Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the deadliest pandemics in history. SARS-CoV-2 not only infects the respiratory tract, but also causes damage to many organs. Organoids, which can self-renew and recapitulate the various physiology of different organs, serve as powerful platforms to model COVID-19. In this Perspective, we overview the current effort to apply both human pluripotent stem cell-derived organoids and adult organoids to study SARS-CoV-2 tropism, host response and immune cell-mediated host damage, and perform drug discovery and vaccine development. We summarize the technologies used in organoid-based COVID-19 research, discuss the remaining challenges and provide future perspectives in the application of organoid models to study SARS-CoV-2 and future emerging viruses.
Similar content being viewed by others
Origin and history of organoid models
Organoids are miniaturized in vitro organ models that are derived from stem cells or adult tissues extracted from patients in a specific three-dimensional (3D) microenvironment. They can self-organize and differentiate into functional cell types, which highly mimic the characteristics of the organs in vivo. Analyzing the formation of organoids can deepen the understanding of the mechanisms of human development and tissue/organ regeneration. In addition, organoids provide powerful platforms for disease modeling and drug discovery.
The origin of organoids can be traced back to 1907 when H. V. Wilson tried to regenerate organisms in vitro and showed that the mechanically separated sponge cells can reassemble and self-organize into new sponge organisms with normal functions1. In early studies of 3D cell culture from 1965, organoids were defined as abnormal cellular growths or intracellular structures2, and it was 1975 when Rheinwald and Green first described the method of long-term culturing human cells3. As 3D culture methods progressed, Emerman and Pitelka cultured mammary epithelial cells by floating collagen gels, with expression of milk protein for a month in 1977 (ref. 4). In 1981, for the first time, embryonic stem cells (ESCs) were isolated from mouse embryos, and were cultured in vitro5,6,7 and from human blastocysts in 1998 (ref. 8).
These advances fueled further developments in the field of stem cell biology and organoids. For example, in 2006, pluripotent stem cells (PSCs) were induced from mouse embryonic or adult fibroblasts by introducing four transcription factors9. In 2008, Eiraku and colleagues were able to generate cortical tissues that displayed apicobasal polarity from ESCs using the 3D aggregation culture system10 and, in 2009, Sato et al. demonstrated that the presence of leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5)-positive adult intestinal stem cells can induce the formation of 3D structures in Matrigel, which can self-organize and differentiate into crypt–villus organoids in the absence of a non-epithelial niche11. Many 3D-cultured organoid systems have since been successfully generated, including gut12, retinal13, brain14, liver15, kidney16, stomach17, lung18,19,20, pancreas21, colon22, cardiac23,24,25 and blood vessel26 organoids derived from human pluripotent stem cells (hPSC-derived organoids) and stomach27, liver28, pancreas29, placenta30 and lung31 organoids derived from adult tissues (adult organoids; Fig. 1).
Organoids to study infectious diseases
Organoids have been applied to model human diseases, including infectious diseases. This animal-free system is easy to use and provides new insight into human physiology and pathology. Early work started during the Zika virus (ZIKV) outbreak. In 2016, using brain organoid (BO) models, ZIKV was found to infect neural progenitor cells resulting in increased cell death and reduced proliferation32,33. Subsequently, ZIKV-infected BOs were used to study viral tropism and strain pathogenicity and in whole-genome analyses34,35. In addition, BOs were used in chemical screening studies, which identified several potential anti-ZIKV drug candidates36. A recent study demonstrated both ZIKV and herpes simplex virus type 1 impaired the growth of BOs, induced distinct morphological defects and transcriptional changes, and eventually led to microcephaly, providing key insights into the pathophysiology of the disease37. Before the COVID-19 pandemic, several studies applied human lung organoids to study respiratory diseases such as respiratory syncytial virus31 and influenza virus38. These studies stimulated the field to apply the organoid platform to study newly emerging viruses, such as SARS-CoV-2.
COVID-19 and SARS-CoV-2
COVID-19 is an infectious disease caused by SARS-CoV-2 that leads to a severe acute respiratory syndrome, a hyperinflammatory response and widespread multi-organ damage (Fig. 2). Common symptoms of COVID-19 include fever, cough, fatigue, shortness of breath and loss of taste and smell.
Many studies have reported the entry factors for SARS-CoV-2. The first-identified and predominant SARS-CoV-2 receptor is angiotensin-converting enzyme 2 (ACE2) present on the cell surface of diverse human cell types39. Viral entry into the cell is further facilitated by the cleavage of the spike protein to promote viral membrane fusion, which can be mediated by co-receptors transmembrane serine protease 2 (TMPRSS2)39 and cathepsin L (CTSL)40. The FURIN cleavage site also plays an important role in SARS-CoV-2 entry41,42. Recent studies identified neuropilin-1 (NRP1) as another factor that can facilitate SARS-CoV-2 entry43,44,45.
As ACE2 is widely distributed in mammalian cells, SARS-CoV-2 can infect the human body through the respiratory tract and spread quickly toward other organ systems. As a result, COVID-19 not only causes severe respiratory diseases, but also induces damage in other organs, including brain, heart, liver, kidney, intestinal tract and pancreas. These complications include mental, cognitive or physical disorders, venous thromboembolism, myocarditis, heart failure, arrhythmia, cardiac arrest, acute kidney injury, liver injury, hyperglycemia and ketosis, acute cerebrovascular disease, and gastrointestinal complications.
Several monolayer culture systems, such as Vero E6 cells or ACE2-overexpressing cancer cells, have been utilized for therapeutic screening and mechanism studies of SARS-CoV-2 infection. But two-dimensional (2D) cell culture models cannot simulate the complexity of the host organisms and often have a different phenotype from the natural tissue. A platform with higher similarity to in vivo physiological and pathological characteristics of human tissues/organs, such as 3D organoids, can serve as an advanced model to study SARS-CoV-2 infection, mimic pathophysiology of injured organs and perform drug discovery.
Organoid models for COVID-19 research
Respiratory organoids
During embryonic development, respiratory epithelial cells are derived from the endodermal layer, followed by a complex series of signal transduction events to generate multiple compartments, including alveoli, proximal and intermediate airways, and respiratory bronchioles46. SARS-CoV-2 mainly targets epithelial cells in the respiratory system and causes severe cough, excessive mucus production, and shortness of breath. In severe COVID-19 cases, individuals develop pneumonia, progress to acute respiratory distress syndrome, and finally develop respiratory failure. Therefore, lung alveolar, lung airway and bronchial organoids have been widely applied to monitor virus infection, explore pathological changes and identify potential therapeutics (Table 1).
Alveolar lung organoids
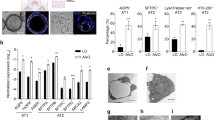
Alveolar lung organoids (ALOs) contain type 1 and 2 alveolar epithelial (AT1 and AT2) cells and mesenchymal cells. ACE2 is mainly expressed in a subpopulation of AT2 cells, while the co-receptor TMPRSS2 is widely distributed47. hPSC-derived ALOs have been applied to model SARS-CoV-2 infection in alveoli, which demonstrated AT2 cells are permissive to SARS-CoV-2 infection47,48. Furthermore, SARS-CoV-2-infected hPSC-AT2 cells were able to recapitulate host immune responses49, leading to an inflammatory phenotype, induction of nuclear factor κB (NF-κB) signaling, and loss of the mature alveolar population50.
hPSC-derived ALOs have also been used to identify drug candidates and antibodies blocking SARS-CoV-2 entry or replication. For example, a high-throughput screen of drugs approved by the US Food and Drug Administration (FDA) found three drug candidates that block SARS-CoV-2 entry47. Another chemical screening identified drugs that reduce ACE2 levels and inhibit SARS-CoV-2 in hPSC-derived cardiac cells and ALOs51. In addition, EK1 peptide blocking of spike protein and TMPRSS2 inhibitors were found to be effective against viral entry in hPSC-ALOs48. Finally, a human neutralizing antibody was found to inhibit SARS-CoV-2 infection in hPSC-ALOs52. In addition to viral entry, several drug candidates were validated to block SARS-CoV-2 replication in hPSC-ALOs, including remdesivir50, a class of 2-phenyl-1,2-benzoselenazol-3-one compounds targeting SARS-CoV-2 main protease (Mpro)53. Functional studies found that 25-hydroxycholesterol (25HC), an interferon-stimulated gene (ISG) and broad viral inhibitor, could inhibit viral entry in hPSC-ALOs. Mechanistic studies found 25HC can activate acyl-CoA: cholesterol acyltransferases and induce the depletion of accessible cholesterol from the plasma membrane54.
Single adult human AT2 or keratin 5 (KRT5)-positive basal cells can generate adult ALOs, which contain AT1 and AT2 cells55 with apical-out polarity of ACE2 expression. Similar to hPSC-ALOs, the major target of SARS-CoV-2 in adult ALOs is AT2 cells. Infected AT2 cells showed similar features of COVID-19 lungs, including type I/III interferon response55,56, interferon-mediated inflammatory responses, loss of surfactant proteins and apoptosis57. Furthermore, mutation analysis calculated that a single SARS-CoV-2 particle was responsible for viral infection in most of the AT2 cells57. Several drugs, including remdesivir and the combination of ciclesonide, nelfinavir and camostat with remdesivir, have demonstrated anti-SARS-CoV-2 activity in adult alveolospheres58. In addition, interferon was also shown to block viral replication in adult alveolospheres59.
Lung airway organoids
hPSC-derived airway organoids (AWOs) consist of functional multiciliated cells, basal cells, mucus-producing secretory cells, CC10-secreting club cells, and so on. Upon SARS-CoV-2 infection, hPSC-AWOs undergo notable metabolic changes, including downregulation of lipid metabolism52 and upregulation of glycolysis60. A high-content chemical screen using hPSC-AWOs identified that GW6471 can block SARS-CoV-2 infection by inhibiting the hypoxia-inducible factor 1-alpha (HIF1α) pathway and glycolysis rates. Several inhibitors of fatty acid biosynthesis, such as xanthohumol, 5-(tetradecyloxy)-2-furoic acid and ND-646, have also been found to block SARS-CoV-2 infection60.
The adult AWOs, which have similar cellular components as hPSC-AWOs31, also support active viral replication49. Hence adult AWOs have been used to study replication dynamics of SARS-CoV-2 variants, including B.1.1.7, B.1.427, B.1.429 and B.1.617.2. In a live virus system, SARS-CoV-2 variant B.1.1.7 showed higher replication efficiency than an ancestral isolate Bavpat-1, and a subsequent study demonstrated variant B.1.617.2 exhibited higher replication efficiency than B.1.1.7. In a pseudovirus system, variants B.1.427 and B.1.429 carrying the mutation p.Leu452Arg in the spike protein, demonstrated a higher entry activity than variants carrying the p.Asp614Gly mutation61. Transcriptomics and biomarker-based analyses identified the upregulation of ISGs62, as well as cytokines and chemokines, after viral infection. Moreover, a humanized decoy antibody (ACE2-Fc) was used to block viral infection through disruption of the binding of ACE2 and viral spike protein, thus blocking entry in adult AWOs63.
Bronchial organoids
In addition to lung ALOs and AWOs, adult bronchial organoids (BCOs) have been used in SARS-CoV-2 studies. Microarray assays identified the change of cytokine/chemokines upon SARS-CoV-2 infection64. By comparing high-throughput expression matrix data in SARS-CoV-2-infected BCOs with those of other cell types, colony stimulating factor 3 (CSF3) was identified as a potential drug target65. Another study found that the microRNA hsa-MIR-5004-3p targets the leader sequence of SARS and SARS-CoV-2, the expression of which decreases after SARS-CoV-2 infection in BCOs66.
In summary, respiratory organoid models have demonstrated that ciliated cells, club cells and a subpopulation of AT2 cells, which are arranged from the proximal to distal ends in the airway and terminal alveoli, are permissive to SARS-CoV-2 infection. These findings are consistent with data from autopsies of individuals with COVID-19 (ref. 67), indicating that respiratory organoid models can recapitulate in vivo SARS-CoV-2 infection.
Intestinal organoids
Individuals with COVID-19 commonly exhibit gastrointestinal symptoms, such as diarrhea, vomiting or belly pain68, and human intestinal organoids (IOs) have been identified as a physiologically relevant model to study intestinal pathophysiology in vitro. IOs contain several cell types including enterocytes, goblet cells, enteroendocrine cells, transit-amplifying cells and stem cells69. hPSC-derived and adult IOs, including small intestinal organoids (SIOs), colonic organoids (COs) and ileal organoids (ILOs), have been used to study SARS-CoV-2 infection.
Both hPSC-derived SIOs and COs have been shown to be permissive to SARS-CoV-2 (refs. 47,70,71). SARS-CoV-2 infection was accompanied by ultrastructural changes in hPSC-SIOs, and induction of a robust transcriptional response, including ISGs71. SARS-CoV-2 induced organoid deterioration, which could be rescued by remdesivir71. In addition, SARS-CoV-2 inhibitors identified using hPSC-ALOs47 and hPSC-AWOs60 were further confirmed using hPSC-COs. One study also found peptides or antibodies against interferon-induced transmembrane proteins (IFITMs), cofactors for efficient SARS-CoV-2 infection, could inhibit SARS-CoV-2 entry and replication in hPSC-SIOs72.
Adult SIOs and COs have also been used to evaluate SARS-CoV-2 infection in the gastrointestinal tract. Studies in adult SIOs containing mutations of 19 host factors previously implicated in coronavirus infection, demonstrated that ACE2, dipeptidyl-peptidase 4 and TMPRSS2 could regulate SARS-CoV-2 entry in SIOs73. Another study showed TMPRSS2 and TMPRSS4 could facilitate SARS-CoV-2 spike fusogenic activity and promote virus entry into SIOs74. In SIOs, enterocytes were demonstrated to be permissive to SARS-CoV-2 infection with an induction of a generic viral response program75. Direct comparison of SARS-CoV-2 and SARS-CoV on SIOs showed the two viruses exhibit distinct dynamics of virus–host interaction. For instance, more robust propagation of SARS-CoV triggers minimal cellular responses, whereas SARS-CoV-2 elicits a stronger cellular response with lower replication capacity76. Single-cell RNA-sequencing (scRNA-seq) analysis of SARS-CoV-2-infected SIOs revealed an induction of pro-inflammatory pathways and the interferon-mediated signaling pathway77. Interestingly, in adult COs, interferon (IFN)-γ treatment promoted the differentiation of mature keratin 20 (KRT20)-positive enterocytes, which showed high expression levels of ACE2, and promoted SARS-CoV-2 infection and production78. Finally, IFN-β1 and IFN-λ were reported to inhibit SARS-CoV-2 replication in SIOs69. Several drugs have also been tested in IOs, such as the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG), which suppresses Middle East respiratory syndrome-related coronavirus and propagation of SARS-CoV-2 in SIOs. It was also found that polyamine supplementation and autophagy induction modulated cellular metabolism and limited SARS-CoV-2 growth in SIOs79.
Kidney organoids
Kidney damage such as acute kidney injury is common in individuals with COVID-19. Thus, hPSC-derived kidney organoids (KOs) have been applied to study SARS-CoV-2 infection in kidney. A recent study comparing ACE2 expression levels in 2D and 3D kidney proximal tubule epithelial cells found twofold higher expression of ACE2 in 3D KOs compared to 2D culture conditions, highlighting the significance of applying 3D organoids for COVID-19 disease modeling80. scRNA-seq of hPSC-derived KOs in 3D suspension culture further revealed ACE2 expression in solute carrier family 3 member 1 (SLC3A1)-positive solute carrier family 27 member 2 (SLC27A2)-positive proximal tubule cells and podocalyxin like (PODXL)-positive nephrin (NPHS1)-positive podocin (NPHS2)-positive podocyte cells81. Accordingly, hPSC-KOs were shown to be permissive to SARS-CoV-2 infection and support viral replication81. Studies also found that both a variant of human soluble ACE2, consisting of 1–618 amino acids fused with an albumin-binding domain82, and clinical-grade human recombinant soluble ACE2 (hrsACE2)81, can neutralize SARS-CoV-2 infectivity in hPSC-KOs. Finally, a human dihydroorotate dehydrogenase inhibitor, MEDS433, was also found to inhibit SARS-CoV-2 infection in hPSC-KOs, highlighting the use of organoids in drug discovery applications83.
Liver organoids
A high prevalence of liver test abnormalities and liver injury have been reported in individuals with COVID-19. The liver-related complications include elevation of total bilirubin and alanine aminotransferase abnormalities. Because hepatocytes and cholangiocytes are two major functional cell types in the liver, both hepatocyte and cholangiocyte organoids containing these cell types have been used to study SARS-CoV-2 infection.
In hPSC-derived liver organoids (hPSC-LOs), ACE2 expression is detected in most albumin-positive hepatocytes. Both SARS-CoV-2 pseudo-entry virus and live SARS-CoV-2 were shown to infect hepatocytes84. This finding was further validated in adult hepatocyte and cholangiocyte organoids. Transcript profile analysis of SARS-CoV-2-infected human LOs revealed several upregulated pathways, including cytokine–cytokine receptor interaction, interleukin (IL)-17 signaling, chemokine signaling, tumor necrosis factor (TNF) signaling and NF-κB signaling pathways, while cellular metabolism was largely downregulated84, which is similar to the transcriptional changes in COVID-19 lung autopsy samples47. In liver ductal organoid models, a subset of cholangiocytes express ACE2, which has been shown as a target of SARS-CoV-2 (ref. 85). In addition, liver ductal organoids generated from individuals with nonalcoholic steatohepatitis showed strongly increased permissiveness to SARS-CoV-2 pseudo-entry virus infection, suggesting that nonalcoholic steatohepatitis may be a risk factor for SARS-CoV-2 infection86. Adult LOs have also been used for drug testing. A combination therapy method using remdesivir and rsACE2 synergistically improved the therapeutic effects against SARS-CoV-2 in primary human liver spheroids63.
Brain organoids
Individuals with COVID-19 experience an array of neurological symptoms, ranging in severity from loss of smell and taste, to memory loss, to life-threatening strokes. hPSC-BOs, including both whole-brain and regional brain-specific BOs87, have been applied in SARS-CoV-2 studies. In hPSC-BOs, SARS-CoV-2 infection was identified in cortical, hippocampal, hypothalamic and midbrain organoids by immunostaining, with limited detection in neurons and astrocytes88. Astrocytes were found to boost SARS-CoV-2 infection in BOs89. A study focused on SARS-CoV-2-infected cerebral organoids found an induction of innate immunity and neurodegeneration genes48. Distribution of the protein Tau from axons to soma and hyperphosphorylation were also detected in infected neuron cells90. In addition, scRNA-seq of infected hPSC-derived BOs showed enriched expression of genes associated with metabolic processes in neurons, including electron transport–coupled proton transport, and mitochondrial electron transport of cytochrome c to oxygen and NADH to ubiquinone91. SARS-CoV-2 infection in BOs can be inhibited by ACE2-blocking antibodies or by administering cerebrospinal fluid from individuals with COVID-19 disease91. Further, DICER1, an isoform of DICER (a protein that cleaves double-stranded RNA), protects BOs from SARS-CoV-2 infection92.
In addition to neuronal lineage, hPSC-derived choroid plexus organoids (CPOs) have been used to study brain damage in individuals with COVID-19 disease93. scRNA-seq revealed ACE2 expression in mature choroid plexus cells, but not in neurons or other cell types in choroid plexus and telencephalic organoids94. In CPOs, few neurons and neural progenitors can be infected93, which is consistent with the studies using 2D hPSC-derived cortical neurons84. Interestingly, if pericyte-like cells (PLCs) are integrated into hPSC-cortical organoids, the organoids form a PLC–basement membrane–astrocyte structure and showed an increased proportion of neuronal cell population. SARS-CoV-2 was able to infect PLCs in integrated organoids, with spreading to astrocytes, which induced type I interferon astrocytic response95. In addition, viral infection induced an inflammatory response and cellular function deficits, followed by cell death, in hPSC-CPOs88. Furthermore, SARS-CoV-2 has been shown to cause damage of the tight junctions between the epithelial cells of the barrier and caused leakage of cerebrospinal fluid in a CPO model94.
Tonsil organoids
Many of current efforts on organoids focus on host response and drug discovery. Recently, human tonsil organoids (TOs) were developed to evaluate humoral immune responses to SARS-CoV-2 vaccine96. The interaction between follicular helper T cells and B cells in specialized germinal centers (GCs) plays a key role in the development of antigen-specific humoral responses. TOs, derived by reaggregating the dissociated primary human tonsils, maintained the tonsil cellular components and supported antigen-specific somatic hypermutation, affinity maturation and class switching of human B cells96. Upon immunization with a live attenuated influenza vaccine, TOs developed GCs with clear light zones and dark zones, a distinction that is of key importance for GC selection. And B cells migrated from the dark zones to the light zones and interacted with follicular dendritic cells and follicular helper T cells to regulate the GC reaction. This facilitated the development of long-lived antibody-secreting plasma cells and high-affinity memory B cells. Finally, TOs derived from different donors were applied to evaluate an adenovirus-based SARS-CoV-2 vaccine. CD8+ T cell activation and specific IgG and IgA antibodies were notably increased in TOs from several donors at day 14 after stimulation96. This highlights the potential to use these TOs to determine the personalized response to vaccine candidates.
Other organoids
About one-third of hospitalized individuals with COVID-19 develop ocular abnormalities, including conjunctivitis97. ACE2 and TMPRSS2 were found to be expressed in hPSC-derived retinal organoids (ROs)98. SARS-CoV-2 pseudovirus98 and live SARS-CoV-2 (ref. 99) can infect hPSC-derived ROs. The infected hPSC-derived ROs and adult human ocular cells showed robust SARS-CoV-2 replication and host immune response, including the NF-κB signaling pathway99.
COVID-19 causes cardiovascular disorders, including acute myocardial injury, myocarditis and arrhythmias by infection, immune cell-mediated damage and the cytokine storm100. Recently, hPSC-derived cardiac organoids (CDOs) co-cultured with endothelial cells were applied to mimic the cytokine storm observed in the hearts of individuals with COVID-19. Pro-inflammatory factors, including IFN-γ, IL-1β and poly(I:C) drove systolic and diastolic cardiac dysfunction, which could be rescued by INCB054329, a bromodomain and extraterminal domain inhibitor101.
Studies have shown that 5 of 29 proteins of SARS-CoV-2 can damage blood vessels by changing the endothelial permeability102. Human capillary organoids (CAPOs) resemble human capillary structure with a lumen, endothelial cells, pericyte coverage and a basal membrane. SARS-CoV-2 was detected by quantitative PCR with reverse transcription in CAPOs and can be blocked by hrsACE2 with no obvious toxicity81.
Organoids to study host genetic variants, viral variants and species differences
Organoids provide a physiologically relevant platform to study SARS-CoV-2 infection. In addition to viral tropism, host response and drug discovery as described above, organoids have also been applied to study the impact of host genetic variants, to examine species differences and to evaluate the properties of different SARS-CoV-2 variants. For example, a single-nucleotide polymorphism (rs4702; AA to GG allele) located in the 3′ untranslated region of FURIN was shown to decrease FURIN expression and further influence SARS-CoV-2 infection in hPSC-derived ALOs70. In addition, organoids from different species can be generated to study the species differences in their permissivenss and immune responses to viral infections. Fox example, SARS-CoV-2 was also shown to infect and replicate in bat SIOs103. Studies have also used organoid models to compare the infection and host response against different SARS-CoV-2 variants using ALOs and SIOs61.
Differences between hPSC-derived and adult organoids
In organoid-based COVID-19 studies, both hPSC-derived and adult organoids have been used. There are pros and cons for hPSC-derived and adult organoids regarding the availability, editability, maturity and diversity. hPSCs, in theory, have unlimited proliferation capacity and developmental potential to produce organoids in all three germ layers104 (Fig. 3). hPSC-derived organoids can be easily scaled up, which allows large-scale studies, such as metabolic profiling52,60 and drug screening47,51. Some organoids, such as brain and cardiac organoids, are only available from hPSCs. In comparison, adult organoids derived from adult tissues, have limited self-renewal capacity for a prolonged period of time, which limits their application for scalable research. Gene-editing technology has been applied to both hPSC-derived and adult organoids to study the role of genes/variants in SARS-CoV-2 infection70,72,73. Compared to adult organoids, the gene knockout and precise gene-editing techniques are better established in hPSCs, which makes it feasible to explore the biological function of single variants in viral infection. One major advantage of adult organoids is the maturity. Different from adult organoids, most hPSC-derived organoids still present fetal or neonatal identities. Additional work still needs to be done to further improve the maturation status of hPSC-derived organoids. Finally, adult organoids can be easily derived from donors from diverse backgrounds. Substantial efforts have been applied to develop induced pluripotent stem cells (iPSCs) from different genetic and disease backgrounds105. Together, organoids with diverse genetic backgrounds can be applied to explore the impact of genetic background on disease progression.
Future perspectives and methodologies
In the last 2 years, important progress has been made to apply organoids to COVID-19 disease modeling. However, additional investigations should be done to further optimize the organoid models for infectious diseases, such as improving organoid complexity by adding niche/immune cells, combining 3D printing and organ-on-a-chip technology to develop platforms that closely resemble the physiology and pathology of human systems, applying single-cell technology to perform in-depth studies of virus–host interactions at high resolution, as well as applying genomic sequencing and gene editing to explore genotype–phenotype correlations during viral infection (Fig. 4). These technologies will strengthen organoid culture methods and accelerate the research process of new antiviral therapeutics and vaccines, which may help investigate other viral outbreaks in the future.
Immune vascular organoids
Most organoid platforms used for COVID-19 studies only contain the cells of the host tissue/organ, which lacks other niche components, such as the immune cells and cells of the vascular system. Immune cells are the key to many aspects of COVID-19 pathophysiology and disease progression. Immune-mediated damage might play an equally or even more important role in host damage than direct infection. Thus, in vitro co-cultured organoids with immune cells can be used to study the pathophysiology of COVID-19 caused by immune cells, in addition to the consequences of direct infection. Furthermore, organoids with immune cells allow a better understanding of the interaction between infected host cells and immune cells, how immunomodulatory molecules released from infected cells affect immune components, and how immune responses in turn affect infected host tissues. Recently, an immune–host co-culture system containing hPSC-derived cardiomyocytes and macrophages showed that, upon SARS-CoV-2 infection, hPSC-derived cardiomyocytes secrete CC motif chemokine ligand 2 (CCL2)106, which is a chemokine that recruits monocytes. The monocytes differentiate into macrophages that damage the cardiac tissues by secreting the pro-inflammatory cytokines TNF and IL-6 (ref. 107). Furthermore, a high-content chemical screen using the immune–host co-culture system identified a janus kinase inhibitor, which blocks macrophage-mediated cardiac cell damage107. Coincidentally, baricitinib, a janus kinase inhibitor, has received emergency-use authorization from the FDA for the treatment of individuals hospitalized with COVID-19. Together, these examples highlight the importance of the immune components in organoid-based models to study viral infection.
Another limitation of many organoid platforms is the lack of blood vessels. Thrombotic complications are frequent in COVID-19 and contribute to mortality and morbidity. Co-culture of organoids with vascular endothelial cells and pericytes to form a blood vessel-containing organoid in a suitable spatial structure provides promise in further developing organoid model systems108. An immune–vascular–organoid model, containing host organoids, immune cells and vascular cells, will advance organoid technology and provide a next-generation model to study the infection of emerging viruses (Fig. 5).
Organoids and bioprinting system
In addition to including different cell types in organoids, it is also important to create organoids with appropriate spatial cellular location and interorgan interactions. 3D bioprinting technologies use tissue-specific cell types laden in bioink and reconstruct human organ-like structures by layered printing technology109. The organ simulation system technology generated by 3D bioprinting has great prospects in the study of human virus infection. For example, 3D-bioprinted lung-like structures can be used to study SARS-CoV-2 infections as they provide an air–cell interface in a complex hollow structure composed of multiple layers and cell types, which cannot be achieved with traditional tissue engineering methods. Considering the many advantages of the 3D-bioprinted human in vitro system in simulating viral infections, efforts should be made to generate more complex 3D lung models that support SARS-CoV-2 infection110.
Organoids on microfluidics system
Another innovative technology merging with organoids is organs-on-chips, which are microfluidic devices for cell and tissue culturing in continuously perfused small chambers. The organ-on-a-chip system allows for the creation of dynamic and controllable microenvironments suitable for studying the virus–host interactions, the evolution of resistance to viral therapy, the development of new antiviral therapies and the underlying viral pathogenesis. Using a vascularized lung-on-a-chip model to study SARS-CoV-2 infection, alveolar epithelial cells showed susceptibility to infection and decreased expression of platelet endothelial cell adhesion molecule (PECAM-1), resulting in the disruption of the barrier and establishment of a pro-coagulatory microenvironment111. Organoids cultured on a chip provide new possibilities to recapitulate the physiological functions at the organ level, which will further enhance the understanding of COVID-19 pathogenesis.
Organoids and single-cell multi-omics analysis
One major advantage of organoids is that they contain multiple types of cells, which can be utilized to explore the interaction between the virus and different host cells, as well as the interaction among host cells upon viral infection. In the early stages of COVID-19 studies, scRNA-seq had been extensively applied to determine the expression of SARS-CoV-2 entry factors in different types of cells within organoids. However, overall ACE2 expression based on scRNA-seq analyses is low. As an example, the percentage of ACE2+ cells in lung AT2 cells varies between 0.3% and 2.4%112, depending on the individual, which reflects the low detection sensitivity of scRNA-seq platforms112. Thus, there is strong need for careful interpretation of scRNA-seq data of SARS-CoV-2 entry factor expression. The scRNA-seq data need to be further validated with independent approaches, such as immunostaining or flow cytometry analyses. Although technically challenging, scRNA-seq started being applied to directly analyze the SARS-CoV-2-infected tissues/organoids in a BSL-3 setting112. These types of studies are expected to provide in-depth understanding of host cell responses and host cell interactions upon SARS-CoV-2 infection at single-cell resolution.
Organoids and genome-wide association studies
Both hPSC and adult organoids can be derived from individuals with diverse genetic backgrounds, which provide a useful model to explore the impact of genetic variants on disease progression, including viral infection. In one way, CRISPR-based gene-editing approaches, which can efficiently knock out a single gene/locus or knock in a single variant113, provide a high-throughput platform for generating isogenic hPSCs/organoids. These isogenic hPSCs/organoids can be used to determine the precise role of a single gene/genetic locus, or even a single-nucleotide polymorphism, in viral infection and host damages. On the other hand, by combining whole-genome sequencing and screening of organoids from different donors, scientists can perform genotype–phenotype studies to identify novel genes/loci/variants associated with viral infection. The identified genes/loci/variants can be further validated using isogenic organoid models developed by genetic editing tools. In addition, these hPSC/organoids derived from different donors can also be applied to investigate individual responses to antiviral drugs, which will facilitate the development of personalized treatment for individuals with COVID-19.
Organoids and deep machine learning
Deep machine learning has been effectively used in molecular and cellular biology, drug discovery, protein structure prediction and translational biomedicine. The utilization of deep machine learning in the field of viral genetics has mainly focused on the prediction of viral mutations related to drug resistance. Recently, DeepNEU, an unsupervised deep machine learning technology based on a fully connected recurrent neural network architecture with a network processing layer for each input, used a set of defined reprogramming transcription factors to simulate the iPSC system. In DeepNEU, AT2 cells can be simulated, and were shown to be permissive to simulated SARS-CoV-2 virus infection114. Infected lung cells and organoids can be used to stimulate and assess the impact of potential functional mutations in the SARS-CoV-2 genome115. These data were consistent with recently published data and provided a new approach to identifying potentially effective anti-SARS-CoV-2 drug combinations.
Conclusions
Studies using organoid platforms have contributed substantially to COVID-19 disease modeling and drug discovery. Compared with animal models, organoid platforms still have some limitations due to the lack of vasculature, immune cells, and interorgan communication. At the current stage, animal models are still required for vaccine and drug development. In the future, these organoid platforms will be used to simulate more complex organs, mimic the intra-organ interaction, and to explore pathogenic mechanisms. Multiple tissues and organoids from healthy or COVID-19 donors can provide reliable molecular assessments of viral susceptibility in individuals of different age, sex or race groups, and provide personalized treatment strategies for current and future pandemics.
References
Wilson, H. V. A new method by which sponges may be artificially reared. Science 25, 912–915 (1907).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Rheinwald, J. G. & Green, H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 (1975).
Emerman, J. T. & Pitelka, D. R. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro 13, 316–328 (1977).
Evans, M. Origin of mouse embryonal carcinoma cells and the possibility of their direct isolation into tissue culture. J. Reprod. Fertil. 62, 625–62 (1981).
Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981).
Martin, G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638 (1981).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Xia, Y. et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat. Cell Biol. 15, 1507–1515 (2013).
Barker, N. et al. Lgr5+ stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Chen, Y. W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
Dye, B. R. et al. In vitro generation of human pluripotent stem cell-derived lung organoids. Elife 4, e05098 (2015).
Gotoh, S. et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Rep. 3, 394–403 (2014).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Crespo, M. et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 23, 878–884 (2017).
Mills, R. J. et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl Acad. Sci. USA 114, E8372–E8381 (2017).
Voges, H. K. et al. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 144, 1118–1127 (2017).
Drakhlis, L. et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 39, 737–746 (2021).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Turco, M. Y. et al. Trophoblast organoids as a model for maternal–fetal interactions during human placentation. Nature 564, 263–267 (2018).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Garcez, P. P. et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 (2016).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Dang, J. et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258–265 (2016).
Gabriel, E. et al. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20, 397–406 (2017).
Zhou, T. et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274–283 (2017).
Krenn, V. et al. Organoid modeling of Zika and herpes simplex virus 1 infections reveals virus-specific responses leading to microcephaly. Cell Stem Cell 28, 1362–1379 (2021).
Zhou, J. et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl Acad. Sci. USA 115, 6822–6827 (2018).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is Bblocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020).
Ou, T. et al. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 17, e1009212 (2021).
Johnson, B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299 (2021).
Cattin-Ortolá, J. et al. Sequences in the cytoplasmic tail of SARS-CoV-2 spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 12, 5333 (2021).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020).
Daly, J. L. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020).
Tang, X. et al. SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metab. 33, 1577–1591 (2021).
Basil, M. C. et al. The cellular and physiological basis for lung repair and regeneration: past, present and future. Cell Stem Cell 26, 482–502 (2020).
Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275 (2021).
Tiwari, S. K., Wang, S., Smith, D., Carlin, A. F. & Rana, T. M. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 16, 437–445 (2021).
Tindle, C. et al. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. Elife 10, e66417 (2021).
Huang, J. et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell 27, 962–973 (2020).
Samuel, R. M. et al. Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell 27, 876–889 (2020).
Pei, R. et al. Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell 12, 717–733 (2021).
Huff, S. et al. Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J. Med. Chem. 65, 2866–2879 (2021).
Wang, S. et al. Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 39, e106057 (2020).
Lamers, M. M. et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type 2-like cells. EMBO J. 40, e105912 (2021).
Salahudeen, A. A. et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675 (2020).
Youk, J. et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 27, 905–919 (2020).
Ebisudani, T. et al. Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening. Cell Rep. 35, 109218 (2021).
Katsura, H. et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27, 890–904 (2020).
Duan, X. et al. An airway organoid-based screen identifies a role for the HIF1α–glycolysis axis in SARS-CoV-2 infection. Cell Rep. 37, 109920 (2021).
Deng, X. et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 184, 3426–3437 (2021).
Cheemarla, N. R. et al. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med. 218, e20210583 (2021).
Huang, K. Y. et al. Humanized COVID-19 decoy antibody effectively blocks viral entry and prevents SARS-CoV-2 infection. EMBO Mol. Med. 13, e12828 (2021).
Fang, K. Y. et al. Exploration and validation of related hub gene expression during SARS-CoV-2 infection of human bronchial organoids. Hum. Genomics 15, 18 (2021).
Fang, C. et al. CSF3 is a potential drug target for the treatment of COVID-19. Front. Physiol. 11, 605792 (2020).
Mohammadi-Dehcheshmeh, M. et al. A transcription regulatory sequence in the 5′ untranslated region of SARS-CoV-2 is vital for virus replication with an altered evolutionary pattern against human inhibitory microRNAs. Cells 10, 319 (2021).
Mulay, A. et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 35, 109055 (2021).
Ramachandran, P. et al. Gastrointestinal symptoms and outcomes in hospitalized coronavirus disease 2019 patients. Dig. Dis. 38, 373–379 (2020).
Stanifer, M. L. et al. Critical role of type III Interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 32, 107863 (2020).
Dobrindt, K. et al. Common genetic variation in humans impacts in vitro susceptibility to SARS-CoV-2 infection. Stem Cell Rep. 16, 505–518 (2021).
Kruger, J. et al. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol. Gastroenterol. Hepatol. 11, 935–948 (2021).
Prelli Bozzo, C. et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 12, 4584 (2021).
Beumer, J. et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 12, 5498 (2021).
Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5, eabc3582 (2020).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
Zhao, X. et al. Human intestinal organoids recapitulate enteric infections of enterovirus and coronavirus. Stem Cell Rep. 16, 493–504 (2021).
Triana, S. et al. Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut. Mol. Syst. Biol. 17, e10232 (2021).
Heuberger, J. et al. Epithelial response to IFN-γ promotes SARS-CoV-2 infection. EMBO Mol. Med. 13, e13191 (2021).
Gassen, N. C. et al. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat. Commun. 12, 3818 (2021).
Xia, S. et al. Long-term culture of human kidney proximal tubule epithelial cells maintains lineage functions and serves as an ex vivo model for coronavirus-associated kidney injury. Virol. Sin. 35, 311–320 (2020).
Monteil, V. et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913 (2020).
Wysocki, J. et al. A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS-CoV-2 infection in human kidney organoids. J. Am. Soc. Nephrol. 32, 795–803 (2021).
Calistri, A. et al. The new generation hDHODH inhibitor MEDS433 hinders the in vitro replication of SARS-CoV-2 and other human coronaviruses. Microorganisms 9, 1731 (2021).
Yang, L. et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27, 125–136 (2020).
Zhao, B. et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 11, 771–775 (2020).
McCarron, S. et al. Functional characterization of organoids derived from irreversibly damaged liver of patients with NASH. Hepatology 74, 1825–1844 (2021).
Kelava, I. & Lancaster, M. A. Stem cell models of human brain development. Cell Stem Cell 18, 736–748 (2016).
Jacob, F. et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell 27, 937–950 (2020).
Wang, C. et al. ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 28, 331–342 (2021).
Ramani, A. et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 39, e106230 (2020).
Song, E. et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 218, e20202135 (2021).
Poirier, E. Z. et al. An isoform of dicer protects mammalian stem cells against multiple RNA viruses. Science 373, 231–236 (2021).
McMahon, C. L., Staples, H., Gazi, M., Carrion, R. & Hsieh, J. SARS-CoV-2 targets glial cells in human cortical organoids. Stem Cell Rep. 16, 1156–1164 (2021).
Pellegrini, L. et al. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood–CSF barrier in human brain organoids. Cell Stem Cell 27, 951–961 (2020).
Wang, L. et al. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat. Med. 27, 1600–1606 (2021).
Wagar, L. E. et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 27, 125–135 (2021).
Wu, P. et al. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 138, 575–578 (2020).
Ahmad Mulyadi Lai, H. I. et al. Expression of endogenous angiotensin-converting enzyme 2 in human induced pluripotent stem cell-derived retinal organoids. Int. J. Mol. Sci. 22, 1320 (2021).
Eriksen, A. Z. et al. SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium. Cell Stem Cell 28, 1205–1220 (2021).
Nishiga, M., Wang, D. W., Han, Y., Lewis, D. B. & Wu, J. C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 17, 543–558 (2020).
Mills, R. J. et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell 184, 2167–2182 (2021).
Rauti, R. et al. Effect of SARS-CoV-2 proteins on vascular permeability. Elife 10, e69314 (2021).
Zhou, J. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083 (2020).
McCauley, H. A. & Wells, J. M. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962 (2017).
Giani, A. M. & Chen, S. Human pluripotent stem cell-based organoids and cell platforms for modelling SARS-CoV-2 infection and drug discovery. Stem Cell Res. 53, 102207 (2021).
Yang, L. et al. Cardiomyocytes recruit monocytes upon SARS-CoV-2 infection by secreting CCL2. Stem Cell Rep. 16, 2274–2288 (2021).
Yang, L. et al. An immuno–cardiac model for macrophage-mediated inflammation in COVID-19 hearts. Circ. Res. 129, 33–46 (2021).
Shi, Y. et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 18, e3000705 (2020).
Ashammakhi, N. et al. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater. Today Bio 1, 100008 (2019).
Chakraborty, J., Banerjee, I., Vaishya, R. & Ghosh, S. Bioengineered in vitro tissue models to study SARS-CoV-2 pathogenesis and therapeutic validation. ACS Biomater. Sci. Eng. 6, 6540–6555 (2020).
Thacker, V. V. et al. Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 22, e52744 (2021).
Hou, Y. J. et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446 (2020).
Li, H. et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target Ther. 5, 1 (2020).
Esmail, S. & Danter, W. Viral pandemic preparedness: a pluripotent stem cell-based machine-learning platform for simulating SARS-CoV-2 infection to enable drug discovery and repurposing. Stem Cells Transl. Med. 10, 239–250 (2021).
Esmail, S. & Danter, W. R. Lung organoid simulations for modelling and predicting the effect of mutations on SARS-CoV-2 infectivity. Comput. Struct. Biotechnol. J. 19, 1701–1712 (2021).
Danilczyk, U. & Penninger, J. M. Angiotensin-converting enzyme 2 in the heart and the kidney. Circ. Res. 98, 463–471 (2006).
Acknowledgements
S.C. is supported by the Bill and Melinda Gates Foundation (INV-037420), NIDDK (R01 DK124463, DP3 DK111907-01, R01 DK116075-01A1 and R01 DK119667-01A1). S.C. is supported by an Irma Hirschl Trust Research Award. Y.H. is an NYSTEM Stem Cell Biology Scholar (DOH01-TRAIN3-2016-00004). L.A.L. is supported by NICHD (1F32HD096810-01) and a Weill Cornell Medicine Research Assistance for Primary Parents Award.
Author information
Authors and Affiliations
Contributions
S.C., Y.H., L.Y. and L.A.L. prepared the manuscript. Y.H. and L.Y. prepared the figures.
Corresponding author
Ethics declarations
Competing interests
S.C. is the cofounder of OncoBeat. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Madeline Lancaster and Robert Zweigerdt for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Han, Y., Yang, L., Lacko, L.A. et al. Human organoid models to study SARS-CoV-2 infection. Nat Methods 19, 418–428 (2022). https://doi.org/10.1038/s41592-022-01453-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01453-y
This article is cited by
-
SARS-CoV-2 biology and host interactions
Nature Reviews Microbiology (2024)
-
Multi-omics analysis of attenuated variant reveals potential evaluation marker of host damaging for SARS-CoV-2 variants
Science China Life Sciences (2024)
-
Development of a screening platform to discover natural products active against SARS-CoV-2 infection using lung organoid models
Biomaterials Research (2023)
-
A beginner’s guide on the use of brain organoids for neuroscientists: a systematic review
Stem Cell Research & Therapy (2023)
-
Vascular organoids: unveiling advantages, applications, challenges, and disease modelling strategies
Stem Cell Research & Therapy (2023)