30% (Extended Data Fig. 6c). To increase statistical power, we repeated these 4 week experiments with increased sample size (n = 10 per group) using only female Apoe−/− mice and focusing only on the nt-LNA-A sequence, referred to herein as nt-LNA (Fig. 5a). We validated that nt-LNA treatments reached the lesion using in situ hybridization (ISH) with an anti-nt-LNA (anti-sense) probe, which revealed deposition within the lesion and surrounding adventitia (Fig. 5b). Most importantly, nt-LNA treatments in this 4 week study were found to reduce atherosclerosis in mice by approximately 33% compared with saline control in the absence of altering plasma lipid levels and minor changes in plasma lipoproteins apolipoprotein AI (APOA1) and APOB (Fig. 5c,d, Supplementary Tables 6 and 7 and Extended Data Fig. 6d–g). Strikingly, neither macrophage content (Adhesion G protein-coupled receptor E1 (F4/80; Fig. 5e,f), Galectin-3 (MAC-2; Fig. 5g,h)), or collagen content (Fig. 5i,j) were found to be significantly altered by nt-LNA treatments within atherosclerotic lesions (Fig. 5g–j). Moreover, treatments with nt-LNA did not result in changes to hepatic lipid metabolism or inflammatory gene expression in the liver, suggesting some specificity for TLR8 antagonism within the lesion (Extended Data Fig. 6h). We repeated this study design in female Apoe−/− mice for 10 weeks to allow development of advanced atherosclerotic lesions (Fig. 5a). Plasma total cholesterol levels increased by >50% after 10 weeks, but were similar between treatment groups (Supplementary Tables 6and 7). Most importantly, weekly injections with nt-LNA reduced lesion development in the aorta by nearly 40% relative to saline controls (Fig. 5k,l). In the aortic sinus (cross-sections), a reduction in necrotic (lesion) core area was also observed with nt-LNA treatments after 10 weeks (Fig. 5m–p). Interestingly, lipid deposition in the aortic sinus (cross-sections) was not altered when normalized to lesion area (Fig. 5q,r), nor was macrophage content (Fig. 5s,t). However, collagen content was found to be increased by nt-LNA treatments (Fig. 5u,v). Collectively, the observed reduction in necrotic core formation coupled with increased collagen deposition support that nt-LNA treatments improve lesion stability and antagonize atherosclerotic progression. To determine if nt-LNA also promotes lesion regression, adult LDL receptor-deficient (Ldlr−/−) mice were fed an atherogenic diet for 14 weeks to develop severe atherosclerotic lesions and then switched to a chow diet for an additional 4 weeks, as previously described48. After conversion to the chow diet, remaining mice were injected weekly with nt-LNA or saline (Extended Data Fig. 7a). At baseline (14 weeks), mice on atherogenic diet showed expected elevations in plasma lipids compared with mice fed a chow diet, which returned to normal levels after 4 weeks of chow diet (Supplementary Tables 6 and 7 and Extended Data Fig. 7b,c). No differences in circulating APOA1 or APOB protein levels were observed between nt-LNA-treated and Ctr-treated mice after 4 weeks of treatments (Extended Data Fig. 7d,e). Yet, nt-LNA treatments were found to significantly reduce atherosclerotic lesion area compared with control mice, which was more pronounced in males than females (Extended Data Fig. 7f–i). Similar to the 4 week study in Apoe−/− mice, we failed to observe a change in fibrosis (Extended Data Fig. 7j,k) and macrophage content between the regression treatment groups (Extended Data Fig. 7l,m). Although a 30% increase in the mean macrophage content was observed in the regression nt-LNA mice relative to saline-treated mice, this was not statistically significant. Importantly, nt-LNA treatment in either progression or regression studies did not increase circulating liver enzymes alanine transaminase (ALT) or aspartate aminotransferase (AST), nor C-reactive protein, suggesting nt-LNA treatment was not hepatotoxic (Supplementary Tables 6 and 7). Furthermore, nt-LNA treatment did not alter circulating levels of serum amyloid A, a marker of systemic inflammation in mice (Supplementary Tables 6 and 7). Nevertheless, these results support that nt-LNA treatments reduce lesion progression and promote lesion regression in mouse models of atherosclerosis.
a, Schematic of progression study design. Apoe−/− mice, fed a Western diet for up to 10 weeks, were injected once weekly with saline Ctr or nt-LNA (30 mg kg−1, intraperitoneal (i.p.)). b–j, early lesions (4 weeks): ISH using anti-sense probes against nt-LNA in the early lesion (representative images of three independent hybridization experiments) (b); lesion AUC, as determined by ORO staining of early lesions (n = 10 mice per group) (c and d); quantification of F4/80+ macrophage content in early lesions by IHC (n = 10 mice per group; normalized to lesion area) (e and f); quantification of Mac2+ macrophage content in early lesions by IF microscopy (n = 8 mice per group; normalized to lesion area) (g and h); and quantification of early lesion fibrosis by Masson’s Trichrome staining (n = 10 mice per group) (i and j). k–v, Advanced lesions (10 weeks of Western diet feeding), weekly injections of saline Ctr or nt-LNA (30 mg kg−1): en face staining (Sudan IV) and quantification in the arch and descending aorta (Ctr: n = 10 mice; nt-LNA: n = 11 mice) (k and l); H&E staining of advanced lesions in the aortic sinus with areas of necrosis indicated with a dotted line (m); quantification of lesion AUC (H&E) and necrotic core AUC throughout the aortic sinus (Ctr: n = 10 mice; nt-LNA: n = 11 mice) (n–p); lipid deposition in the aortic sinus visualized by ORO and normalized by total lesion area (Ctr: n = 10 mice; nt-LNA: n = 11 mice) (q and r); quantification of Mac2+ macrophage content in advanced lesions by IF microscopy (Ctr: n = 10 mice; nt-LNA: n = 11 mice) (s and t); and quantification of total collagen content in advanced lesions by Picrosirius Red staining (Ctr: n = 10 mice; nt-LNA: n = 11 mice) (u and v). Scale bar, 500 μm. Data are mean ± s.e.m. In d, f, h, j, l, o, p, r, t and v, Mann–Whitney U test (two-sided) was performed; in n, two-way ANOVA with Sidak’s multiple comparison test were carried out. *P < 0.05, **P < 0.01. Numerical source data, statistics, exact P values and q values are provided.
nt-LNA reduces inflammatory macrophage phenotypes in lesions
To better understand the mechanisms through which nt-LNA reduced atherosclerosis in mice, single-cell suspensions were collected from atherosclerotic aortas and scRNA-seq was performed (Fig. 6a). CD45+CD3- immune cells were isolated by fluorescence-activated cell sorting (Extended Data Fig. 8) from aortic lesions of atherosclerotic mice (Apoe−/− mice on atherogenic diet) administered with nt-LNA or a saline control (pool of n = 8 treatment−1) for 4 weeks (Extended Data Fig. 9a). In total, 2,742 and 4,871 quality cells (representing 92% of total input cells) were obtained from control and nt-LNA-treated lesions, respectively. This approach identified 20 distinct clusters that were broadly identified as myeloid cells (9/20), B cells (4/20), T cells (6/20) and natural killer (NK) cells (1/20), based on gene enrichment analysis (Fig. 6b,c, Extended Data Fig. 9b–d and Supplementary Table 8). The number of total myeloid cell counts was comparable for both treatment groups; however, myeloid cells accounted for only a third of the cells detected in the nt-LNA lesions (Fig. 6c). Nevertheless, differential gene expression analyses between treatment groups identified relatively few differences among lymphocytic groups (Supplementary Table 9), whereas robust changes were detected in myeloid cells between treatments (Fig. 6d and Supplementary Table 10). Among the genes suppressed by nt-LNA treatments in myeloid cells were critical inflammasome mediators IL1b and Nlrp3 (NLR Family Pyrin Domain Containing 3), as well as regulators of foam cell formation (Cebpb and Ctsd (Cathepsin D)) (Fig. 6e)49,50,51. Gene Ontology analysis of all transcripts altered by nt-LNA in predicted myeloid cells identified an enrichment of genes associated with immune responses, and more specifically leucocyte activation, supporting the hypothesis that nt-LNA’s anti-atherosclerotic effects are due to reduced macrophage activation (Fig. 6f and Supplementary Table 11). To further investigate this point, myeloid cells were re-clustered to enhance phenotypic resolution (Fig. 7a,b). On the basis of enrichment of putative marker genes, specific macrophage phenotypes were identified, including inflammatory (C3), foamy (C6), cavity (C8), proliferating (C0), repair (C14) and two clusters of resident-like macrophages (C1 and C4) (Fig. 7c,d and Supplementary Table 12). Remarkably, inflammatory and foamy macrophages clusters were found to be predominant in lesions from saline-treated mice, whereas proliferating, cavity and repair macrophages were more prevalent in lesions of nt-LNA-treated mice (Fig. 7c,d). Classical monocytes (C7), aortic intimal resident macrophages (C9), mixed monocyte/macrophage/dendritic cells (cDC2; C2) and a neutrophil-like cluster of cells (C11) were roughly equal between treatments, whereas mature (C12), plasmacytoid (C13) and cDC1 (C10) dendritic cells, as well as resident-like (C1/C4) and transitioning (C5) macrophages were skewed towards nt-LNA-treated mouse lesions. Repair macrophages (C14) were found to be approximately 80% derived from nt-LNA-treated mice and showed high expression of Adgre1 (F4/80), thus supporting immunohistochemistry (IHC) data (Fig. 5e,f). Collectively, these results support that nt-LNA treatments promote the re-organization of macrophage phenotypes from pro-inflammatory and lipid-storage macrophages towards expansion of resident and repair macrophages that correlate with reduced disease burden.
a, Flow chart depicting (1) the digestion of aortas dissected from Apoe−/− mice fed a Western diet for 4 weeks and treated weekly with saline (Ctr: n = 8 mice) or nt-LNA (30 mg kg week−1; n = 8 mice) and (2) single-cell suspensions from individual mice pooled before fluorescence-activated cell sorting to isolate a CD45+ and CD3− population of immune cells used for (3) 10× chromium RNA single-cell partitioning and barcoding for (4) downstream scRNA-seq. b, Uniform manifold approximation and projection (UMAP) of individual cellular transcriptomes obtained from Ctr- and nt-LNA-treated mice. c, Absolute (histogram) and relative (pie) quantification of immune cell types obtained from b. d, Differential expression analysis between myeloid cells obtained from each treatment group. FDR (α = 0.05) adjusted P values after genewise negative binomial generalized linear models with quasi-likelihood tests. log2FC >0.5, adjusted P < 0.05). e, Violin plots of selected transcripts found to be differentially altered in d. f, Voronoi plots of Gene Ontology analysis (left) and detailed expansion of key segments of the immune system process (right). FDR (α = 0.05) adjusted P values calculated on the basis of EASE score, a modified Fisher’s exact test (one-sided).
a,b, Uniform manifold approximation and projection (UMAP) projections of myeloid cells identified by scRNA-seq analysis following selection and re-clustering analysis. Visualized as unbiased clusters (a) or by source (b). c, Relative expression (colour) and percentage of cells reaching the threshold of detection (size) for selected transcripts enriched for each cluster of a. d, Relative contribution of each treatment to clusters obtained from a. e, Proposed mechanism for LDL-msRNA in atherosclerosis-associated inflammation: (1) nLDL with associated msRNA cargo is internalized and catabolized within the endosomal network; (2) released msRNA cargo during catabolism is recognized by endosomal TLR8, which can be outcompeted by chemically modified oligonucleotides (for example, nt-LNA); (3) activation of TLR8 promotes translocation of NF-κB to the nucleus and upregulation of target genes (for example, TNF, IL6 and IL1B); and (4) release of cytokines that initiate an innate immune response and (5) sustained LDL-msRNA-driven inflammation promotes atherosclerotic plaque progression and impedes resolution.
Discussion
Our findings support that a diverse collection of non-host sRNA are present on LDL and, upon uptake by macrophages, trigger inflammatory activation through TLR8 to promote atherosclerosis (Fig. 7e). The mechanisms and route by which LDL acquire msRNA remains entirely unexplored; however, phagocytotic cells within mucosal tissues, which serve as the interface to the microbial environment, probably contribute to the acquisition and loading of msRNA on circulating LDL. We postulate that acquisition and transport of msRNA by LDL probably highlights a physiological role of lipoproteins that is at least partially independent of lipid trafficking and may serve to maintain mucosal homeostasis. Our results support a model in which impaired lipoprotein clearance and subsequent deposition of LDL within the vascular wall promotes non-saturable uptake by macrophages, which in turn propagate vascular inflammation following recognition of LDL-msRNA cargo. Critical support of this model lies in our finding that removal of sRNA cargo from LDL depletes its inflammatory potential, as well as our observations that antagonizing TLR8 impairs nLDL-induced macrophage polarization and reduces atherosclerotic burden in hypercholesterolemic mice. Of note, we have not been successful in reloading or repopulating LDL’s sRNA cargo to physiological levels ex vivo, which would greatly expand our ability to investigate the biological functions of LDL-sRNAs and further support our overall conclusions. Future studies will be needed to address technical issues in LDL-sRNA reloading approaches. Nevertheless, the identification of nLDL-msRNA as an inciting inflammatory factor in atherosclerosis is a critical advance as it overcomes a major barrier in our understanding of sterile inflammation that frequently accompanies cardiometabolic diseases. This study also highlights that not all macrophages (for example, CD14+ monocytes, alternatively activated macrophages) respond to nLDL with inflammatory activation and effect sizes are often modest. This, however, may explain the oscillating and subacute nature of inflammation in CVD and its slow progression over decades in humans. While all macrophage subtypes are probably exposed to high concentrations of nLDL in the lesion, not all macrophages are polarized towards inflammatory phenotypes.
Further studies are needed to gain a greater understanding of the impact of genetic, environmental, lifestyle and socio-economic factors on nLDL-msRNA content, as such investigation may identify risk factors for human disease, particularly ASCVD. We showed that the lipoprotein sRNA profile of GF mice was not altered relative to control SPF mice. Consistent with these observations, work from other groups has shown that plasma endotoxin, which also circulates on lipoproteins, is only modestly reduced in GF mice relative to SPF and conventional mice52. Bacterial products in GF mice are probably attributable to exposure to non-viable microbes present in food, water, bedding and air, which the host is undoubtedly still exposed to, and probably does not distinguish from viable microbes from an immunological standpoint. Thus, our data, and overall model, are consistent with reports from other groups that the absence of a viable microbiome does not prevent murine atherogenesis;53,54 however, further investigation is required in this space. So far, the overall abundance of total RNA content on LDL, including msRNA, have not been quantified within a human population, or between subtypes of LDL (for example, small dense LDL) within individuals. We observed a range of macrophage inflammatory responses to independent preparations of nLDL, which could be attributable to variations in sRNA abundance or unknown factors co-purified with density-gradient ultracentrifugation (DGUC)-LDL. Although many TLR-modulating agents have been considered for combating CVD and other metabolic diseases, strategies to selectively antagonize TLR8 are underdeveloped55. The identification of nt-LNA as competitive antagonists of TLR8 highlights a new avenue for therapeutic development, one that is likely to be well tolerated in human subjects56. The development of therapeutic strategies to mitigate LDL-msRNA signalling may hold great potential for the treatment and prevention of CVD, as well as many other diseases that feature hyperlipidaemia and chronic inflammation.
Methods
Animal protocols were approved by and performed according to the regulations of Vanderbilt University Medical Center’s Institutional Animal Care and Usage Committee and animal studies complied with all relevant ethical regulations for vertebrate animal research (number M1600252-01). Studies requiring acquisition of blood were obtained from healthy human donors able to give informed consent under approved Vanderbilt University Medical Center Institutional Review Board protocol (IRB number 170046). Human subjects were compensated ($20.00).
Materials
Coding sequences of individual toll-like receptors (TLR) were amplified from complementary DNA of human and mouse macrophages using Phusion high-fidelity DNA polymerase (ThermoFisher) and cloned to the multiple-cloning site of the pAdTrack-CMV vector (a gift from Bert Vogelstein (Addgene plasmid number 16405; http://n2t.net/addgene:16405; RRID: Addgene_16405), which expresses green fluorescent protein (GFP) from an independent CMV promoter57. The coding sequence of the TLR chaperone protein UNC93B1 and co-receptor CD14 were similarly amplified from human macrophage cDNA and inserted to the pMSCV-neo and pMSCV-puro vectors, respectively (Clontech). The NanoLuc Reporter Vector with NF-κB response element (pNL3.2.NF-κB-RE) is commercially available (Promega). Transfections of plasmid DNA were performed using Lipofectamine 3000 (Life Technologies), per the manufacturer’s instructions. In vivo grade nt-LNA (catalogue number 339203, Qiagen) was purchased in individual lots of 100 mg, reconstituted in sterile saline, aliquoted and stored at −20 °C. ssRNA oligonucleotides were synthesized with a phosphorothioate backbone (Integrated DNA Technologies). Nucleoside analogues CL075 and Resiquimod (R848) were purchased commercially (Sigma). Bafilomycin A1 and EGTA-AM were purchased commercially (Sigma). C29 was purchased from Cayman Chemical (catalogue number 27029) and solubilized in dimethyl sulfoxide (DMSO) according to the manufacturer’s recommendations. For oligoribonucleotide transfections, nt-LNA, ssRNA and sRNA oligonucleotides were complexed with N-(2,3-Dioleoyloxy-1-propyl)trimethylammonium methyl sulfate (DOTAP; Sigma) according to the manufacturer’s instructions. Ligands for TLR1/2 (Pam3CSK4), TLR2 (heat-killed Listeria monocytogenes), TLR3 (Poly(I:C)), TLR4 (LPS Escherichia coli K12), TLR5 (Flagellin Salmonella typhimurium), TLR6/2 (FSL-1) and TLR9 (ODN2006) were purchased commercially (Invivogen) and diluted according to the manufacturer’s recommendations. Antibodies used for immunoblotting and flow cytometry in this study are provided in Supplementary Table 13.
Mouse studies
Animals and models of atherosclerosis
Wild-type (RRID: IMSR_JAX:000664), Tlr7−/− (RRID: IMSR_JAX:008380), Ldlr−/− (RRID: IMSR_JAX:002207) and Apoe−/− (RRID: IMSR_JAX:002052) mice of the C57BL/6 background were purchased (The Jackson Laboratory). Mice were maintained on a chow diet (NIH-31) or an atherogenic diet containing 42% kcal from milk fat and 0.2% cholesterol added (Envigo; TD.88137) in a 12 h:12 h light–dark cycle with unrestricted access to food and water. For atherosclerosis progression studies, 10 week-old female Apoe−/− mice were converted from a chow diet to an atherogenic diet for 4 weeks or 10 weeks where indicated, receiving weekly intraperitoneal (i.p.) injections of saline (n = 10–12) or non-targeting locked nucleic acid (nt-LNA; 30 mg kg−1; n = 10–12) that were randomized within cages to limit cage effects. For atherosclerotic regression studies, male and female Ldlr−/− mice were fed the atherogenic diet for 14 weeks to promote hypercholesterolaemia and lesion development (n = 25 per sex). Ldlr−/− mice fed a chow diet for this time period were used as an age-matched control. After 14 weeks of atherogenic diet feeding, mice from each cage were randomly assigned to either a baseline group that was killed at this time (n = 7–8 per sex), or one of two treatment groups that were converted to a chow diet for 4 additional weeks with weekly i.p. injections of saline (n = 9 per sex) or nt-LNA (n = 8–9 per sex), as previously described48. Results of the regression study are pooled from two independent cohorts of mice staggered by 4 weeks. Upon killing, blood was collected by cannulating the vena cava using an EDTA-coated syringe and plasma was obtained by centrifugation at 2,500g for 10 min. Isolated plasma was stored at −80 °C before analysis. Unless otherwise indicated, tissues were flash frozen in liquid nitrogen and stored at −80 °C.
Plasma obtained upon killing from facility-matched 129SvEv SPF and GF mice was purchased from the National Gnotobiotic Rodent Resource Center through collaboration with Dr Susan Tonkonogy.
Lipid analysis
Plasma collected during animal killing was assayed directly for total cholesterol and triglycerides using colorimetric kits (Pointe Scientific). For assessment of lipid/lipoprotein distribution, 200 µl of mouse plasma was diluted in SEC running buffer and injected in the ÄKTA Pure SEC system (GE Healthcare) composed of three tandem Superdex-200 Increase columns with collection of 1.5 ml fractions58. Individual fractions were then assessed for total protein (BCA; Pierce) and lipids (Pointe Scientific). For quantification of cellular cholesterol following nLDL treatment, cells were washed once with PBS and scraped from the plate in ice-cold methanol and transferred to a 12 ml borosilicate tube. Subsequently, a Bligh–Dyer lipid extraction was performed with a final ratio of 1:1:0.9 chloroform:methanol:saline59. Extracted lipids were solubilized with 1% Triton-X 100 and assayed for cellular cholesterol by a colorimetric kit (Pointe Scientific). The aqueous phase was evaporated, residual protein solubilized with heated 1 M NaOH and total protein assayed by a colorimetric kit (BCA; Pierce).
Assessment of circulating liver enzymes and systemic inflammation
ALT and AST activity in mouse plasma (one to two freeze–thaws) were assessed by colorimetric kits according to the manufacturer’s recommendations (Abcam; catalogue numbers ab105134 and ab105135, respectively). Circulating mouse C-reactive protein levels in plasma were measured by ELISA (RayBiotech; ELM-CRP-1) according to the manufacturer’s recommendations. Murine serum amyloid A levels in plasma were measured by ELISA (Abcam; ab215090) according to the manufacturer’s recommendations. For ALT/AST activity assays, as well as ELISAs, samples with extensive haemolysis were not assayed owing to the potential for non-specific activity.
Quantification of atherosclerosis
For each experiment, mice were fasted for 4 h at the end of the dark phase and killed with isoflurane. Blood was collected by cannulating the vena cava and mice were perfused with PBS until vasculature was clear. The heart, with aortic root attached, was collected, embedded with optimal cutting temperature compound (OCT) and frozen on dry ice. As detailed by Paigen et al.60, 30 × 10 µm sections were taken from the beginning of the aortic root (where valve cusps first appear) using a Leica CM3050s cryostat by the same technician for all studies. For each root, every other slide for the 300 µm span was stained with ORO to identify the neutral lipid deposition. To quantify lesion area, the ORO-positive area was imaged using the KS300 imaging system (Kontron Elektronik GmbH), as previously described61,62,63, and reported as ORO μm2. The area under the curve (AUC) was calculated from the lesion area of serial sections. All other stains were conducted on sections 40–60 mm distal of the aortic sinus. Masson’s Trichrome staining and IHC for F4/80 (Novus Biologicals; secondary by Vector Laboratories) were performed by the Vanderbilt Translational Pathology Shared Resource core using four sections per animal and quantified using the KS300 imaging system applying methodologies previously described64. For immunofluorescence (IF) studies, frozen aortic root sections were dried at 55 °C for 10 min and then fixed in formalin for 20 min. Slides then underwent 3× 30 s dip rinses in de-ionized water. Slides then underwent antigen retrieval (H-3300, VectorLabs) in a pressure cooker for 10 s. Slides were then rinsed (30 dips) three times in PBS. In a humidified chamber, slides were blocked using 1% BSA for 30 min and then aspirated and incubated overnight in primary antibody (Mac2, 1:10,000, Cedarlane). Slides were then rinsed (30 dips) three times in 1× PBS and then probed with secondary antibody (goat anti-rat AF488, 1:200, ThermoFisher) and sealed with a coverslip. Fluorescence microscopy was completed using a Nikon STORM scope and images were recorded for subsequent analysis in ImageJ (Fiji v1.51). For ISH experiments, 10 µm frozen sections were fixed overnight in 10% neutral-buffered formalin and rinsed three times in PBS for 3 min each the following day. Slides were then digested with Proteinase K at 37 °C for 10 min and washed in PBS. Hybridization was performed with 40 nM double-FAM anti-sense LNA probes against nt-LNA (Qiagen) according to the manufacturer’s recommendations (miRCURY LNA miRNA in situ hybridization kit; Qiagene). Alternatively, the 10 week progression study was processed using 7 µm sections from the same starting points, but ORO staining was performed on five sets of two sequential sections spaced 70 µm from when the valve cusps first appear. Adjacent sections were then used for haematoxylin and eosin (H&E) staining (Abcam), Picrosirius Red (Polysciences) or IHC. In these studies, H&E staining was used to quantify lesion area (ImageJ (Fiji v1.51)) and necrosis, fibrosis and lipid deposition were defined as a percentage of the total lesion area. Aortas collected for en face presentation were fixed in neutral-buffered formalin, washed in PBS, pinned and stained with Sudan IV.
Cell culture studies
Tissue culture
HEK293T cells were maintained in DMEM supplemented with sodium pyruvate (110 mg l−1), l-glutamine, 10% heat-inactivated FBS and 1% penicillin–streptamycin (GIBCO). THP-1 cells were maintained in RPMI 1640 supplemented with 25 mM HEPES, l-glutamine, 10% heat-inactivated FBS and 1% penicillin–streptamycin (GIBCO) on suspension culture dishes (Corning). To generate THP-1 macrophages, THP-1 cells were suspended in fresh medium spiked with 5 ng ml−1 PMA and transferred to non-tissue-culture-treated plates for 48 h. After 48 h, adherent cells were washed twice with PBS and given fresh media without PMA for 24 h (ref. 65). Best results were obtained from THP-1 cells in early passages, with phenotypic drift and lack of normal responses observed in cells with extended culture. For siRNA experiments, non-differentiated THP-1 cells were concentrated ten-fold in Opti-MEM and incubated with 300 nM scrambled or siTLR8 siRNA (ON TARGETplus; Horizon Discovery) or without siRNA and electroporated using program Y-001 on a Nucleofector 2b device (Lonza). Cells were immediately diluted ten-fold in RPMI containing 5 ng ml−1 PMA and differentiated, as above. Primary human CD14+ peripheral blood mononuclear cells (PBMC) were isolated from whole blood obtained from self-reported healthy male and female donors using the Ficoll–Paque method (GE Healthcare) followed by CD14+ selection with magnetic-activated cell sorting (Miltenyi Biotec) following the manufacturer’s protocols. Cells isolated by positive selection were then seeded directly to tissue culture plates with or without cytokine treatments where indicated. Cells not treated with cytokines were allowed to attach to plastic overnight, non-adherent cells washed away, and then treated with nLDL for 24 h. To generate M1-like macrophages, CD14+ cells were first seeded with GM-CSF (20 ng ml−1) for 5 days without medium change. On the sixth day, non-adherent cells were removed and mediumcontaining GM-CSF (20 ng ml−1; PeproTech) and IFNγ (20 ng ml−1; Sino Biological) was added to remaining adherent cells, as previously described. Similarly, M2-like macrophages were differentiated from CD14+ monocytes by incubating with M-CSF (Sino Biological), IL-4 (PeproTech) and IL-13 (Tonbo Bioscience; 20 ng ml−1 each) for 9 days with a single medium change on the sixth day. On the ninth day, M1-like and M2-like cells were treated as indicated. Primary BMDMs were generated by incubating total bone-marrow aspirates isolated from the tibia and femur of C57BL/6 male and female mice in DMEM containing sodium pyruvate (110 mg l−1), l-glutamine, 10% heat-inactivated FBS and 1% penicillin–streptamycin (GIBCO) supplemented with murine GM-CSF (50 ng ml−1; Tonbo Bioscience) for 72 h on non-treated tissue culture plates66,67. After 72 h, non-adherent cells were washed with PBS and provided additional fresh medium containing GM-CSF with indicated treatments.
NF-κB activity (Luciferase) reporter assay
HEK293T cells were seeded in 24- or 96-well plates and allowed to attach overnight in full medium. The following day, cells were transfected with 1 μg ml−1 plasmid DNA (35% pAdTrack-CMV, 30% pMSCV-puro-CD14, 30% pMSCV-neo-UNC93B1 and 5% pNL3.2.NF-κB-RE) using Lipofectamine 3000. After 3 h, transfection medium was carefully aspirated and replaced with fresh media for 24 h. The following day, treatments were performed as indicated for 20–24 h, at which time media was removed and a passive lysis buffer (Nano-Glo Luciferase Assay Buffer; Promega) was added, and lysates were incubated on a horizontal shaker for 10 min at 25 °C with 500 reads per million total reads (RPM). Lysates were then transferred to an opaque 96-well plate for measurement of GFP (co-expressed with gene-of-interest (GOI) from pAdTrack-CMV) fluorescence as an indicator of cell density and transfection efficiency on a Synergy Mx plate reader. Subsequently, the NanoLuc substrate Furimazine was diluted in lysis buffer, added to lysates with a multichannel pipette and incubated with agitation for 2 min at room temperature. Luminescence was then quantified using a Synergy Mx plate reader. Luciferase activity was calculated as luminescence divided by GFP fluorescence and reported as a relative fold change (FC).
Methods
RNA isolation and real-time PCR
For cells, total RNA was isolated using Total RNA Purification kits (Norgen), as per the manufacturer’s instructions. For tissue, total RNA was isolated using miRNeasy Mini kits (Qiagen), as per the manufacturer’s instructions. For lipoproteins, total RNA was isolated using miRNeasy Mini kits with slight modifications to the manufacturer’s instructions58. Briefly, Qiazol was added at 10× the initial volume of lipoprotein concentrate, vortexed for 30–60 s and allowed to incubate for 5 min at room temperature before the addition of 0.2 volumes of chloroform. After phase separation, the aqueous phase was mixed with an equal volume of 100% ethanol, vortexed and stored overnight at −80 °C. The following day, the RNA sample was applied to the mini column, per the manufacturer’s instructions. For targeted isolation of human sRNAs, 1 × 107 THP-1 cells were pelleted by centrifugation and lysed in Qiazol and processed according to Appendix A of the manufacturer’s instructions (Qiagen). For targeted isolation of bacterial sRNA, an overnight culture of E. coli (DH5α; Invitrogen, catalogue number 18265-017) was pelleted at 4,000g, washed twice with PBS and incubated with 1 µg ml−1 lysozyme for 5 min at room temperature with agitation. Qiazol was then added and the partial cell lysate was pulverized with Zirconium beads (Sigma) in an oscillating tissue homogenizer for 30 s. Subsequently, the sample was processed following the Appendix A protocol of the miRNeasy Mini kit (Qiagen) for the separate isolation of long and small RNAs. Total RNA, sRNA and LDL-sRNA were assessed by PicoChip on a 2100 Bioanalyzer instrument (Agilent). Endotoxin was detectable in both E. coli total RNA (95 EU mg−1 RNA) and sRNA (34.1 ± 7.6 EU mg−1 RNA), but levels used for cell culture were below 0.5 EU ml−1 medium. cDNA was generated from total RNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher) for mRNA targets. For sRNA targets, the miRCURY LNA RT kit was used for cDNA synthesis (Qiagen). Real-time PCR was performed for targeted transcripts and sRNAs using the Power SYBR 2× Master Mix (ThermoFisher) on QuantStudio 6 or QuantStudio 12 instruments (Life Technologies). RPOLII served as a housekeeper for human cells and Rplp01 (36b4) served as a housekeeper for murine cells and tissues. For LDL-sRNAs, an arbitrary Ct of 35 was used for relative quantitative values calculations. All primer sequences are available upon request. Relative quantitative values were calculated by the delta-Ct method and reported as FC.
High-throughput RNA-seq
Individual RNA-seq libraries were generated using Ovation Human FFPE RNA-Seq Multi-plex System (NuGen) kits, per the manufacturer’s instructions. Individual libraries were assessed for quality using the Agilent 2100 Bioanalyzer and quantified with a Qubit Fluorometer. Paired-end sequencing of multiplexed libraries was performed on the NovaSeq6000 (Illumina) platform by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core (Vanderbilt University, Nashville, TN). Reads were trimmed to remove adaptor sequences using Cutadapt v1.1668 and aligned to the human hg19 genome using STAR v2.5.3a69. GENCODE human v19 gene annotations were provided to STAR to improve the accuracy of mapping. Quality control on both raw reads and adaptor-trimmed reads was performed using FastQC (v0.11.9) www.bioinformatics.babraham.ac.uk/projects/fastqc). featureCounts v1.15.2 (ref. 70) was used to count the number of mapped reads to each gene. Significantly differential expressed genes with false discovery rate (FDR)-adjusted P value <0.05 and absolute FC >2.0 were detected by DESeq2 (v1.18.1) (ref. 71). Gene Ontology and transcription factor analyses were assessed from differentially expressed mRNAs using the MetaCore (v5.0) software suite (Clarivate Analytics).
sRNA-seq
Individual sRNA libraries were generated from LDL-sRNA using the NEXTFlex Small RNA Library Preparation Kits v3 for Illumina Platforms (PerkinElmer), per the manufacturer’s instructions with the following modifications: (1) 3′- and 5′-adaptors were diluted 1:8, (2) 3′-adaptor ligation was performed overnight in multiple steps –25 °C for 2 h, 20 °C for 4 h and 16 °C overnight, (3) following cDNA synthesis and before barcoding PCR, the supplementary modified step F protocol was followed (that is, no size selection protocol), (4) PCR amplification was 20 cycles. Following PCR amplification, individual libraries were size-selected (136–200 bp product) using pre-cast 3% agarose gel cassettes with a Pippin Prep (Sage Sciences) and quantified on a Qubit Fluorometer. Single-end sequencing (SE-75) of equimolar multiplexed libraries was performed on the NextSeq500 (Illumina) platform by the VANTAGE core (Vanderbilt University, Nashville, TN). Bioinformatic processing was performed using an in-house data analysis pipeline (TIGER (v20210422), previously described in detail42. Briefly, Cutadapt (v1.16) (ref. 68) was used to trim 3′ adaptors for raw reads. All reads with fewer than 16 nucleotides (nts) were designated as ‘too short’ and discarded. The adaptor-trimmed reads were mapped to the hg19 genome, with additional rRNA and tRNA reference sequences, by Bowtie1 (v1.1.2) (ref. 72) allowing only one mismatch (1MM). Reads <20 nts that failed to be annotated as sRNA without perfect alignment to human genome were discarded. Remaining unmapped reads were then mapped in parallel to non-host structural RNA databases and curated microbial genome databases allowing no mismatches (perfect alignment). Reads that failed to align to any strategy were categorized as ‘unknown.’
scRNA-seq
After collection of the heart and aortic root, as described above (n = 8 per treatment), the aortic arch and thoracic aorta were removed, cleaned and placed into 1 ml of RPMI 1640 containing 10% FBS on ice, as previously described73. Once all aortas were collected, aortas were minced to small pieces using vigorous scissor action and digested by shaking at 37 °C for 30 min in 1 ml of RPMI 1640 containing 2.6 U ml−1 Liberase TM (Sigma), 60 U ml−1 hyaluronidase (Sigma), 0.1 mg ml−1 Dnase I (Sigma), 1 µg ml−1 actinomycin D (Sigma), 0.744 U ml−1 Elastase (Worthington Biochemical). Digested samples were placed in fresh tubes and spun at 500g for 10 min at 4 °C. Pellets were pooled and filtered through a 70 mm cell strainer using RPMI + 10% FBS. Fc receptors were blocked with anti-mouse CD16/32 (BioLegend) for 10 min at 4 °C before surface marker staining. Cells were stained with Pacific Blue anti-CD45 (BioLegend), FITC anti-Ter119/RBC (BD Biosciences) and PerCP-Cy5.5 anti-CD3 (BD Pharmingen) for 30 min at 4 °C. Cells were washed twice with RPMI + 10% FBS, filtered and sorted using a four-laser FACSAria III by the VUMC Flow Cytometry Shared Research Core. Dead cells (labelled with 7-amino-actinomycin D) (Biolegend)) and red blood cells were excluded and CD45+CD3− cells were collected using a 100 μm nozzle into RPMI 1640 + 10% FBS. Each sample (saline, 13,055 cells; nt-LNA, 19,980 cells) was processed for single-cell 3′ RNA-seq utilizing the 10× Chromium system through the VANTAGE core (Vanderbilt University, Nashville, TN). Libraries were prepared using P/N 1000006, 1000080 and 1000020 following the manufacturer’s protocol. The libraries were sequenced using the NovaSeq6000 with 150 bp paired-end reads. RTA (version 2.4.11; Illumina) was used for base calling and analysis was completed using 10× Genomics Cell Ranger software v2.1.1. scRNABatchQC (v0.10.3) software package was used for assessment of scSEQ data quality control, as previously described74. Clustering analysis was generated by Seurat (v3.2.3) (ref. 75). Initial cell type classification (that is, myeloid/macrophage, B cells, T cells and NK cells) of complete datasets was based on cell activity database76. Refined annotation of clusters was conducted manually following recommendations of a meta-analysis of similar sequencing approaches;17 however, discrepancies between predicted clusters based on the previously reported meta-analysis and our data were observed. These differences were probably attributable to the early atherosclerosis model employed in our study. Hence, we observed two resident-like macrophage clusters distinguishable by the expression of B-cell markers Cd79a, Ms4a1, Fcamr and Ighm (C1 versus C4), which may be the result of pre/pro-B-cell differentiation to macrophages77. Aortic intimal resident macrophages (C9) were categorized by their expression of Cx3cr1 and Itgax, as well as genes in lipid metabolism (Abcg1, Scarb1, Pparg and Cebpb)78. Interestingly, this population also had selective expression of Cd300e, as well as enriched expression of Nr4a1, a transcription factor associated with resident macrophage differentiation79. Repair macrophages (C14) were categorized by their expression of Mlxipl (carbohydrate response-element binding protein; ChREBP), Alox15 and Fn1 (fibronectin)80. Transitioning cells (C5) showed expression of proliferation genes (Top2a, Mki67 and Pclaf), as well as differentiation modulators Celf1 and Nr4a1. EdgeR (v3.15) software was used for differential expression analysis between treatments and within cell types81. Gene Ontology and transcription factor analyses were assessed from differentially expressed mRNAs using the MetaCore software suite (Clarivate Analytics).
Lipoprotein isolation and preparation of rLDL
Human whole blood was collected from fasted male and female subjects age 18 years or older (IRB number 170046) in K2EDTA tubes after informed written consent was obtained. In total, 55 blood samples were obtained from self-reported healthy donors (confirmed to have total cholesterol <240 mg dl−1) (Supplementary Table 14). Owing to recruitment challenges during the coronavirus disease 2019 pandemic, four individuals contributed 14 samples over the course of the study. Aliquoted plasma was either used fresh or stored at −80 °C and used after one freeze–thaw. In many instances, nLDL for experiments were obtained from pooled plasma of multiple donors. Sequential DGUC using a Beckman Coulter Optima XPN-80 Ultracentrifuge with either SW32Ti or SW41Ti swinging bucket rotors was used to isolate VLDL (1.006–1.018), LDL (1.019–1.063) and HDL (1.064–1.021) by adjusting sample density with potassium bromide, as previously described58. Isolated lipoproteins were immediately dialysed in 1× PBS for at least four changes of 200-fold excess buffer. After (up to) 48 h dialysis, lipoproteins were concentrated using Amicon Ultra Centrifugal Filters (3 kDa molecular weight cut-off; Millipore) and then immediately sterile-filtered (0.22 µm) by syringe. Total protein concentration was measured by BCA Protein Assay Kit (Pierce). Isolated lipoproteins were stored in the dark at 4 °C and monitored for aggregation and loss of colour. Best results were obtained from freshly collected plasma and newly processed nLDL. As quality control, LDL isolated and stored under these conditions for up to 1 month were assayed for oxidation using the thiobarbituric acid reactive substance (TBARS) method and compared against matched LDL oxidized by indicated concentrations and exposure time to copper sulfate82. Typical results are shown in Extended Data Fig. 2b. Additionally, nLDL quality was assessed by electrophoresis in 0.5% agarose gels with albumin and copper-sulfate oxidized LDL (oxLDL) serving as positive controls for electrophoretic mobility (Extended Data Fig. 2a) or fractionated by size-exclusion chromatography (SEC) on two tandem Superose 6 gel filtration columns (Cytiva) with HDL, VLDL and albumin size references (Extended Data Fig. 2c). Endotoxin levels were monitored by Chromogenic Endotoxin Quant Kit (Pierce) and found to be less than 0.5 EU mg−1 total protein. In a survey of seventeen DGUC-purified LDL samples, Lp(a) was detected by ELISA (Mercodia) in six samples. In samples with detectable signal Lp(a) content averaged 0.122 U mg−1 total protein. rLDL was prepared as previously described with minimal modifications43. Briefly, de-salted nLDL was mixed with potato starch (Sigma) by vortexing and flash frozen in liquid nitrogen before lyophilization for 6 h. Neutral lipids were selectively removed by sequential heptane extractions at −10 °C, before re-constitution with cholesteryl linoleate (NuChek-Prep) spiked with a 1:20 ratio of fluorescent 18:2 TopFluor cholesteryl ester (Avanti Polar Lipids) solubilized in heptane. This mixture was evaporated under a nitrogen stream until powder dry. Formation of rLDL particles was solubilized in 10 mM tricine, pH 8.4 for 108 h at 4 °C. Insoluble starch was pelleted by centrifugation at 2,000g and solubilized rLDL was extracted and sterile-filtered. In parallel, nLDL and rLDL samples were analyzed by size-exclusion chromatography in running buffer (10 mM Tris–HCl, 0.15 M NaCl and 0.2% NaN3) using two tandem Superose 6 gel filtration columns (Cytiva) on an ÄKTA SEC system (Cytiva). The distribution of fluorescent cholesteryl ester was measured by a fluorometer using the Synergy Mx plate reader (Biotek). Total protein and lipid composition of LDL samples were determined by colorimetric kits: total cholesterol (Pointe Scientific), triglycerides (Pointe Scientific), phospholipid C (Wako) and protein by BCA (Pierce). In all experiments, LDL (nLDL, oxLDL and rLDL) treatments were normalized by total protein concentration. Most experiments presented in this manuscript were performed using pools of two or more donors.
Immunoblotting
Cell lysates were collected on ice in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris–HCl pH 8.0) containing Halt Protease and Phosphatase Inhibitors Cocktail (ThermoFisher) and rotated at 4 °C for 1 h. Insoluble cell debris was pelleted at 12,000g for 10 min in a pre-chilled centrifuge (4 o°C), and soluble protein lysates were transferred to new tubes on ice. For immunoblots of cells, 12 µl of cell lysate (180 µl per well of a six-well plate) was mixed with 4 µl of 4× Protein Sample Loading Buffer (LI-COR), denatured at 70 °C for 5 min or 48 °C for 10 min and immediately placed on ice. For immunoblots of mouse plasma, 1.5 μl of plasma was mixed with 10.5 μl RIPA buffer containing Halt Protease Inhibitor Cocktail (ThermoFisher) and 4 μl of 4× Protein Sample Loading Buffer (LI-COR), denatured at 70 °C for 5 min and immediately placed on ice. Denatured protein samples were then run on NuPAGE 4–12% Bis-Tris or 3–8% Tris-acetate gels (Life Technologies) at 125 V on ice with Precision Plus Protein Kaleidoscope Pre-Stained Protein Standards (BioRad) or HiMark Pre-Stained Protein Standards (Life Technologies) as molecular weight references. After electrophoresis, gels were transferred to nitrocellulose membranes using the iBlot system (Life Technologies). Blots were then blocked in Intercept TBS Blocking Buffer (LI-COR) for 30 min before dilution of the primary antibody. Primary antibodies were incubated for at least 1 h at room temperature or overnight at 4 °C, washed three times with Tris-buffered saline containing 0.05% Tween-20 for 10 min. Subsequently, this process was repeated for secondary antibodies. Blots were assayed using the LI-COR Odyssey Infrared Imaging system, and analyzed using Image Studio Lite and ImageJ software suites.
Single-plex and multi-plex ELISA
Media supernatants of cell culture experiments were assayed for indicated cytokine concentrations by MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel (Millipore) on the Luminex MAGPIX System by the Vanderbilt Hormone Assay and Analytical Services Core, per the manufacturer’s recommendations. For individual assays, cytokine concentration in medium was assayed by conventional ELISA for human and mouse IL-6 and TNF-α (DuoSet; R&D Systems).
Formulation of PLGA-CU-CPT9a microparticles
The selective TLR8 inhibitor CU-CPT9a is not water soluble, requiring alternative methods for effective drug delivery in cell culture systems. Hence, drug-loaded microparticles (MPs) were formulated using a conventional oil-in-water emulsion technique. Poly(lactide-co-glycolide) (PLGA, 50:50 lactide:glycolide, 10 kDa, Sigma) was dissolved at 100 mg ml−1 in dichloromethane (DCM) with vortexing and brief sonication. CU-CPT9a (Sigma) was re-suspended at 10 mg ml−1 in DCM. Then 0.5 m of the drug solution was added to 0.5 ml PLGA solution, at which point the drug fully dissolved. The polymer/drug O phase was added dropwise to 5 ml 1.5% poly(vinyl alcohol) (30–70 kDa, Sigma) in water and homogenized for 30 s at 10,000 rpm with an IKA T18 Digital Ultra-Turrax homogenizer. The oil-in-water emulsion was transferred to a 100 ml round-bottom flask, and DCM was removed via rotary evaporation to complete MP hardening. The aqueous MP suspension was centrifuged at 7,500g for 10 min and re-suspended in de-ionized water to remove excess poly(vinyl alcohol), and the re-suspended MPs were aliquoted for lyophilization and subsequently stored at −20 °C. Immediately before experiments, lyophilized MP were equilibrated to room temperature and re-suspended with PBS to 4 mg ml−1 before dilution at the indicated doses. Resulting particles were deposited on a glass coverslip, imaged on a Nikon Eclipse Ti inverted scope and the diameters measured using Nikon Ti software. Drug loading was quantified by dissolving in DMSO and measuring the ultraviolet–visible absorbance spectra relative to free CU-CPT9a on a Varian UV–VIS spectrophotometer. Resulting particles were 3.39 ± 1.93 μm in diameter with 84.8 μg drug per milligram MP (encapsulation efficiency 84.8%).
Statistics and reproducibility
Data shown for in vitro experiments are representative experiments performed at least three times with independent donors/isolations of LDL. In vitro experiments were performed with three or more technical replicates. Unless otherwise specified, data are mean ± standard error of the mean (s.e.m.). For animal studies, all treatments were randomized within individual cages to limit cage effects. For atherosclerosis studies, quantification of lesion area, fibrosis, IHC and IF were assessed in a blinded fashion by at least two researchers. Collection and preparation of heart and aortas for histology was also performed blinded by a single technician. Where two treatments were compared with a sample size >3 for each treatment a Mann–Whitney U test (two-sided) was performed with α = 0.05. Where three or more groups were compared, a one-way analysis of variance (ANOVA) was performed using a Benjamini, Krieger and Yekutieli post-hoc test to control the false discovery rate (q = 0.05; Figs. 1 and 4) or Sidak’s post-hoc test to control type I errors (Fig. 5). Where more than one test was performed for three or more groups a two-way ANOVA was performed using Benjamini, Krieger and Yekutieli post-hoc test to control the FDR (q = 0.05), or when multiple comparisons were made to a single group, Dunnett’s test was performed. To determine whether differences existed between treatments in dose–response curves a non-linear regression with extra sum-of-squares F test was performed. Schematics and aspects of some figures were created with BioRender.com, Tableau and R Studio (v4.1.2). Microsoft Excel (v16.62) and GraphPad Prism 9 (GraphPad Software; v9.0.1) were used to create graphs and calculate statistical significances. Images were assembled in Affinity Designer (Serif; v1.8.4) to generate figures.
Data
RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession series GSE206859. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Informatics tools used for sequencing analysis in this manuscript are available for public use via GitHub (https://github.com/shengqh). Additional support is available through the corresponding authors.
References
Danesh, J., Collins, R., Appleby, P. & Peto, R. Association of fibrinogen, C-reactive protein, albumin, or leukocyte count with coronary heart disease: meta-analyses of prospective studies. JAMA 279, 1477–1482 (1998).
Ridker, P. M., Rifai, N., Rose, L., Buring, J. E. & Cook, N. R. Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. N. Engl. J. Med. 347, 1557–1565 (2002).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Moore, K. J. & Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 145, 341–355 (2011).
Moore, K. J. & Freeman, M. W. Scavenger receptors in atherosclerosis: beyond lipid uptake. Arterioscler. Thromb. Vasc. Biol. 26, 1702–1711 (2006).
Babaev, V. R. et al. Reduced atherosclerotic lesions in mice deficient for total or macrophage-specific expression of scavenger receptor-A. Arterioscler. Thromb. Vasc. Biol. 20, 2593–2599 (2000).
Suzuki, H. et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature 386, 292–296 (1997).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Febbraio, M. et al. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Invest. 105, 1049–1056 (2000).
Stewart, C. R. et al. CD36 ligands promote sterile inflammation through assembly of a toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11, 155–161 (2010).
Miller, Y. I. Toll-like receptors and atherosclerosis: oxidized LDL as an endogenous toll-like receptor ligand. Future Cardiol. 1, 785–792 (2005).
Steinbrecher, U. P. & Lougheed, M. Scavenger receptor-independent stimulation of cholesterol esterification in macrophages by low density lipoprotein extracted from human aortic intima. Arterioscler. Thromb. J. Vasc. Biol. 12, 608–625 (1992).
Steinberg, D. & Witztum, J. L. Is the oxidative modification hypothesis relevant to human atherosclerosis? Do the antioxidant trials conducted to date refute the hypothesis? Circulation 105, 2107–2111 (2002).
Kim, Kyeongdae et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Fernandez, D. M. et al. Single-cell immune landscape of human atherosclerotic plaques. Nat. Med. 25, 1576–1588 (2019).
Zernecke, Alma et al. Meta-analysis of leukocyte diversity in atherosclerotic mouse aortas. Circ. Res. 127, 402–426 (2020).
Spann, N. J. et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152 (2012).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Estruch, M. et al. CD14 and TLR4 mediate cytokine release promoted by electronegative LDL in monocytes. Atherosclerosis 229, 356–362 (2013).
Lamarche, B., Lemieux, I. & Després, J. P. The small, dense LDL phenotype and the risk of coronary heart disease: epidemiology, patho-physiology and therapeutic aspects. Diabetes Metab. 25, 199–211 (1999).
Smith, E. B. & Ashall, C. Low-density lipoprotein concentration in interstitial fluid from human atherosclerotic lesions. Relation to theories of endothelial damage and lipoprotein binding. Biochim. Biophys. Acta 754, 249–257 (1983).
Kruth, H. S., Huang, W., Ishii, I. & Zhang, W.-Y. Macrophage foam cell formation with native low density lipoprotein. J. Biol. Chem. 277, 34573–34580 (2002).
Goldstein, J. L., Ho, Y. K., Basu, S. K. & Brown, M. S. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl Acad. Sci. USA. 76, 333–337 (1979).
Anzinger, J. J. et al. Native low-density lipoprotein uptake by macrophage colony-stimulating factor-differentiated human macrophages is mediated by macropinocytosis and micropinocytosis. Arterioscler. Thromb. Vasc. Biol. 30, 2022–2031 (2010).
Jackson, W. D., Weinrich, T. W. & Woollard, K. J. Very-low and low-density lipoproteins induce neutral lipid accumulation and impair migration in monocyte subsets. Sci. Rep. 6, 20038 (2016).
Goulopoulou, S., McCarthy, C. G. & Webb, R. C. Toll-like receptors in the vascular system: sensing the dangers within. Pharmacol. Rev. 68, 142–167 (2016).
Seimon, T. A. et al. Atherogenic lipids and lipoproteins trigger CD36-TLR2-dependent apoptosis in macrophages undergoing endoplasmic reticulum stress. Cell Metab. 12, 467–482 (2010).
Mistry, P. et al. Inhibition of TLR2 signaling by small molecule inhibitors targeting a pocket within the TLR2 TIR domain. Proc. Natl Acad. Sci. USA. 112, 5455–5460 (2015).
Kadl, A. et al. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ. Res. 107, 737–746 (2010).
Vaeth, M. et al. Ca2+ signaling but not store-operated Ca2+ entry is required for the function of macrophages and dendritic cells. J. Immunol. 195, 1202–1217 (2015).
Heil, F. et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529 (2004).
Tanji, H. et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 22, 109–115 (2015).
Sarvestani, S. T. et al. Sequence-dependent off-target inhibition of TLR7/8 sensing by synthetic microRNA inhibitors. Nucleic Acids Res. 43, 1177–1188 (2015).
Hornung, V. et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 (2005).
Zhang, S. et al. Small-molecule inhibition of TLR8 through stabilization of its resting state. Nat. Chem. Biol. 14, 58–64 (2018).
Jurk, M. et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 3, 499 (2002).
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004).
Liu, J. et al. A five-amino-acid motif in the undefined region of the TLR8 ectodomain is required for species-specific ligand recognition. Mol. Immunol. 47, 1083–1090 (2010).
Ishii, N., Funami, K., Tatematsu, M., Seya, T. & Matsumoto, M. Endosomal localization of TLR8 confers distinctive proteolytic processing on human myeloid cells. J. Immunol. 193, 5118–5128 (2014).
Gorden, K. K. B., Qiu, X. X., Binsfeld, C. C. A., Vasilakos, J. P. & Alkan, S. S. Cutting edge: activation of murine TLR8 by a combination of imidazoquinoline immune response modifiers and polyT oligodeoxynucleotides. J. Immunol. 177, 6584–6587 (2006).
Allen, R. M. et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J. Extracell. Vesicles 7, 1506198 (2018).
Krieger, M., Brown, M. S., Faust, J. R. & Goldstein, J. L. Replacement of endogenous cholesteryl esters of low density lipoprotein with exogenous cholesteryl linoleate. Reconstitution of a biologically active lipoprotein particle. J. Biol. Chem. 253, 4093–4101 (1978).
Jonsson, A. L. & Bäckhed, F. Role of gut microbiota in atherosclerosis. Nat. Rev. Cardiol. 14, 79–87 (2017).
Robbins, M. et al. 2’-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. J. Am. Soc. Gene Ther. 15, 1663–1669 (2007).
Sioud, M., Furset, G. & Cekaite, L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 361, 122–126 (2007).
Mitchell, S. et al. An NFκB activity calculator to delineate signaling crosstalk: type I and II interferons enhance NFκB via distinct mechanisms. Front. Immunol. 10, Article 1425 (2019).
Rayner, K. J. et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 121, 2921–2931 (2011).
Rahman, S. M. et al. CCAAT/Enhancer-binding protein β (C/EBPβ) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem. 287, 34349–34360 (2012).
Zernecke, A. & Weber, C. Chemokines in atherosclerosis: proceedings resumed. Arterioscler. Thromb. Vasc. Biol. 34, 742–750 (2014).
Duewell, P. et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010).
Sugiyama, Y., Sugiyama, F., Yagami, K., Miyaji, S. & Kurosawa, T. Comparison of plasma endotoxin levels in germ-free, SPF and conventional laboratory animals (mice and rats). Jikken Dobutsu 42, 89–92 (1993).
Wright, S. D. et al. Infectious agents are not necessary for murine atherogenesis. J. Exp. Med. 191, 1437–1442 (2000).
Stepankova, R. et al. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. J. Atheroscler. Thromb. 17, 796–804 (2010).
Patinote, C. et al. Agonist and antagonist ligands of toll-like receptors 7 and 8: ingenious tools for therapeutic purposes. Eur. J. Med. Chem. 193, 112238 (2020).
Janssen, H. L. A. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013).
He, T. C. et al. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA 95, 2509–2514 (1998).
Michell, D. L. et al. Isolation of high-density lipoproteins for non-coding small rna quantification. J. Vis. Exp. https://doi.org/10.3791/54488 (2016).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem Physiol. 37, 911–917 (1959).
Paigen, B., Morrow, A., Holmes, P. A., Mitchell, D. & Williams, R. A. Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis 68, 231–240 (1987).
Linton, M. F., Atkinson, J. B. & Fazio, S. Prevention of atherosclerosis in apolipoprotein E-deficient mice by bone marrow transplantation. Science 267, 1034–1037 (1995).
Makowski, L. et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat. Med. 7, 699–705 (2001).
Babaev, V. R. et al. Macrophage EP4 deficiency increases apoptosis and suppresses early atherosclerosis. Cell Metab. 8, 492–501 (2008).
Chen, Y., Yu, Q. & Xu, C.-B. A convenient method for quantifying collagen fibers in atherosclerotic lesions by ImageJ software. Int J Clin Exp Med. 10, 14904–14910 (2017).
Park, E. K. et al. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56, 45–50 (2007).
Fleetwood, A. J., Lawrence, T., Hamilton, J. A. & Cook, A. D. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage csf-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178, 5245–5252 (2007).
Na, Y. R., Jung, D., Gu, G. J. & Seok, S. H. GM-CSF grown bone marrow derived cells are composed of phenotypically different dendritic cells and macrophages. Mol. Cells 39, 734 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Galkina, E. et al. Lymphocyte recruitment into the aortic wall before and during development of atherosclerosis is partially L-selectin dependent. J. Exp. Med. 203, 1273–1282 (2006).
Liu, Q. et al. scRNABatchQC: multi-samples quality control for single cell RNA-seq data. Bioinformatics 35, 5306–5308 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 1–9 (2019).
Audzevich, T. et al. Pre/pro-B cells generate macrophage populations during homeostasis and inflammation. Proc. Natl Acad. Sci. USA 114, E3954–E3963 (2017).
Williams, J. W. et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat. Immunol. 21, 1194–1204 (2020).
Tacke, R. et al. The transcription factor NR4A1 is essential for the development of a novel macrophage subset in the thymus. Sci. Rep. 5, 10055 (2015).
Sarrazy, V. et al. Maintenance of macrophage redox status by ChREBP limits inflammation and apoptosis and protects against advanced atherosclerotic lesion formation. Cell Rep. 13, 132–144 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Greenwald, R. A. CRC Handbook of Methods for Oxygen Radical Research (CRC Press, 1987).
Acknowledgements
The authors thank W. Reichard, M. Kuzmich, S. Landstreet, A. Ifram, L. Sedgeman, C. Wiese and V. Babaev for technical assistance and helpful discussions. We also thank Q. Liu of the Vanderbilt Center for Quantitative Sciences for consultation on single-cell sequencing analysis, and A. Jones of VANTAGE at VUMC for expertise in high-throughput sequencing technologies, the Vanderbilt Flow Cytometry Shared Resource and Translational Pathology Shared Resource. This work is supported by American Heart Association awards 19CDA34660280 (R.M.A.) and 18IPA34180005 (R.M.A.), W.M. Keck Research Foundation Grant (K.C.V., R.M.A., M.F.L. and Q.S.) and National Institutes of Health grants P01HL116263 (M.F.L.) and R01HL128996 (K.C.V.).
Author information
Authors and Affiliations
Contributions
R.M.A.: conceptualization, methodology, investigation, formal analysis, visualization and writing—original draft. D.L.M.: methodology, investigation, formal analysis and writing—reviewing and editing. A.B.C.: investigation and formal analysis. N.M.: investigation and formal analysis. E.M.S.: formal analysis and investigation. D.M.C.: resources and formal analysis. W.Z.: resources. C. DeJulius: resources. M.C.: resources. Y.Z.: resources. C.A.R.: formal analysis. M.R.-S.: software and visualization. S.Z. software and visualization. C. Duvall: methodology. A.C.D.: methodology and writing—reviewing and editing. Q.S.: methodology, software, visualization and writing—reviewing and editing. M.F.L.: methodology, supervision and writing—reviewing and editing. K.C.V.: conceptualization, methodology, supervision, formal analysis, visualization and writing—reviewing and editing.
Corresponding authors
Ethics declarations
Competing interests
M.F.L. has received research funding from Amgen, Regeneron, Ionis, Merck, REGENXBIO, Sanofi and Novartis, and has served as a consultant for Esperion, Alexion Pharmaceuticals and REGENXBIO. All other authors have no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Jeffrey Kroon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 nLDL induces inflammatory activation of macrophages.
(a) mRNA expression determined by qPCR of THP-1 macrophages treated with indicated doses of nLDL (matched to Fig. 1f) for 24 h (n = 3 biological replicates). (b) Quantification of immunoblots presented in Fig. 1f (n = 3 biological replicates). (c) Secreted cytokines in the media of THP-1 macrophages stimulated with 0.5 mg/ml nLDL for 3 h, 6 h, 24 h relative to cells receiving no treatment for 24 h (Ctr) (matched to Fig. 1e; n = 3 biological replicates) (d) Primary mouse bone-marrow derived macrophages (BMDM) differentiated with GM-CSF were treated with nLDL (0.5 mg/ml) for 24 h and assayed for mRNA expression by qPCR (n = 3 biological replicates), (e) cytokine secretion by ELISA (n = 3 biological replicates), and (f) protein expression in cell lysates by immunoblot (n = 2 biological replicates). Data are mean ± SEM. (a-c) One-way ANOVA and (d) Two-way ANOVA with Benjamini, Krieger and Yekutieli FDR (Q = 0.05), *q < 0.05, **q < 0.01, ***q < 0.001, ****q < 0.0001. e, Student’s t-test (unpaired, two-sided), **p < 0.01. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 2 nLDL and macrophage TLR responses quality control.
(a) Total protein (top) and neutral lipid (bottom) of ox LDL, bovine serum albumin (BSA), and 10 human nLDL of independent donors resolved by agarose gel electrophoresis. Image represents two independent experiments. (b) TBARS assay of nLDL samples, or matched LDL samples treated with copper sulfate as indicated (limit of detection = 0.625 μM) (n = 10 independent preparations). (c) Quantification of total protein and lipids of DGUC-VLDL, -LDL, -HDL, or BSA following fractionation with 2x-Superose-6 columns. Lipoprotein data are matched to a single donor representative of >10 independent experiments. (d) mRNA expression of primary human macrophages (CD14 + ; GM-CSF/IFNγ) pre-treated with C29 (200 μM) for 30 min, then stimulated with PAM3CSK4 (2 ng/mL) for 4 h (n = 3 biological replicates(BR)). (e) Normalized NF-κB-driven luciferase activity of HEK293T cells over expressing an empty vector, hTLR7 or hTLR8 following treatments with vehicle (Ctr, n = 4 BR), nLDL (0.5 mg/ml, n = 4 BR), ssRNA40 (TLR8 ligand; 2 μg/mL, n = 4 BR), R848 (TLR7 ligand; 10 μM, n = 3 BR), or CL075 (TLR8 ligand; 2.5 μg/mL, n = 3 BR). (f-h) mRNA expression of THP-1 macrophages electroporated with siRNA against TLR7,TLR8, or no siRNA (Control, n = 4 BR) and then treated with (f) nLDL (0.5 mg/mL, n = 6 BR), (g) R848 (10 μM, n = 6 BR) or (h) ssRNA40 (0.5 μg/mL, n = 6 BR). (i) Relative NF-κB-driven luciferase activity of HEK293T cells over expressing an empty vector (n = 7–8), mTLR7 (n = 7–8) or mTLR8 (n = 6-8) treated with mock transfection, R848 (10 μM), ssRNA40 (2 μg/mL), or nLDL (0.5 mg/ml) for 24 h. (j) mRNA expression in wild-type (WT) and Tlr7-/- BMDMs following treatment with 0.5 mg/ml nLDL, or 1 μg/mL ssRNA40 for 24 h (n = 3 BR). (k) Relative NF-κB-driven luciferase activity of HEK293T cells over expressing human or mTLR8 pre-treated with CU-CPT9a and exposed to nLDL for 24 h (n = 4 BR). Data are mean ± SEM. (d) Two-way ANOVA; Sidak’s multiple comparisons test, ***p < 0.001, ****p < 0.0001. (e, i-k) Two-way ANOVA; Benjamini, Krieger and Yekutieli FDR (Q = 0.05), *q < 0.05, **q < 0.01, ***q < 0.001). (f-h) One-way ANOVA; Dunnett’s multiple comparisons test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 3 The small RNA on LDL is predominantly exogenous and removed by LDL re-constitution.
a) Normalized abundance of taxa identified upon alignment of LDL-sRNA to non-host tRNA database (tRNA-db). RPM, reads per million total reads. b) Normalized abundance of bacterial phyla (human microbiome database) contributing sRNA to LDL. c) Normalized abundance of fungal sRNA and representative genomes present on LDL. d) Normalized abundance of algal and protist sRNA and representative genomes present on LDL. RPM, reads per million total reads. Matched nLDL and rLDL samples were fractionated by size-exclusion chromatography (SEC) using two superose-6 columns in tandem and assessed for e) phospholipid and protein content by colorimetric kit (representative data of three independent experiments), f) fluorescence (TopFluor Cholesteryl ester), and g) APOB protein by immunoblot (representative image of three independent experiments). h) Relative expression of exogenous sRNA in matched rLDL and nLDL of a single preparation relative to buffer controls. i) Oil-Red-O staining and fluorescence microscopy (TopFluor Cholesterol ester) (representative images of three biological replicates). Scale bar = 200 μm. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 4 Microbial small RNA on lipoproteins is not depleted in germ-free mice.
a) Plasma from two cohorts of adult mice - specific pathogen free (SPF; n = 6 mice total) and facility-matched germ-free (GF; n = 17 mice total) fed a chow diet were harvested at the National Gnotobiotic Rodent Resource Center (NGRRC; North Carolina, USA) for lipoprotein sRNA-seq b) Plasma was fractionated by size-exclusion chromatography (SEC) and cholesterol-rich fractions corresponding with HDL were selected for sRNA-seq. c) Relative percentage of reads aligned to host and non-host databases, as well as reads too short for analysis or reads that failed to align to either database (unmapped). d) Percentage of sRNA reads aligned to host miRNA, host tRNA and host rRNA transcripts. e) Percentage of reads aligned to the non-host rRNA database and tRNA database. f) Percentage of reads aligned to genomes of fungi and algae. g) Percentage of reads aligned to bacterial genomes associated with a human microbiome (HMB) database. h) Reads per million total reads (RPM) mapped to indicated bacterial phyla within the HMB database. i) Percentage of reads aligned to bacterial genomes within an environmental bacteria (ENV) database. j) Differential abundance (log2) of bacterial sRNA (dots represent individual genomes of the HMB and ENV databases) between GF and SPF mice categorized by phyla. Gray bar represents a 1.5 fold change. Data are mean ± SEM. (d-g, i) Statistical differences between GF and SPF were assessed by Mann-Whitney U-test, but no evaluations were statistically significant. j) Differences in abundance of individual genomes within each database were assessed between groups by the Wald Test, but applying False Discovery Rate correction (α = 0.05) resulted in no differentially abundant genomes. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 5 Locked nucleic acid (LNA) bases mediate antagonism of single-stranded RNA ligands of TLR8.
a) HEK293T cells over-expressing human TLR8, UNC93B1 and CD14 were pre-treated with vehicle (DOTAP) or corresponding DNA/LNA oligonucleotides (2.5 μg/mL) for 30 min and then stimulated with TLR8 nucleoside analogue agonist CL075 or TLR8 ORN agonists ssRNA40 or ORN06 (2 μg/mL) for 24 h (n = 5 biological replicates). Two-way ANOVA with Dunnett’s multiple comparison test (statistical significance relative to untreated within each group; **p < 0.0001). b) THP-1 macrophages were pre-treated + /- nt-LNA (1 µg/mL) for 45 min and treated with LPS (500 ng/mL), Poly I:C (1 µg/mL), CL075 (2.5 μg/ml), or ssRNA40 at 1 µg/mL (1:1, nt-LNA:ssRNA40) or 0.2 µg/ml (5:1) for 24 h (n = 3 biological replicates). Relative mRNA expression of IL1B, IL6 and TNF were then assessed by qPCR. For each treatment, the relative fold change of each treatment in the presence of nt-LNA was expressed as a percentage of the relative fold change of each treatment without nt-LNA pre-treatment (% inhibition). Two-way ANOVA, Benjamini, Krieger and Yekutieli FDR (Q = 0.05), **q < 0.01. c-d) Primary human CD14 + PBMC differentiated with GM-CSF and IFNγ were pre-treated with 2.5 μg/mL nt-LNA or vehicle (DOTAP) for 30 minutes and then stimulated with or without ssRNA40 (0.5 μg/mL) for 24 h. c) mRNA expression was quantified by qPCR (n = 4 biological replicates) d) Cytokine (IL-6) secretion was quantified by ELISA (n = 4 biological replicates). One-way ANOVA with Dunnett’s multiple comparison test. **p < 0.01. ***p < 0.001, ****p < 0.0001 (e) mRNA expression (n = 4 biological replicates), (f) cytokine secretion (n = 3 biological replicates) and (g) immunoblotting (representative image of three independent experiments) of BMDMs following up to 24 h treatment with IFNγ (100 U/mL) +/- 0.5 mg/ml nLDL in the presence or absence of 2.5 μg/mL nt-LNA. Two-way ANOVA, Benjamini, Krieger and Yekutieli FDR (Q = 0.05), *q < 0.05, **q < 0.01. Data are mean ± SEM. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 6 nt-LNA treatment reduces atherosclerosis without altering lipid or lipoprotein metabolism in Apoe-/- mice.
a) Female and male Apoe-/- mice fed a western diet were administered saline (Ctr; n = 4 mice per sex), nt-LNA-A (20 mg/kg; n = 4 mice per sex) or nt-LNA-B (20 mg/kg; n = 4 mice per sex) by intraperitoneal injection once weekly for four weeks. Treatments for each were randomized between cohabitating animals separated by sex. At sacrifice, the aortic sinus was serially sectioned and stained with Oil-Red O to identify atherosclerotic lesions. Scale bar = 500 μm. b) Quantification of lesion area in serial sections and c) sex-normalized, relative lesion area under the curve (n = 8 mice per treatment). d) Plasma of female Apoe-/- mice treated for 4 weeks with saline (Ctr; n = 10) or nt-LNA (n = 10) were fractionated by size-exclusion chromatography and assessed for total cholesterol (TC) e) Plasma protein levels were assessed by immunoblot of individual cages receiving either Saline/Ctr (n = 5 mice) or nt-LNA (n = 5 mice) treatments (representative images of two independent assessments). f) Quantification of independent immunoblots by densitometry (n = 10 mice per treatment) normalized to C3. g) Lesion area (Oil-red O) of matched sections of the aortic root following treatment with saline (Ctr) or nt-LNA for 4 weeks (n = 10 mice per treatment). h) Hepatic mRNA expression determined by qPCR (n = 10 mice per treatment). Data are mean ± SEM. (c) One-way ANOVA, Sidak’s multiple comparison test, *p < 0.05. **p < 0.01. (f-h) Two-way ANOVA with Benjamini, Krieger and Yekutieli FDR (Q = 0.05), *q < 0.05, **q < 0.01,***q < 0.001. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 7 nt-LNA treatment promotes atherosclerotic regression in Ldlr-/- mice.
(a) Schematic for regression study design. Male(M) and female(F) Ldlr-/- mice were fed a chow diet (n = 6 mice) or an atherogenic diet (n = 50 mice) for 14 weeks. After 14 weeks, chow-fed mice and a subset of mice from the atherogenic diet group (baseline; n = 7 M/8 F mice). Remaining diet-fed mice were then switched to a chow diet to allow lesion regression (Reg.) and were injected once weekly with saline control (Reg. Ctr; n = 9 M/9 F mice) or Reg. nt-LNA (30 mg/kg; n = 9 M/8 F mice). b) Plasma total cholesterol (TC) or c) triglycerides (TG) following fractionation by SEC; chow (n = 6; 3 M/3 F), baseline (n = 8; 4 M/4 F), Reg. Ctr; (n = 10; 5 M/5 F) and Reg. nt-LNA (n = 10; 5 M/5 F) d) Immunoblots of plasma proteins in Reg. Ctr (n = 10) or Reg. nt-LNA (n = 9) groups. Representative images of two independent experiments are shown. e) Quantification of immunoblots by densitometry. f-g) Lesion area of serial sections of the aortic root in baseline (n = 15 mice; 7 M/8 F), Reg. Ctr(n = 18 mice; 9 M/9 F) or nt-LNA (n = 17; 9 M/8 F) groups. h) Lesion area under the curve (AUC) for both sexes of mice as determined by Oil Red O staining in the aortic root (Baseline: n = 15; Reg. Ctr: n = 18; Reg. nt-LNA; n = 17). One-way ANOVA; Dunnett’s multiple comparison test, **p < 0.01, ***p < 0.001. i) Lesion AUC for mice of each group separated by sex. Two-way ANOVA; Dunnett’s multiple comparison test, **p < 0.01. j-k) Masson’s Trichrome staining and quantification of fibrosis in aortic roots of baseline (n = 7 mice; 3 M/4 F) Reg. Ctr (n = 9 mice; 4 M/5 F) or Reg. nt-LNA (n = 10 mice; 5 M/5 F) groups. l-m) MAC2 (green) immunofluorescence and quantification within aortic roots obtained of at baseline (n = 7 mice; 3 M/4 F), Reg. Ctr (n = 10 mice; 5 M/5 F), or Reg. nt-LNA (n = 10 mice; 5 M/5 F) groups. Two-way ANOVA; Tukey’s multiple comparison test, **p < 0.01, ***p < 0.001. Data are mean ± SEM. Scale bar = 500 μm. Numerical source data, statistics, exact p values and q values are provided.
Extended Data Fig. 8 Gating strategy of leukocytes from mouse aortas for single-cell RNA sequencing.
Sequential gating fluorescent activated cell sorting for single and live cells, followed by non-red blood cells. Cells were then sorted that were CD45 + but CD3−.
Extended Data Fig. 9 Single-cell RNA sequencing of the atherosclerotic lesion to identify anti-atherosclerotic mechanisms of nt-LNA treatment.
a) Apoe-/- mice fed an atherogenic diet for 4 weeks were injected once weekly with saline control (Ctr; n = 8) or nt-LNA (30 mg/kg; n = 8). b) UMAP projection of unbiased clusters obtained from atherosclerotic lesions. c) Relative contribution of cells from saline (Ctr) and nt-LNA treated mice to each cluster of (b). d) Relative expression (color) and % of cells reaching threshold of detection (size) of transcripts pertaining to T cell and NK cell phenotypes (top) or B-cell phenotypes (bottom) in atherosclerosis for each cluster. Numerical source data, statistics, exact p values and q values are provided.
Supplementary information
Source data
Source Data Fig. 1
Numerical source data.
Source Data Fig. 1
Uncropped Western blots.
Source Data Fig. 2
Numerical source data.
Source Data Fig. 2
Uncropped Western blots.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Numerical source data.
Source Data Fig. 4
Uncropped Western blots.
Source Data Fig. 5
Numerical source data.
Source Data Fig. 6
Numerical source data.
Source Data Fig. 7
Numerical source data.
Source Data Extended Data Fig. 1
Numerical source data.
Source Data Extended Data Fig. 1
Uncropped Western blots.
Source Data Extended Data Fig. 2
Numerical source data.
Source Data Extended Data Fig. 2
Uncropped Western blots.
Source Data Extended Data Fig. 3
Numerical source data.
Source Data Extended Data Fig. 3
Uncropped Western blots.
Source Data Extended Data Fig. 4
Numerical source data.
Source Data Extended Data Fig. 5
Numerical source data.
Source Data Extended Data Fig. 5
Uncropped Western blots.
Source Data Extended Data Fig. 6
Numerical source data.
Source Data Extended Data Fig. 6
Uncropped Western blots.
Source Data Extended Data Fig. 7
Numerical source data.
Source Data Extended Data Fig. 7
Uncropped Western blots.
Source Data Extended Data Fig. 9
Numerical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Allen, R.M., Michell, D.L., Cavnar, A.B. et al. LDL delivery of microbial small RNAs drives atherosclerosis through macrophage TLR8. Nat Cell Biol 24, 1701–1713 (2022). https://doi.org/10.1038/s41556-022-01030-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-01030-7