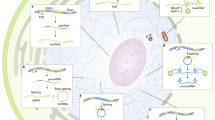
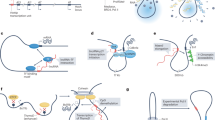
Abstract
Low expression levels and stoichiometric imbalances of long noncoding RNAs (lncRNAs) are often used as evidence for their probable lack of function or for limiting the scope of their potential influence. Recent advances in our understanding of the substoichiometric functions of lncRNAs challenge these notions and suggest routes through which unabundant lncRNAs can affect cellular functions and gene regulatory networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wu, M., Yang, L.-Z. & Chen, L.-L. Long noncoding RNA and protein abundance in lncRNPs. RNA 27, 1427–1440 (2021).
Travers, A. Transcriptional switches: the role of mass action. Phys. Life Rev. 1, 57–69 (2004).
Chong, S. et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018).
Reisser, M. et al. Single-molecule imaging correlates decreasing nuclear volume with increasing TF–chromatin associations during zebrafish development. Nat. Commun. 9, 5218 (2018).
Statello, L., Guo, C.-J., Chen, L.-L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Henninger, J. E. et al. RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225.e24 (2021).
Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021).
Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021).
Bhat, P., Honson, D. & Guttman, M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 22, 653–670 (2021).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Garcia-Jove Navarro, M. et al. RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 10, 3230 (2019).
Wang, M. et al. Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 24, 1713–1721.e4 (2018).
Hur, W. et al. CDK-regulated phase separation seeded by histone genes ensures precise growth and function of histone locus bodies. Dev. Cell 54, 379–394.e6 (2020).
Guillén-Boixet, J. et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 (2020).
Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020).
Aillaud, M. & Schulte, L. N. Emerging roles of long noncoding RNAs in the cytoplasmic milieu. Noncoding RNA 6, 44 (2020).
Yamazaki, T. et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018).
Fox, A. H., Nakagawa, S., Hirose, T. & Bond, C. S. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124–135 (2018).
Pessina, F. et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21, 1286–1299 (2019).
Li, R.-H. et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid–liquid phase separation to promote oncogenic YAP signaling. Cell Res. 31, 1088–1105 (2021).
Daneshvar, K. et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol. 22, 1211–1222 (2020).
Huo, X. et al. The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol. Cell 77, 368–383.e7 (2020).
McSwiggen, D. T., Mir, M., Darzacq, X. & Tjian, R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 33, 1619–1634 (2019).
Lee, S. et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016).
Elguindy, M. M. & Mendell, J. T. NORAD-induced Pumilio phase separation is required for genome stability. Nature 595, 303–308 (2021).
Tichon, A. et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 7, 12209 (2016).
Simon, M. D. et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469 (2013).
Sunwoo, H., Wu, J. Y. & Lee, J. T. The Xist RNA–PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl Acad. Sci. USA 112, E4216–E4225 (2015).
Pacini, G. et al. Integrated analysis of Xist upregulation and X-chromosome inactivation with single-cell and single-allele resolution. Nat. Commun. 12, 3638 (2021).
Engreitz, J. M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013).
Markaki, Y. et al. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 184, 6174–6192.e32 (2021).
Monfort, A. et al. Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561 (2015).
Lu, Z. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016).
Jachowicz, J. W. et al. Xist spatially amplifies SHARP/SPEN recruitment to balance chromosome-wide silencing and specificity to the X chromosome. Nat. Struct. Mol. Biol. 29, 239–249 (2022).
Pandya-Jones, A. et al. A protein assembly mediates Xist localization and gene silencing. Nature 587, 145–151 (2020).
Wutz, A. & Jaenisch, R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5, 695–705 (2000).
Brockdorff, N., Bowness, J. S. & Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 34, 733–744 (2020).
Wang, C.-Y., Colognori, D., Sunwoo, H., Wang, D. & Lee, J. T. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10, 2950 (2019).
Colognori, D., Sunwoo, H., Wang, D., Wang, C.-Y. & Lee, J. T. Xist repeats A and B account for two distinct phases of X inactivation establishment. Dev. Cell 54, 21–32.e5 (2020).
Weidmann, C. A., Mustoe, A. M., Jariwala, P. B., Calabrese, J. M. & Weeks, K. M. Analysis of RNA–protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 39, 347–356 (2021).
Frank, L. & Rippe, K. Repetitive RNAs as regulators of chromatin-associated subcompartment formation by phase separation. J. Mol. Biol. 432, 4270–4286 (2020).
Ziv, O. et al. Structural features within the NORAD long noncoding RNA underlie efficient repression of Pumilio activity. Preprint at bioRxiv https://doi.org/10.1101/2021.11.19.469243 (2021).
Wutz, A., Rasmussen, T. P. & Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174 (2002).
Lu, Z. et al. Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 11, 6163 (2020).
Chu, C. et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015).
Quinodoz, S. A. et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 184, 5775–5790.e30 (2021).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Schertzer, M. D. et al. lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG Island DNA. Mol. Cell 75, 523–537.e10 (2019).
Nagano, T. et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 (2008).
Pintacuda, G. et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–969 (2017).
Isono, K. et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 26, 565–577 (2013).
Hacisuleyman, E. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 (2014).
Nozawa, R.-S. et al. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 169, 1214–1227.e18 (2017).
Reichholf, B. et al. Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol. Cell 75, 756–768.e7 (2019).
Kingston, E. R. & Bartel, D. P. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 29, 1777–1790 (2019).
Gebert, L. F. R. & MacRae, I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2019).
Shi, C. Y. et al. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370, eabc9359 (2020).
Han, J. et al. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 370, eabc9546 (2020).
Kleaveland, B., Shi, C. Y., Stefano, J. & Bartel, D. P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174, 350–362.e17 (2018).
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011).
Cazalla, D., Yario, T. & Steitz, J. A. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 328, 1563–1566 (2010).
Ghini, F. et al. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 9, 3119 (2018).
Bitetti, A. et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 25, 244–251 (2018).
Denzler, R. et al. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell 64, 565–579 (2016).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Li, L. et al. Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins. Genes Dev. 35, 1595–1609 (2021).
Calo, E. et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253 (2015).
Wu, M. et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 373, 547–555 (2021).
Xing, Y.-H. et al. SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169, 664–678.e16 (2017).
Wang, X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 (2008).
Liu, Z. et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 27, 363–372 (2020).
Jia, C. et al. Different heat shock proteins bind α-synuclein with distinct mechanisms and synergistically prevent its amyloid aggregation. Front. Neurosci. 13, 1124 (2019).
Docter, B. E., Horowitz, S., Gray, M. J., Jakob, U. & Bardwell, J. C. A. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res. 44, 4835–4845 (2016).
Bevilacqua, P. C., Williams, A. M., Chou, H.-L. & Assmann, S. M. RNA multimerization as an organizing force for liquid–liquid phase separation. RNA 28, 16–26 (2022).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Turoverov, K. K. et al. Stochasticity of biological soft matter: emerging concepts in intrinsically disordered proteins and biological phase separation. Trends Biochem. Sci. 44, 716–728 (2019).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Peeples, W. & Rosen, M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021).
Brockdorff, N. Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Noncoding RNA 4, 28 (2018).
Langdon, E. M. et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 (2018).
Spitale, R. C. et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20 (2013).
Ares, P. et al. High resolution atomic force microscopy of double-stranded RNA. Nanoscale 8, 11818–11826 (2016).
Uroda, T. et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell 75, 982–995.e9 (2019).
Unfried, J. P. et al. Long noncoding RNA NIHCOLE promotes ligation efficiency of DNA double-strand breaks in hepatocellular carcinoma. Cancer Res. 81, 4910–4925 (2021).
Geiger, F. et al. Liquid–liquid phase separation underpins the formation of replication factories in rotaviruses. EMBO J. 40, e107711 (2021).
Merdanovic, M. et al. Activation by substoichiometric inhibition. Proc. Natl Acad. Sci. USA 117, 1414–1418 (2020).
Golden, R. J. et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202 (2017).
Bosson, A. D., Zamudio, J. R. & Sharp, P. A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 56, 347–359 (2014).
Denzler, R., Agarwal, V., Stefano, J., Bartel, D. P. & Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 (2014).
Robert-Finestra, T. et al. SPEN is required for Xist upregulation during initiation of X chromosome inactivation. Nat. Commun. 12, 7000 (2021).
Rodermund, L. et al. Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 372, eabe7500 (2021).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 (2018).
Acknowledgements
J.P.U. is a recipient of the Azrieli International Postdoctoral Fellowship from the Azrieli Foundation, the Excellence Fellowship Program for International Postdoctoral Researchers from the Council for Higher Education & Israel Academy of Sciences & Humanities, and an EMBO Non-Stipendiary Postdoctoral Fellowship from the European Molecular Biology Organization.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Unfried, J.P., Ulitsky, I. Substoichiometric action of long noncoding RNAs. Nat Cell Biol 24, 608–615 (2022). https://doi.org/10.1038/s41556-022-00911-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00911-1
This article is cited by
-
Targeting and engineering long non-coding RNAs for cancer therapy
Nature Reviews Genetics (2024)
-
Transcription regulation by long non-coding RNAs: mechanisms and disease relevance
Nature Reviews Molecular Cell Biology (2024)
-
METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling
Cell Death & Disease (2023)
-
The NONRATT023402.2/rno-miR-3065-5p/NGFR axis affects levodopa-induced dyskinesia in a rat model of Parkinson’s disease
Cell Death Discovery (2023)
-
Long non-coding RNA SNHG17 contributes to the progression of pancreatic adenocarcinoma by modulating miR-32-5p/EZH2/STAT3 signaling
Molecular Biology Reports (2023)