Abstract
Salivary glands act as virus reservoirs in various infectious diseases and have been reported to be targeted by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the mechanisms underlying infection and replication in salivary glands are still enigmatic due to the lack of proper in vitro models. Here, we show that human induced salivary glands (hiSGs) generated from human induced pluripotent stem cells can be infected with SARS-CoV-2. The hiSGs exhibit properties similar to those of embryonic salivary glands and are a valuable tool for the functional analysis of genes during development. Orthotopically transplanted hiSGs can be engrafted at a recipient site in mice and show a mature phenotype. In addition, we confirm SARS-CoV-2 infection and replication in hiSGs. SARS-CoV-2 derived from saliva in asymptomatic individuals may participate in the spread of the virus. hiSGs may be a promising model for investigating the role of salivary glands as a virus reservoir.
Similar content being viewed by others
Main
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and spreads quickly worldwide. The main target organs of SARS-CoV-2 are considered to belong to the respiratory system, including the lungs and upper respiratory tract, but accumulating evidence has shown that multiple organs, such as the heart, kidneys, liver, spleen and gastrointestinal tract, can be affected1. SARS-CoV-2-infected individuals are a potential source of virus transmission, which occurs through aerosol droplets, close contact and facial–oral transmission2. The majority of infected individuals (approximately 80%) are asymptomatic and have the ability to spread the virus3. It was recently reported that saliva from asymptomatic individuals with COVID-19 can be a potential source of viral transmission4. SARS-CoV-2 has been identified in the oral mucosa and salivary glands of patients with COVID-19. The viral entry factors angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) are widely expressed in the oral mucosa and salivary glands. Consistent with this, salivary glands have been reported to be reservoirs of viruses5. Therefore, it is postulated that salivary glands can serve as a reservoir of COVID-19 and play a role in transmitting the virus in asymptomatic individuals.
To examine the possibility that SARS-CoV-2 infects multiple organoids, in vitro culture models involving organoids have been used. Many organoids, such as lung, gut, brain, pancreas and liver organoids, have already been applied for this purpose6,7,8,9,10. Therefore, an organoid culture model is expected to be suitable for demonstrating the SARS-CoV-2 infection of salivary glands in vitro. We recently generated functional salivary gland organoids derived from mouse embryonic stem cells11. Here, we succeeded in generating functional salivary gland organoids from human induced pluripotent stem cells (hiPSCs) by modifying the method used for the induction of mouse embryonic stem cell-derived salivary gland organoids. The induced human salivary gland organoids showed similar morphological features and physiological functions to salivary glands. These organoids were composed of salivary gland-specific cell lineages. We then showed that SARS-CoV-2 infection and replication occurred in the salivary gland organoids. Thus, we reproduced the SARS-CoV-2 infection of salivary glands using an in vitro model.
Induction of human salivary gland organoids from hiPSCs
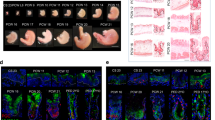
We sought to induce oral ectodermal cells from hiPSCs using a modified three-dimensional culture system for the induction of mouse salivary gland organoids. Oral primitive ectoderm was induced using bone morphogenetic protein 4 (BMP4) and SB431542 (an inhibitor of transforming growth factor-β). hiPSCs were dissociated into single cells and plated onto V-bottom 96-well plates to create uniform cell aggregates (Fig. 1a and Extended Data Fig. 1a). We then transferred the aggregates to new U-bottom plates containing growth factor-reduced Matrigel with BMP4 or SB431542 on day 2. This culture system induced the gene expression of ectodermal markers (DLX3 and TFAP2A)12 regardless of the addition of SB431542 or BMP4 on day 2 (Fig. 1b). SB431542 treatment significantly increased the gene expression of PITX2 (a head ectodermal epithelial marker) compared with the results observed under other differentiation conditions on day 12 after the onset of differentiation (Fig. 1b). The aggregates without SB431542 formed pancytokeratin-positive (Pan-CK+) epithelial cysts with thin walls, whereas the SB431542-treated aggregates formed solid Pan-CK+ epithelial aggregates on day 12 (Fig. 1c–e). SRY-box transcription factor 9 (SOX9) is one of the essential transcription factors involved in salivary gland development and is regulated by fibroblast growth factors (FGFs) secreted by the salivary gland mesenchyme. In our culture system, FGF7 and FGF10 treatment starting on day 12 upregulated SOX9 expression in Pan-CK+ epithelial cells in the aggregates on day 20 (Fig. 1e,f). In vivo human salivary gland development begins in embryonic weeks 6–8, and the formation of a branching structure with clearly defined end buds is observed by week 16 (ref. 13). Thus, the culture was continued for 60 d to allow the ectodermal epithelial cells to reach a branching stage. On day 60, salivary gland-like buds appeared within the aggregates (Fig. 1g). The aggregates expressed salivary gland markers, including SOX9, FOXC1, CK19, AQP5, ACTA2 and muscarinic receptor-3 (CHRM3) (Extended Data Fig. 1b). The centres of the aggregates were composed of Pan-CK+, SOX9− epithelium without branching structures, and the peripheral branching structures consisted of Pan-CK+, SOX9+ and FOXC1+ epithelial cells. The distal cells were aquaporin 5-positive (AQP5+) acinar-like cells, CK18+ ductal-like cells, CK5+ basal cells and alpha-smooth muscle actin-positive (α-SMA+) myoepithelial-like cells (Fig. 1h and Extended Data Fig. 1c). To examine whether the combination of FGF7 and FGF10 was necessary for salivary gland differentiation, hiPSC-derived ectodermal epithelial cells were treated with or without FGF7 and FGF10. Consistent with a previous mouse study14,15,16, branching structures were induced only in the presence of FGF7 and FGF10. Treatment with either FGF7 or FGF10 induced the appearance of Pan-CK+ epithelial cells but not SOX9 or AQP5 expression (Extended Data Fig. 2a,b).
a, Culture protocol of salivary gland differentiation from hiPSCs. TGF-β-i, inhibitor of transforming growth factor-β1. b, Real-time RT-PCR analysis of oral ectoderm genes in the aggregates on day 12 with or without BMP4 and SB431542 (SB). The data are normalized to ribosomal protein L27 and are presented as means ± s.d. (n = 3 biological replicates). Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test. c, Phase-contrast images of aggregates on days 2 (left) and 12 (right) without SB431542. Scale bars, 200 μm. The images are representative of three independent experiments. d, Phase-contrast images of aggregates on days 12 (left) and 20 (right) with SB431542 treatment. Scale bars, 200 μm. The images are representative of 18 independent experiments. e, Pan-CK and SOX9 were detected via immunofluorescence analysis. The images are representative of three samples showing similar results. Scale bars, 200 μm. f, Real-time RT-PCR analysis of SOX9 in the aggregates on days 12, 16 and 20. The data are normalized to ribosomal protein L27 and are presented as means ± s.d. (n = 4 biological replicates). Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test. g, Representative phase-contrast images of branching structures on day 60. Scale bars, 200 μm. h, Immunofluorescence for Pan-CK, SOX9 and AQP5 in the branching structures of day 60 aggregates. The images are representative of six independent experiments. Scale bars, 400 μm (top) and 50 μm (bottom).
Human salivary gland organoids recapitulate salivary gland features and functions
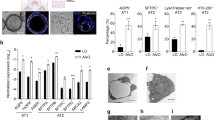
To further characterize the observed branching structures, the organoids were isolated on day 60 and cultured with FGF7 and FGF10 until day 80 (Fig. 2a,b). Immunofluorescence analysis revealed that Pan-CK+ epithelial cells expressed SOX9 and forkhead box C1 (FOXC1) and included AQP5+ acinar-like cells, cytokeratin 19-positive (CK19+) ductal-like cells, α-SMA+ myoepithelial-like cells, CK14+ basal cells and ZO-1+ luminal structures, as observed in embryonic mouse salivary glands (Fig. 2c). In addition, AQP5 expression was distributed in the cytosol and basolateral membrane but did not accumulate in the apical membrane. Some AQP5 cells were CK19 and AQP5 double positive. Thus, the hiPSC-derived organoids exhibited similarities to embryonic salivary glands on the basis of their morphology. Hence, the generated structures are referred to as human induced salivary glands (hiSGs). We compared the gene expression profiles of the hiSGs with the deposited expression profiles of human embryonic and adult salivary glands17 via bulk RNA sequencing (RNA-seq) (Supplementary Table 1). A heatmap of salivary gland-specific gene expression data indicated that the organoids expressed most salivary gland markers, including AQP5, ACTA2, SOX9, SOX10 and CHRM3, and embryonic salivary gland markers, including ETV5, SPRY1 and SPRY2, but not pluripotency markers (SOX2 or NANOG) or other branching organ markers (NKX2.1 (lung) or PDX1 (pancreas)) (Fig. 2d). These data indicated that the hiSGs showed similar gene expression patterns to embryonic human salivary glands. Fluid secretion is one of the fundamental functions of salivary glands and is induced by acetylcholine through muscarinic acetylcholine receptor pathways18. Treatment with carbachol transiently induced an increase in the intracellular calcium concentration of the hiSGs in a dose-dependent manner (Fig. 2e). An increase in intracellular calcium was observed, which was blocked by the muscarinic receptor antagonist atropine. Next, we investigated whether the salivary glands could be orthotopically transplanted using a previously reported method and developed them in vivo19. Briefly, hiSGs were cocultured with salivary gland mesenchymal tissue from E13.5 mice for 24 h. On the recipient side, parotid gland-defective severe combined immunodeficient (SCID) mice were prepared, and the mesenchyme combined with the hiSGs was transplanted into parotid gland-defective mice (Fig. 3a,b). Eighty percent of the transplanted organoids (20 of 27 xenografts; two experiments) were engrafted at transplant sites (Fig. 3c). The other transplanted organoids were lost due to technical issues. Histological analysis using haematoxylin and eosin staining revealed that the engrafted hiSGs showed mature salivary gland structures, such as ducts and acinar glands (Fig. 3c). Immunofluorescence analysis revealed that the glandular tissues showed a positive signal of human mitochondria and that the organoid duct was linked to the host excretory duct (Fig. 3d). The distributions of salivary gland-specific markers, such as SOX9, FOXC1, AQP5, CK5 and α-SMA, in transplanted organoids were similar to those in normal salivary glands (Fig. 3e). AQP5 was localized at the apical membrane of acinar cells, as observed in mature acinar cells of salivary glands. The acinar cells in engrafted organoids expressed the serous acinar cell marker α-amylase 1A (AMY1A) (Fig. 3e). Among other genes reported to be expressed in salivary glands, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) results showed that the gene expression levels of MUC5B and MUC7 were higher in the hiSGs than in undifferentiated hiPSCs, while the expression levels of both genes were much higher in the transplanted hiSGs than in untransplanted hiSGs. In addition, the gene expression level of SLC12A2 was much higher in both untransplanted and transplanted hiSGs than in undifferentiated hiPSCs, while the expression level was quite similar between untransplanted and transplanted hiSGs (Extended Data Fig. 3a). These results suggest that transplantation induces hiSG maturation. In addition, mouse cluster of differentiation 31-positive endothelial cells and tubulin beta 3 class III-positive neuronal fibres were found at positions close to acinar cells (Extended Data Fig. 3b). These data show that orthotopically transplanted organoids grow into mature salivary glands penetrated by host blood vessels and nerves.
a, Phase-contrast image of an aggregate on day 60 before isolation. The image is representative of 18 independent experiments. Scale bar, 200 μm. b, Left, representative phase-contrast image of an isolated hiSG on day 80. Middle and right, representative stereomicroscopic images of an hiSG on day 80. The images are representative of 18 independent experiments. Scale bars, 200 μm. c, Immunofluorescence images of hiSGs on day 80. The images are representative of four independent experiments showing similar results. Scale bars, 200 μm (top left) and 50 μm. d, Heatmap of the gene sets of adult salivary glands (SOX9, FOXC1, SOX10, AQP5, ACTA2 and CHRM3), embryonic salivary glands (ETV5, SPRY1 and SPRY2), pluripotent stem cells (SOX2 and NANOG) and other branching organs (PDX1 and NKX2.1). Data are shown for hiSGs on day 80 (n = 3 biological replicates), human embryonic SMGs, adult SMGs, adult parotid glands (PARs) and adult sublingual glands (SLGs). e, Representative data from hiSG calcium release analysis. Fluo-4 was used to measure the calcium concentrations inside cells. hiSGs were stimulated with two concentrations of carbachol (CCh) (100 μM or 10 μM) or pretreated with atropine (ATR), followed by 100 μM CCh. Changes in the Fluo-4 fluorescence intensity were recorded. This experiment was replicated three times with similar results. a.u., arbitrary unit.
a, Schematic of the orthotopic transplantation of hiSGs into mice with defective parotid glands. Mes, mesenchymal tissues. b, Representative stereomicroscopic image of a hiSG cocultured with normal embryonic mouse-derived salivary gland mesenchymal tissues containing a PGA monofilament guide (n = 27 samples; two individual experiments). c, Left, representative stereomicroscopic image of hiSG transplantation in a salivary gland-defective immunodeficient mouse on day 30 after transplantation (n = 27 samples; two individual experiments). Right, haematoxylin and eosin staining of the transplantation site. The image is representative of three experiments showing similar results. Scale bars, 1 mm (left), 50 μm (right). d, Immunofluorescence images of the transplantation site. Left, human mitochondria+ (hMitochondria) cells formed gland-like structures. Scale bar, 400 μm. Right, higher-magnification images of the connection between the mouse Epcam+ (mEpcam) host duct and human mitochondria+ duct of the salivary gland organoid. Scale bars, 50 μm The images are representative of three samples showing similar results. e, Immunofluorescence staining of the salivary gland markers SOX9, FOXC1, AQP5, α-SMA and AMY1A in transplanted hiSGs. The images are representative of three samples showing similar results. Scale bars, 50 μm.
Single-cell RNA-seq reveals the cellular components of hiSGs
To gain insight into human salivary gland cell lineages, we performed single-cell RNA sequencing (scRNA-seq) of hiSG cells on day 80 using the 10X Genomics platform. Unbiased clustering identified ten subtypes, which we manually annotated into six major cell groups using Seurat20 (Fig. 4a and Extended Data Fig. 4a). The mesenchymal group contained COL1A1+ and COL1A2+ cells that did not express FGF7 or FGF10 (Fig. 4b). Therefore, the small mesenchymal population in the organoids did not show features of salivary gland mesenchyme. The epithelial group was separated into eight clusters that were identified based on the expression of EPCAM (all epithelium), KRT14 and KRT5 (basal duct), KRT19 (duct), AQP5 (end bud and proacinar), ACTA2 (myoepithelial) and MKI67 (cycling cells). The epithelial group was annotated into five cell groups: acinar, basal, ductal, myoepithelial and actively cycling cells (Fig. 4c,d). Most recently, a method for the induction of hiPSC-derived lacrimal glands was reported21, but PAX6—a master regulator for the lacrimal gland—was not expressed in hiSGs (Fig. 4d). In addition, no cell clusters expressed pluripotent markers, such as NANOG and OCT4 (Extended Data Fig. 4b). A bulk RNA-seq analysis of human salivary glands reported previously identified MTRNR2L8 and LYZ as salivary gland-specific genes and TOP2 and HEY1 as genes involved in gland development17. These genes are also expressed in hiSGs, resulting in an expression pattern similar to the human foetal salivary gland gene expression pattern (Extended Data Fig. 4c). In addition, uniform manifold approximation projection (UMAP) analysis showed that the epithelial cells were divided into two large populations: basal/myoepithelial and ductal/myoepithelial. Using epithelial cells, we constructed a transcriptional trajectory to examine whether organoids recapitulate the process of salivary gland organogenesis. For trajectory analysis, the epithelial subsets were annotated using Monocle 3 (ref. 22) based on the previous results (Extended Data Fig. 5a,b). Trajectory analysis revealed that there were two epithelial lineages in the organoids (Extended Data Fig. 5c). We manually selected cycling cells as the origin of the trajectory because a previous mouse scRNA-seq study showed the localization of mitotic cells at the branching point from which all other postnatal populations are derived. In addition, the study revealed that from embryonic day 16 onward, myoepithelial cells and basal duct cells belong to terminal branches with low pseudotime scores, probably because of their early specification23. Consistent with these findings, our pseudotime analysis results suggested a distinct lineage of basal–myoepithelial cells from the ductal–acinar lineage (Fig. 4e). When the two lineages were analysed individually, the expression levels of differentiation genes were found to increase along the pseudotime axis (Extended Data Fig. 5d–i). These results demonstrate the occurrence of cell differentiation in hiSGs and are consistent with previous findings of mouse lineage analyses showing that myoepithelial cells are supplied by basal cells24,25.
a, UMAP plots of hiSGs on day 80. The data represent 7,190 cells. Ten organoids were pooled for one scRNA-seq experiment. These cells were annotated into nine cell clusters. b, Expression of epithelial (EPCAM and CDH1) and mesenchymal markers (COL1A1 and COL1A2) for the classification of cells in whole hiSGs. The mesenchymal cell cluster did not express FGF7 and FGF10. c, UMAP plots of epithelial cells in hiSGs. These cells were separated into eight clusters and annotated into five cell types. d, Feature plots for the expression of selected marker genes: SOX9, FOXC1 and SOX10 (salivary gland transcription factor), KRT18, KRT19 and KRT8 (ductal marker), AQP5 (acinar marker), ACTA2 (myoepithelial marker), KRT14, KRT5 and TP63 (basal cell marker), MKI67, CCNB1 and CDK1 (cycling cell marker) and PAX6 (lacrimal gland marker). The colour scale represents the expression level. e, Pseudotime trajectory projected onto a UMAP embedding of ductal–acinar lineage (left) or basal–myoepithelial lineage (right). Pseudotime values are colour coded.
Modelling of human salivary gland development in organoids
As reported in previous mouse studies, Sox9 is an important transcription factor that regulates salivary gland development. In Sox9 conditional knockout mice, the salivary glands are unable to undergo branching morphogenesis14. To elucidate whether hiSGs recapitulate the SOX9-mediated developmental process, we suppressed SOX9 during hiSG induction. To this end, we generated inducible SOX9 knockdown hiPSCs using the CRISPR interference (CRISPRi) system26. We generated inducible CRISPRi hiPSC clones expressing dCas9-KRAB driven by the inducible TetO promoter at the AAVS1 locus using the transcription activator-like effector nuclease (TALEN) system (Fig. 5a). To test the efficacy of guide RNAs (gRNAs) in CRISPRi, we designed three gRNAs targeting the transcription start site of SOX9 or SOX2 (positive control) and generated CRISPRi hiPSC clones expressing each gRNA (Fig. 5b). These clones expressed dCas9 in a doxycycline (Dox)-dependent manner (Fig. 5c). We found that one of three gRNAs significantly inhibited SOX9 expression in day 80 organoids following the addition of Dox starting on day 12 (Fig. 5c). Branching structures were induced in the Dox-treated wild-type clone and the CRISPRi-SOX9 clone without Dox treatment but not in the Dox-treated CRISPRi-SOX9 clone (Fig. 5d,e). Consistent with in vivo studies conducted in mice, SOX9 knockdown inhibited branch formation in hiPSCs, suggesting that SOX9 is an important factor in human salivary gland development. Importantly, the established model may be able to replace animal models that have been used as substitutes for the analysis of human salivary gland development.
a, A subset of putative clones and a pooled population from each condition were analysed using junction PCR and confirmed on-target integration of the cassette into the AAVS1 locus. b, The clones from each condition were tested containing a SOX2-specific gRNA. Samples were cultured in the presence or absence of Dox (2 μM) for 4 d and analysed using real-time RT-PCR. The data are normalized to ribosomal protein L27 and are presented as means ± s.d. (n = 3 biological replicates). Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test. c, The clones from each condition were tested containing SOX9-specific gRNAs. Samples were cultured in the presence or absence of Dox (2 μM) from differentiation day 80 and analysed for dCas9 (left) and SOX9 (right) expressions using RT-qPCR. The data are normalized to ribosomal protein L27 and are presented as means ± s.d. (n = 3 biological replicates). Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test. d, Salivary gland-like branching structures observed in the aggregate. Arrowheads indicate salivary gland-like structures under the control conditions (top and middle). Salivary gland-like structures were not detected under Dox-dependent SOX9 knockdown conditions (bottom). Scale bars, 500 μm. e, Efficiency of salivary gland-like structure formation under each condition (n = 24 organoids (per treatment); two independent experiments).
Infection and replication of SARS-CoV-2 in hiSGs
We generated hiPSC-derived salivary gland organoids to model SARS-CoV-2 infection. Similar to the results of the 10X Genomics scRNA-seq, scRNA-seq using random displacement amplification sequencing (RamDA-seq) data revealed six subtypes in hiSGs (Fig. 6a and Extended Data Fig. 6a). The ductal cell group showed specific expression of the SARS-CoV-2 receptor ACE2 and processing protease TMPRSS2 messenger RNAs (mRNAs) (Fig. 6b and Extended Data Fig. 6b,c). Immunostaining for ACE2 and TMPRSS2 on hiSGs showed that ACE2 was strongly expressed on the apical side of ductal cells, while TMPRSS2 was expressed on ductal cells and acinar cells (Extended Data Fig. 6d). Similar to the scRNA-seq data, double-positive cells were identified among ductal cells. Human salivary gland expression has also been examined for some other entry factors, including tissue-specific proteases (TMPRSS4 and TMPRSS11D) and endosomal proteases (cathepsin B (CTSB), cathepsin L (CTSL), Basigin (CD147) and FURIN)4. The hiSG scRNA-seq data revealed that these other entry factors are broadly expressed in hiSG cells, similar to previous reports (Extended Data Fig. 6e). To facilitate the access of SARS-CoV-2 to epithelial cells, branching structures isolated from organoids were infected with SARS-CoV-2 and analysed at 24 and 72 h post-infection (hpi). SARS-CoV-2 genomic RNA was detected in cell lysates and culture supernatants at 24 hpi (Fig. 6c). SARS-CoV-2 genomic RNA levels were increased in both the supernatant and cell lysates at 72 hpi relative to 24 hpi. In addition, infected organoids showed replication-specific SARS-CoV-2 spliced subgenomic RNA (Orf7a) (Fig. 6d). These data suggest that SARS-CoV-2 can infect hiSGs and that SARS-CoV-2 replication occurs within 24 hpi. Immunofluorescence analysis revealed that CK19-positive ductal luminal cells showed a positive SARS-CoV-2 nucleoprotein signal (Fig. 6e). Consistent with the observation of apparent virus replication at 24 and 72 hpi, intracellular virus-like particles were identified within enlarged membranous compartments using transmission electron microscopy (Fig. 6f). These membranous structures containing virus-like particles were not observed in mock-infected hiSG cells. While there are no available electron microscopy images of SARS-CoV-2 virus particles in salivary gland tissue either in vivo or in vitro, spherical virus-like particles have been observed to cluster within a membrane separating them from the cytoplasm and to contain heterogeneous, electron-dense interior granules postulated to be ribonucleoprotein complexes typically seen in virus-infected cells. Furthermore, evaluation of the 50% tissue culture infectious dose (TCID50) using culture supernatants showed that viral release peaked at 24 h but later declined (Fig. 6g). Importantly, these results are consistent with a previous report regarding alveolospheres infected with SARS-CoV-2 (ref. 27). These data show that SARS-CoV-2 productively replicates in hiSG cells.
a, UMAP of RamDA-seq samples of hiSGs on day 80. The cell types are colour coded. b, Dot plot of the gene set for each salivary gland cell type and ACE2 and TMPRSS2. The dot size represents the fraction of cells within that type expressing a given gene and the colour intensity represents the binned count-based expression amount among expressing cells. c, Real-time RT-PCR using primers and probes against the NIID N2 amplicon of SARS-CoV-2 in infected hiSGs (left; n = 3, 4 and 3 biological replicates for the mock, 24 hpi and 72 hpi, respectively) and culture supernatants (right; n = 3 biological replicates). The data are presented as means ± s.d. Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test. d, RT-PCR data using primers against replication-specific SARS-CoV-2 spliced subgenomic RNA (sgRNA Orf7a) and L27 in infected hiSGs and 293T-ACE2-TMPRSS2 cells. e, Immunofluorescence for CK19 and SARS-CoV-2 nucleoprotein (NP) in the uninfected (mock) and infected hiSGs. The images are representative of three samples showing similar results. Scale bars, 100 μm. f, Transmission electron microscopy images of uninfected (mock) and infected hiSGs. The vesicles in infected cells were filled with virions. The images are representative of three samples showing similar results. Scale bars, 500 nm (left and middle) and 100 nm (right). g, TCID50 assay for SARS-CoV-2 in culture supernatant (n = 4, 5 and 4 biological replicates for 1, 24 and 72 hpi, respectively). The data are presented as means ± s.d. Statistical significance was determined by one-way ANOVA followed by Tukey’s post-hoc test.
Discussion
In the present study, we successfully generated hiSGs from hiPSCs. The hiSGs showed similar characteristics to human embryonic salivary glands, including morphological, protein marker expression and gene expression characteristics. The hiSGs exposed to SARS-CoV-2 showed viral infection and replication within their cells. Thus, hiSGs are expected to serve as a proper model for reproducing heterogeneous cell populations in human salivary glands.
We induced primitive oral epithelium from hiPSCs by modifying the methods described in our previous report11. However, in this case, there was no need to transduce the SOX9 or FOXC1 genes into the induced oral epithelium because the expression of these genes was induced only by FGF7 and FGF10. Crucially, no branching structure was formed by the inhibition of SOX9 gene expression using the CRISPRi system, suggesting that SOX9 is a common important gene in both mouse and human salivary gland development14. The hiSGs consisted of ductal, acinar, basal and myoepithelial cells and showed an increase in the intracellular calcium level after stimulation with the muscarinic acetylcholine receptor agonist carbachol, suggesting that hiSGs morphologically and functionally mimic normal salivary glands. scRNA-seq also showed that hiSGs were composed of independent lineages of basal–myoepithelial cells from the ductal–acinar lineage, and pseudotime analysis distinguished the lineage of basal–myoepithelial cells from the ductal–acinar lineage. In addition, orthotopically transplanted hiSGs were engrafted at the recipient site and presented the phenotype of mature salivary glands over time.
Studies based on autopsy and outpatient samples have reported that the oral cavity is an important site of SARS-CoV-2 infection and implicated saliva as a potential route of SARS-CoV-2 transmission. ACE2 and TMPRSS family members, including TMPRSS2, are used as host entry factors for SARS-CoV-2 (refs. 28,29). These factors are expressed on human salivary gland ductal cells4,30,31,32,33. Accordingly, scRNA-seq and immunofluorescence analyses revealed that the ductal cell group within hiSGs expressed ACE2 and TMPRSS2, and the SARS-CoV-2 nucleoprotein was detected in the ductal luminal cells under infected conditions. In addition, the TCID50 assay indicated that infectious viruses replicated in hiSGs. These data indicate that organoids accurately reflect the in vivo phenomenon of viral replication in salivary glands. In vitro models of salivary gland SARS-CoV-2 infection, which is thought to play an important role in person-to-person transmission, will be important for facilitating SARS-CoV-2 research and controlling COVID-19 as the pandemic continues.
The ability of SARS-CoV-2 to spread via saliva is one of the factors that determines the status of the COVID-19 pandemic and is an important factor, especially in asymptomatic individuals with COVID-19. However, it is unclear how SARS-CoV-2 infects salivary glands and is secreted into saliva. Organoids derived from adult human salivary gland tissue stem progenitor cells may be a promising model for analysing these mechanisms34,35. However, hiSGs show several advantages compared with these organoids because hiPSCs can be expanded relatively well in culture and are easily genetically modified using the CRISPR–Cas9 gene editing tool. Therefore, hiSGs can be a promising tool for investigating SARS-CoV-2 infection in salivary glands at the molecular level.
Methods
Animal experiments
All of the animal experiments were conducted with the approval of the institutional animal care and use committees at Showa University and the RIKEN Center for Biosystems Dynamics Research (permit numbers 13047 and 2014-02-13, respectively). C57BL/6J mouse embryos and CB-17/lcr-scid/scidJcl mice were purchased from CLEA Japan.
hiPSC culture for maintenance and differentiation
hiPSCs (201B7: HPS4290) were purchased from the RIKEN BioResource Research Center and maintained in StemFit AK02N (REPROCELL). hiPSCs were used for experimentation until passage 45. Cells were cultured on six-well plates coated with iMatrix-511 (Nippi). The medium was replaced every day. Cells were passaged with StemPro Accutase Cell Dissociation Reagent (Thermo Fisher Scientific) and seeded in StemFit AK02N medium containing 10 μM Y27632 (Nacalai Tesque). One day after passaging, the medium was replaced with StemFit AK02N medium without Y27632. For differentiation, hiPSCs were dissociated with StemPro Accutase, resuspended in StemFit AK02N medium containing 20 μM Y27632 and plated in a volume of 100 μl per well (5,000 cells per well) in PrimeSurface 96-well V-bottom plates (Sumitomo Bakelite). On day 2, hiPSC-derived aggregates were transferred to differentiation medium and plated in a volume of 100 μl per well (one aggregate per well) in Nunclon Sphera 96-well U-bottom low-cell-adhesion plates (Thermo Fisher Scientific). The differentiation medium consisted of Iscove’s modified Dulbecco’s medium/Ham’s F-12 1:1 (Thermo Fisher Scientific) containing 1% chemically defined lipid concentrate (Thermo Fisher Scientific), 450 μM monothioglycerol (Sigma–Aldrich), 5 mg ml−1 bovine serum albumin (97% purity achieved through crystallization; Sigma–Aldrich), 2% growth factor-reduced Matrigel (BD Biosciences) and 10 μM SB431542 (Stemgent). On days 4 and 8, 50 μl of the medium was added to each well. On day 12 of differentiation, the aggregates were washed with maturation medium and transferred to 24-well low-cell-adhesion plates (Nunc) in maturation medium. The maturation medium consisted of Advanced DMEM/F-12 (Thermo Fisher Scientific) containing 0.5% N2 Supplement (Thermo Fisher Scientific), 1% B27 Supplement (Thermo Fisher Scientific), 1 mM penicillin–streptomycin (Sigma–Aldrich), 1 mM GlutaMAX (Thermo Fisher Scientific), 1% (vol/vol) growth factor-reduced Matrigel, 100 ng ml−1 FGF7 (PeproTech) and 200 ng ml−1 FGF10 (PeproTech). On day 60, the branching structures were manually isolated using a 29G syringe needle and then transferred to 24-well low-cell-adhesion plates (Thermo Fisher Scientific) in maturation medium. The maturation medium was changed every other day during floating culture.
Bulk RNA-seq analysis
Total RNA was extracted from tissue using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s instructions. Library preparation from total RNA was performed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) following the manufacturer’s instructions. The DNA libraries were sequenced on an Illumina HiSeq 2500 platform. Sequence data were analysed using the CLC Genomics Workbench (version 11.0.1). Published RNA-seq data of embryonic and adult human salivary glands (GSE143702) were downloaded as Sequence Read Archive files using the SRA toolkit, and the overall gene expression profiles were compared with those of hiSGs in this study.
Calcium release analysis
Fluo-4 (Thermo Fisher Scientific) was suspended in 0.8% pluronic acid (Thermo Fisher Scientific) and mixed with Hanks’ balanced salt solution (HBSS) without calcium (Invitrogen). The epithelial branching structures of hiSGs were manually isolated using a 29G syringe needle and incubated with Fluo-4 solution for 30 min at 37 °C. After 30 min, the hiSGs were washed twice with HBSS and incubated for 30 min in HBSS. The changes in fluorescence were captured via image acquisition using a confocal microscope system (Nikon A1R), followed by treatment of the hiSGs with 10 or 100 μM carbachol (Sigma–Aldrich). Atropine (Tanabe; 0.1 μM) was added 15 min before carbachol treatment.
Immunofluorescence analysis
Frozen tissue sections were fixed in 4% paraformaldehyde (Wako) and antigen retrieval was then performed by heating at 100 °C in a citrate-buffered solution at pH 6.4. The sections were labelled for 60 min at room temperature with different primary antibodies, including rabbit anti-α-SMA (1:200; ab5694; Abcam), mouse antiAMY1A (clone 2D4) (1:200; WH0000276M4; Sigma–Aldrich), rabbit antiaquaporin 5 (1:200; AQP005; Alomone Labs), rabbit anti-cytokeratin 5 (EP1601Y) (1:200; ab52635; Abcam), rabbit anti-cytokeratin 14 (LL002) (1:200; ab7800; Abcam), mouse antikeratin 18 (Ks18.04) (1:200; 65028; PROGEN), rat anti-Krt19 (1:200; TROMA-III; DSHB), rabbit anti-FOXC1 (EPR20685) (1:200; ab227977; Abcam), mouse anti-human mitochondria (113-1) (1:100; ab92824; Abcam), mouse anti-cytokeratin pan (mixture) (clones C-11, PCK-26, CY-90, KS-1A3, M20 and A53-B/A2) (1:200; C2562; Sigma–Aldrich), rabbit anti-SARS-CoV nucleoprotein/NP (1:100; 40143-RP01; Sino Biological), rabbit anti-Sox9, (1:500; AB5535; Chemicon), rabbit anti-SOX10 (EPR4007) (1:200; ab155279; Abcam), rabbit anti-ZO-1 (ZO1-1A12) (1:200; 33-9100; Thermo Fisher Scientific), rabbit anti-CD31 (1:400; ab124432; Abcam) and rabbit anti-TUBB3 (EP1569Y) (1:200; ab52623; Abcam). The slides were then incubated with fluorescent secondary antibodies (1:200 dilution; Invitrogen). Nuclei were counterstained with DAPI (Nacalai Tesque). Images were acquired using a BZ-9000 fluorescence microscope (Keyence) or a Nikon A1 confocal microscope (Nikon). Submandibular glands (SMGs) of a 70-year-old male were used as a positive control (Extended Data Fig. 7). The SMGs removed during cervical dissection surgery for oral cancer were used. This study was approved by the Ethics Committee of Showa University Hospital (number 3034).
RNA purification, reverse transcription and qPCR
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Reverse transcription was performed using SuperScript VILO (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed using the generated complementary DNA samples and a 7500 detection system (Invitrogen). The primer sequences are shown in Supplementary Table 1. Quantification of the samples was performed according to threshold cycle values using the ΔΔCt method. The values presented in the graphs represent means ± s.d. For SARS-CoV-2 detection, RNA from SARS-CoV-2-infected hiSGs was extracted by adding 1000 μl TRIzol (Thermo Fisher Scientific), followed by incubation at room temperature for 5 min and subsequent addition of 200 μl chloroform. After mixing each sample for 15 s, the samples were incubated at room temperature for 3 min and then centrifuged at 10,000g for 15 min at 4 °C. The aqueous layer was carefully removed from each sample, mixed with two volumes of 100% ethanol and purified using Ethachinmate (Nippon Gene) according to the manufacturer’s instructions. QuantiTect Probe RT-PCR Kit (Qiagen) Master Mix was used to convert RNA into complementary DNA and to amplify specific RNA regions. The reverse transcription reaction was performed for 30 min at 50 °C and 15 min at 95 °C, followed by two-step qPCR at 95 °C for 15 s and 60 °C for 60 s for a total of 45 cycles. RNA copy numbers were estimated from the simultaneously evaluated standard curve. Synthetic standard RNA for NIID_2019-nCOV_N2 primer sets (Nihon Gene Research Laboratories) was purchased. The applied primers and probes included NIID_2019-nCOV_N_F2 (5ʹ-AAATTTTGGGGACCAGGAAC-3ʹ), NIID_2019-nCOV_N_R2 (5ʹ-TGGCAGCTGTGTAGGTCAAC-3ʹ) and NIID_2019-nCOV_N_P2 (5ʹ-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3ʹ), where FAM and BHQ denote Fluorescein amidites and Black Hole Quencher, respectively.
Transmission electron microscopy
The hiSGs were refixed in 2% glutaraldehyde in phosphate buffer and washed three times with 0.2 M sodium cacodylate. The iSGs were subsequently post-fixed with 1% osmium tetroxide for 60 min, dehydrated through an ethanol series and then embedded in epoxy resin. Ultrathin sections (70 nm thick) were then stained with uranyl acetate and lead citrate and observed under a transmission electron microscope (H-7600; Hitachi).
Transplantation
The epithelial branching structures of hiSGs were manually isolated using a 29G syringe needle and transferred to collagen drops, and a polyglycolic acid (PGA) monofilament (Kono Seisakusho) was inserted into each hiSG. For mouse embryonic day 13 SMG mesenchymal tissue separation, embryonic SMGs were incubated in Dispase (Corning) for 2 min at room temperature. Using a pair of 29G syringe needles, the epithelial rudiments and mesenchymal tissues were gently separated. The hiSGs and mouse SMG mesenchymal tissues were transferred to collagen drops, and PGA monofilaments (Kono Seisakusho) were inserted into both types of tissue. To prepare parotid gland-defective mice, the parotid glands of 7-week-old CB-17/lcr-scid/scidJcl female mice were extracted under deep anaesthesia. An hiSG containing a PGA monofilament was then transferred to the masseter muscle. To create connections between the host parotid duct and the hiSGs, a PGA monofilament guide was inserted into the host parotid duct, and the collagen gel and masseter muscle were fixed using nylon thread (8–0 black nylon 4 μm 1/2 R; Bear Medic Corporation).
SARS-CoV-2 infection
SARS-CoV-2 infection studies were performed in a biosafety level 3 facility at the National Institute of Infectious Diseases, Japan. The SARS-CoV-2 WK-521 strain (2019-nCoV/Japan/TY/WK-521/2020; GenBank Accession LC522975) was expanded in VeroE6/TMPRSS2 cells and harvested to prepare a virus inoculum stock. Viral infectivity was assessed by evaluation of the cytopathic effect on VeroE6/TMPRSS2 cells and the determination of endpoint titres. The epithelial branching structures of hiSGs were manually isolated using a 29G syringe needle and transferred to virus medium or an equal volume of mock medium at 1 × 105 plaque-forming units per hiSG. The hiSGs were then incubated at 37 °C under 5% CO2 for 16 h. Virus medium was subsequently replaced with maturation medium. At the indicated time points, hiSGs were washed with maturation medium and resuspended in TRIzol LS (Thermo Fisher Scientific). Viral RNA was extracted from the culture supernatants with a QIAamp Viral RNA Mini Kit (Qiagen). All SARS-CoV-2 work was performed under biosafety level 3 conditions.
TCID50 assay
The viral titre of SARS-CoV-2 was determined through a TCID50 assay in VeroE6/TMPRSS2 cells. The hiSGs were incubated with the SARS-CoV-2 JPN/TY/WK-521 strain at 1 × 105 times the TCID50 in a 96-well plate. After 1 d, these organoids were transferred to fresh phosphate-buffered saline followed by fresh medium. After 1 h, 1 d and 3 d, tenfold serially diluted culture supernatants in wells were transferred to new wells (1 × 104 Vero cells per well) in DMEM (Nacalai Tesque) supplemented with 2% foetal bovine serum (Cytiva HyClone), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (FUJIFILM Wako Pure Chemical Corporation) in a 96-well plate. The day before infection, VeroE6/TMPRSS2 cells were seeded in 96-well plates and cultured in DMEM supplemented with 2% foetal bovine serum at 37 °C until they reached 100% confluence. After culture for 1 d at 37 °C under 5% CO2, the culture supernatants were replaced with fresh medium. After 2 d, these cells were fixed with 20% formalin (FUJIFILM Wako Pure Chemical Corporation) and stained with crystal violet solution (Sigma–Aldrich). The TCID50 was assessed by the evaluation of cytopathic effects to determine the endpoint titres. The lower limit of detection was 1:10.
Single-cell RNA-seq
On day 80, hiSG organoids were dissociated with TrypLE Select Enzyme (Thermo Fisher Scientific) at 37 °C for 30 min. Live propidium iodide-negative single cells were purified with a FACSAria II system (BD Biosciences) and subjected to droplet-based scRNA-seq on the 10X Genomics Chromium Single Cell 3′ platform (version 2) according to the manufacturer’s protocol. Library sequencing was performed via paired-end sequencing with a single sample index on an Illumina NextSeq 500 system. Data preprocessing was carried out with Cell Ranger version 1.2. The subsequent analysis was performed using Seurat (version 4.0.3) and Monocle 3 (version 0.2.2). For the RamDA-seq experiments, sorted cells were collected in 1 µl cell lysis buffer in a 96-well PCR plate (Nippon Genetics). Total RNA was diluted or lysed in 1 μl cell lysis buffer containing 1 U RNasin plus (Promega), 10% RealTime ready Cell Lysis Buffer (Roche Life Science), 0.3% NP-40 (Thermo Fisher Scientific) and RNase-Free Water (Thermo Fisher Scientific). The lysate solution was immediately centrifuged and mixed using a ThermoMixer C (Eppendorf) for 1 min at 4 °C. The cell lysate solution was stored at −80 °C until use. RNA and sequencing library preparation was performed according to a previous study36.
Generation of stable CRISPRi hiPSC lines
hiPSCs were dissociated with StemPro Accutase (Thermo Fisher Scientific), resuspended in AK02N supplemented with Y27632 (10 μM) and counted. To generate the CRISPRi hiPSC lines, hiPSCs were nucleofected with the CRISPRi knockin vector (pAAVS1-NDi-CRISPRi (Gen1); plasmid 73497; 5 μg) and each AAVS1-TALEN pair (AAVS1-TALEN-L (plasmid 59025) and AAVS1-TALEN-R (plasmid 59026)) (2 μg) using Neon (1,050 V; 30 ms; two pulse; 100 μl Neon tip). Cells were then seeded in six-well plates in serial dilutions of AK02N supplemented with Y27632 (10 μM). Geneticin (Thermo Fisher Scientific) was applied at 200 μg ml−1 2 d postnucleofection. To knock in the gRNA constructs, hiPSCs were nucleofected with the gRNA expression vector (pgRNA-CKB; plasmid 73501; 5 μg). Puromycin (Thermo Fisher Scientific) was applied at 0.5 μg ml−1 2 d postnucleofection. The gRNA sequences are shown in Supplementary Table 2. Selection was maintained for 10 d until stable colonies appeared. Colonies with a diameter greater than 500 μm were manually picked using a P10 pipette tip and transferred to individual wells of a 24-well plate containing AK02N medium supplemented with Y27632 (10 μM). Clones were then expanded into larger formats.
Statistics and reproducibility
Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test for multiple comparisons. All of the data were analysed using RStudio or CLC Genomics Workbench. All of the immunofluorescence experiments were performed independently at least three times with reproducible results. No statistical method was used to pre-determine sample size. The experiments were not randomized. The investigators were not blinded to allocation during the experiments or outcome assessment.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The RNA-seq data that support the findings of this study have been deposited in the DDBJ Sequence Read Archive under the accession code DRA014744. Previously published RNA-seq data that were re-analysed here are available from the Gene Expression Omnibus under the accession code GSE143702. All other data supporting the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
No custom codes were developed and used in this manuscript. All codes are available upon request from the corresponding author.
Change history
21 February 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41556-023-01107-x
References
Schurink, B. et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe 1, e290–e299 (2020).
Meyerowitz, E. A., Richterman, A., Gandhi, R. T. & Sax, P. E. Transmission of SARS-CoV-2: a review of viral, host, and environmental factors. Ann. Intern. Med. 174, 69–79 (2021).
Singh, R. et al. COVID-19: Current knowledge in clinical features, immunological responses, and vaccine development. FASEB J. 35, e21409 (2021).
Huang, N. et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27, 892–903 (2021).
Chen, T. & Hudnall, D. Anatomical mapping of human herpesvirus reservoirs of infection. Mod. Pathol. 19, 726–737 (2006).
Salahudeen, A. A. et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675 (2020).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
Yang, L. et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27, 125–136 (2020).
Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275 (2021).
Wang, L. et al. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat. Med. 27, 1600–1606 (2021).
Tanaka, J. et al. Generation of orthotopically functional salivary gland from embryonic stem cells. Nat. Commun. 9, 4216 (2018).
Ealy, M., Ellwanger, D. C., Kosaric, N., Stapper, A. P. & Heller, S. Single-cell analysis delineates a trajectory toward the human early otic lineage. Proc. Natl Acad. Sci. USA 113, 8508–8513 (2016).
Kumagai, M. & Sato, I. Morphological changes in collagen fiber during development of human fetal parotid and submandibular glands. Ann. Anat. 185, 45–51 (2003).
Chatzeli, L., Gaete, M. & Tucker, A. S. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 144, 2294–2305 (2017).
Jaskoll, T. et al. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev. Biol. 5, 11 (2005).
Steinberg, Z. et al. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development 132, 1223–1234 (2005).
Saitou, M. et al. Functional specialization of human salivary glands and origins of proteins intrinsic to human saliva. Cell Rep. 33, 108402 (2020).
Oh, Y. S. & Turner, R. J. Effect of γ-secretase inhibitors on muscarinic receptor-mediated calcium signaling in human salivary epithelial cells. Am. J. Physiol. Cell Physiol. 291, 76–82 (2006).
Ogawa, M. et al. Functional salivary gland regeneration by transplantation of a bioengineered organ germ. Nat. Commun. 4, 2498 (2013).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Hayashi, R. et al. Generation of 3D lacrimal gland organoids from human pluripotent stem cells. Nature 605, 126–131 (2022).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Hauser, B. R., Aure, M. H., Kelly, M. C., Hoffman, M. P. & Chibly, A. M. Generation of a single-cell RNAseq atlas of murine salivary gland development. iScience 23, 101838 (2020).
May, A. J. et al. Diverse progenitor cells preserve salivary gland ductal architecture after radiation-induced damage. Development 145, dev166363 (2018).
Song, E. A. C. et al. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci. Rep. 8, 14043 (2018).
Mandegar, M. A. et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 18, 541–553 (2016).
Katsura, H. et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27, 890–904 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020).
Zhang, H., Penninger, J. M., Li, Y., Zhong, N. & Slutsky, A. S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46, 586–590 (2020).
Descamps, G. et al. ACE2 protein landscape in the head and neck region: the conundrum of SARS-CoV-2 Infection. Biol. (Basel). 9, 235 (2020).
Hamming, I. et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004).
Song, J. et al. Systematic analysis of ACE2 and TMPRSS2 expression in salivary glands reveals underlying transmission mechanism caused by SARS-CoV-2. J. Med. Virol. 92, 2556–2566 (2020).
Soares, C. D., Mosqueda-Taylor, A., Hernandez-Guerrero, J. C., de Carvalho, M. G. F. & de Almeida, O. P. Immunohistochemical expression of angiotensin-converting enzyme 2 in minor salivary glands during SARS-CoV-2 infection. J. Med. Virol. 93, 1905–1906 (2021).
Song, Y. et al. Development of a functional salivary gland tissue chip with potential for high-content drug screening. Commun. Biol. 4, 361 (2021).
Pringle, S. et al. Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cells 34, 640–652 (2016).
Hayashi, T. et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 9, 619 (2018).
Acknowledgements
We thank T. N. Sato, K. Urayama, K. Tejima and S. Kozawa (all from the Advanced Telecommunications Research Institute International) for advice. We also thank T. Suzuki (Director at the Department of Pathology, National Institute of Infectious Diseases) and M. Ohnishi (Deputy Director General at the National Institute of Infectious Diseases) for providing support for the SARS-CoV-2 infection assay. This work was supported by JSPS KAKENHI (grant numbers 19H03838 (to J.T.) and 19H03827 (to K.M.)).
Author information
Authors and Affiliations
Contributions
J.T. designed and performed most of the experiments and analysed the data. H.S. supported and performed the SARS-CoV-2 infection assay. M.O. and T. Tsuji supported and performed the transplantation assay. K.T., S.O., S.I., R.Y., T. Shirota and T. Shimane supported and performed the cell differentiation procedure. T.W. and Y.M. performed the scRNA-seq. S.N. and T.I. supported and performed the calcium release assay. T. Sadaoka and T. Takaki supported and performed the transmission electron microscopy. T. Sakai, M.M. and I.S. supervised and supported the SARS-CoV-2 infection assay. K.M. designed the research plan, conceptualized the idea and interpreted the results. J.T. and K.M. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Jie Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of hiPSC-derived branching structures.
a, Phase-contrast representative images of aggregates were differentiated in chemically defined medium, treated with SB431542 on Day 2 and monitored during Days 0–10. Representative of 18 independent experiments. Scale bars, 200 μm. b, RT–PCR revealed the gene expression levels of salivary gland cell lineage markers in organoids on Day 60 and undifferentiated human iPSCs (201B7). c, Immunofluorescence images of the salivary gland organoids on Day 60. The epithelial marker pan-CK, ductal marker CK18, basal cell marker CK5, myoepithelial cell marker α-SMA, and salivary gland transcription factor FOXC1 were detected. Scale bars, 200 μm.
Extended Data Fig. 2 Induction of surface ectoderm and salivary gland rudiments in organoid cultures with FGFs.
a, Treatment with FGF7 alone on Day 12 promoted solid epithelial components but was not sufficient to produce branching structures by Day 80 of differentiation. Representative phase-contrast images and immunofluorescence revealed that the Pan-CK+ epithelial cells did not express AQP5 and SOX9 on Day 80. Scale bars, 500 μm (left panel), 400 μm (second panel), 50 μm (third and forth panels). n = 8 organoids, 2 independent experiments. b, Treatment with FGF10 alone on Day 12 promoted solid epithelial components but was not sufficient to produce salivary gland organoids by Day 80 of differentiation. Representative phase-contrast images (left) and immunofluorescence revealed that the pan-CK+ epithelial cells did not express AQP5 and SOX9 on Day 80. Scale bars, 500 μm (left panel), 400 μm (second panel), 50 μm (third and forth panels). n = 8 organoids, 2 independent experiments.
Extended Data Fig. 3 Orthotopic transplantation of human salivary gland organoids.
a, Real-time RT–PCR analysis of salivary gland marker genes. Data are normalized to ribosomal protein L27 and presented as mean values + /− SD (n = 3 biological replicates). Statistically significant changes as calculated by one-way ANOVA followed by Tukey’s post hoc test. b, Immunofluorescence images of the transplantation site. Mouse CD31+ vessels (left) and TUBB3+ nerve fibres (right) were observed in the mesenchyme around the Pan-CK+ parenchyma derived from hiSGs. Scale bars, 50 μm. Representative of three independent experiments.
Extended Data Fig. 4 scRNA-seq reveals cell type heterogeneity in salivary gland organoids.
a, Heatmap revealing the scaled expression of differentially expressed genes for each cluster defined in Fig. 3a. Top marker genes for cell types by scRNA-seq from hiSGs on Day 80. Row-normalized heat map of the top 10 markers over all cells. b, Dot plot expression matrix with selected genes for cluster annotation displayed on the x axis and cluster on the y axis. c, Feature plots for the expression of human salivary gland-related genes in hiSGs. a-c,Ten organoids were pooled for one scRNA-seq experiment.
Extended Data Fig. 5 Trajectory analysis based on scRNA-seq data.
a, Monocle-generated annotated uniform manifold approximation projection (UMAP) plots of hiSGs on Day 80. b, Feature plots for the expression of mesenchymal, epithelial, and salivary gland cell type markers in hiSGs. c, Pseudotime trajectory projected onto a UMAP in all clusters. d, Feature plots for the gene expression of ductal, acinar, and cycling cell markeres based UMAP plots of ductal-acinar lineage. e, Pseudotime trajectory projected onto a UMAP in the ductal-acinar lineage. The colour of a cell shows its cluster identity (left) and pseudotime values (right). f, Pseudotime-ordered expression of selected marker genes: AQP5, KRT8, KRT19 (ductal acinar), and MKI67 (cycling). Lines denote the relative expression of each marker in pseudotime. g, Feature plots for the gene expression of basal, myoepithelial, and cycling cell markeres based UMAP plots of basal-myoepithelial lineage. h, Pseudotime trajectory projected onto a UMAP in the basal-myoepithelial lineage. The colour of a cell shows its cluster identity (left) and pseudotime values (right). i, Pseudotime-ordered expression of selected marker genes: ACTA2, KRT14, KRT5 (basal-myoepithelial), and MKI67 (cycling). Lines denote the relative expression of each marker in pseudotime. a-h,Ten organoids were pooled for one scRNA-seq experiment.
Extended Data Fig. 6 SARS-CoV-2 infects salivary gland organoids.
a, Feature plots of RamDA-seq samples of hiSGs on Day 80. The key gene markers for the classification of cell in hiSG. Ten organoids were pooled for one scRNA-seq experiment. b, Violin plots of ACE2 and TMPRSS2 in hiSGs. ACE2 and TMPRSS2 expressing cells were detected in ductal cell cluster. c, Feature plots of ACE2 and TMPRSS2 in hiSGs. ACE2 and TMPRSS2-positive cells were detected in the ductal cll cluster. d, Immunofluorescence for ACE2 and TMPRSS2 in hiSGs. Images are representative of three independent samples showing similar results. Scale bars, 100 μm. e, Feature plots of SARS-CoV-2 entry factors in hiSGs.
Extended Data Fig. 7 Immunofluorescence of human salivary glands as a positive control.
Immunofluorescence of the submandibular gland in a 70-year-old male. Scale bars, 50 μm.
Supplementary information
Supplementary Table 1
Supplementary Table 1.
Supplementary Table 2
Supplementary Table 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 5
Unprocessed gel.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Unprocessed gel.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed gel.
Source Data Extended Data Fig. 3
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tanaka, J., Senpuku, H., Ogawa, M. et al. Human induced pluripotent stem cell-derived salivary gland organoids model SARS-CoV-2 infection and replication. Nat Cell Biol 24, 1595–1605 (2022). https://doi.org/10.1038/s41556-022-01007-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-01007-6
This article is cited by
-
Induced pluripotent stem cells (iPSCs): molecular mechanisms of induction and applications
Signal Transduction and Targeted Therapy (2024)
-
Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy
Molecular Cancer (2023)