Abstract
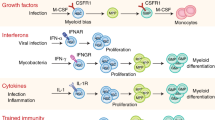
How are haematopoietic stem cells (HSCs) protected from inflammation, which increases with age and can deplete HSCs? Adiponectin, an anti-inflammatory factor that is not required for HSC function or haematopoiesis, promotes stem/progenitor cell proliferation after bacterial infection and myeloablation. Adiponectin binds two receptors, AdipoR1 and AdipoR2, which have ceramidase activity that increases upon adiponectin binding. Here we found that adiponectin receptors are non-cell-autonomously required in haematopoietic cells to promote HSC quiescence and self-renewal. Adiponectin receptor signalling suppresses inflammatory cytokine expression by myeloid cells and T cells, including interferon-γ and tumour necrosis factor. Without adiponectin receptors, the levels of these factors increase, chronically activating HSCs, reducing their self-renewal potential and depleting them during ageing. Pathogen infection accelerates this loss of HSC self-renewal potential. Blocking interferon-γ or tumour necrosis factor signalling partially rescues these effects. Adiponectin receptors are thus required in immune cells to sustain HSC quiescence and to prevent premature HSC depletion by reducing inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data associated with this paper are accessible in the NCBI Sequence Read Archive, BioProjects PRJNA699097 (associated with Fig. 1a–c), PRJNA729963 (associated with Fig. 3a) and PRJNA765672 (associated with Extended Data Fig. 4f). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Berg, A. H., Combs, T. P., Du, X., Brownlee, M. & Scherer, P. E. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 7, 947–953 (2001).
Yamauchi, T. et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 7, 941–946 (2001).
Fruebis, J. et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc. Natl Acad. Sci. USA 98, 2005–2010 (2001).
Scherer, P. E., Williams, S., Fogliano, M., Baldini, G. & Lodish, H. F. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 270, 26746–26749 (1995).
Maeda, K. et al. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1). Biochem. Biophys. Res. Commun. 221, 286–289 (1996).
Nakano, Y., Tobe, T., Choi-Miura, N. H., Mazda, T. & Tomita, M. Isolation and characterization of GBP28, a novel gelatin-binding protein purified from human plasma. J. Biochem. 120, 803–812 (1996).
Hu, E., Liang, P. & Spiegelman, B. M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 271, 10697–10703 (1996).
Masamoto, Y. et al. Adiponectin enhances quiescence exit of murine hematopoietic stem cells and hematopoietic recovery through mTORC1 potentiation. Stem Cells 35, 1835–1848 (2017).
DiMascio, L. et al. Identification of adiponectin as a novel hemopoietic stem cell growth factor. J. Immunol. 178, 3511–3520 (2007).
Tikhonova, A. N. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019).
Cawthorn, W. P. et al. Bone marrow adipose tissue is an endocrine organ that contributes to increased circulating adiponectin during caloric restriction. Cell Metab. 20, 368–375 (2014).
Ohashi, K. et al. Adiponectin promotes macrophage polarization toward an anti-inflammatory phenotype. J. Biol. Chem. 285, 6153–6160 (2010).
Yamaguchi, N. et al. Adiponectin inhibits Toll-like receptor family-induced signaling. FEBS Lett. 579, 6821–6826 (2005).
Wilk, S. et al. Adiponectin modulates NK-cell function. Eur. J. Immunol. 43, 1024–1033 (2013).
Surendar, J. et al. Adiponectin limits IFN-γ and IL-17 producing CD4 T cells in obesity by restraining cell intrinsic glycolysis. Front Immunol. 10, 2555 (2019).
Maeda, N. et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat. Med. 8, 731–737 (2002).
Masamoto, Y. et al. Adiponectin enhances antibacterial activity of hematopoietic cells by suppressing bone marrow inflammation. Immunity 44, 1422–1433 (2016).
Holland, W. L. et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat. Med. 17, 55–63 (2011).
Vasiliauskaite-Brooks, I. et al. Structural insights into adiponectin receptors suggest ceramidase activity. Nature 544, 120–123 (2017).
Yamauchi, T. et al. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat. Med. 13, 332–339 (2007).
Hug, C. et al. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl Acad. Sci. USA 101, 10308–10313 (2004).
Hannun, Y. A. & Obeid, L. M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018).
Xie, S. Z. et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell 25, 639–653 (2019).
Golan, K. et al. S1P promotes murine progenitor cell egress and mobilization via S1P1-mediated ROS signaling and SDF-1 release. Blood 119, 2478–2488 (2012).
Juarez, J. G. et al. Sphingosine-1-phosphate facilitates trafficking of hematopoietic stem cells and their mobilization by CXCR4 antagonists in mice. Blood 119, 707–716 (2012).
Massberg, S. et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell 131, 994–1008 (2007).
Ergen, A. V., Boles, N. C. & Goodell, M. A. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 119, 2500–2509 (2012).
Yamashita, M. & Passegue, E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 25, 357–372 (2019).
Chambers, S. M. et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5, e201 (2007).
Valletta, S. et al. Micro-environmental sensing by bone marrow stroma identifies IL-6 and TGFβ1 as regulators of hematopoietic ageing. Nat. Commun. 11, 4075 (2020).
Young, K. et al. Decline in IGF1 in the bone marrow microenvironment initiates hematopoietic stem cell aging. Cell Stem Cell 28, 1473–1482 (2021).
Henry, C. J. et al. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J. Clin. Invest. 125, 4666–4680 (2015).
Baldridge, M. T., King, K. Y., Boles, N. C., Weksberg, D. C. & Goodell, M. A. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature 465, 793–797 (2010).
Essers, M. A. et al. IFNα activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908 (2009).
Matatall, K. A. et al. Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep. 17, 2584–2595 (2016).
Pietras, E. M. et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016).
Pietras, E. M., Warr, M. R. & Passegue, E. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 (2011).
Morrison, S. J., Wandycz, A. M., Akashi, K., Globerson, A. & Weissman, I. L. The aging of hematopoietic stem cells. Nat. Med. 2, 1011–1016 (1996).
Fujisaki, J. et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature 474, 216–219 (2011).
Hirata, Y. et al. CD150high bone marrow Tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell 22, 445–453 (2018).
Luo, Y. & Liu, M. Adiponectin: a versatile player of innate immunity. J. Mol. Cell. Biol. 8, 120–128 (2016).
Wu, X. et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 172, 423–438 (2018).
Pearl-Yafe, M. et al. Tumor necrosis factor receptors support murine hematopoietic progenitor function in the early stages of engraftment. Stem Cells 28, 1270–1280 (2010).
Pronk, C. J., Veiby, O. P., Bryder, D. & Jacobsen, S. E. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J. Exp. Med. 208, 1563–1570 (2011).
Ishida, T. et al. Pre-transplantation blockade of TNF-α-mediated oxygen species accumulation protects hematopoietic stem cells. Stem Cells 35, 989–1002 (2017).
Donnelly, R. P., Freeman, S. L. & Hayes, M. P. Inhibition of IL-10 expression by IFN-γ up-regulates transcription of TNF-α in human monocytes. J. Immunol. 155, 1420–1427 (1995).
Vila-del Sol, V., Punzon, C. & Fresno, M. IFN-γ-induced TNF-α expression is regulated by interferon regulatory factors 1 and 8 in mouse macrophages. J. Immunol. 181, 4461–4470 (2008).
Palucka, A. K., Blanck, J. P., Bennett, L., Pascual, V. & Banchereau, J. Cross-regulation of TNF and IFN-α in autoimmune diseases. Proc. Natl Acad. Sci. USA 102, 3372–3377 (2005).
Raju, R. et al. A new cecal slurry preparation protocol with improved long-term reproducibility for animal models of sepsis. PLoS ONE 9, e115705 (2014).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003).
Lee, S. H. et al. Identifying the initiating events of anti-Listeria responses using mice with conditional loss of IFN-γ receptor subunit 1 (IFNGR1). J. Immunol. 191, 4223–4234 (2013).
Kajimura, D. et al. Adiponectin regulates bone mass via opposite central and peripheral mechanisms through FoxO1. Cell Metab. 17, 901–915 (2013).
Yue, R., Shen, B. & Morrison, S. J. Clec11a/osteolectin is an osteogenic growth factor that promotes the maintenance of the adult skeleton. eLife 5, e18782 (2016).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Noguchi, K., Gel, Y. R., Brunner, E. & Konietschke, F. nparLD: an RSoftware package for the nonparametric analysis of longitudinal data in factorial experiments. J. Stat. Softw. 50, 1–23 (2012).
Acknowledgements
S.J.M. is a Howard Hughes Medical Institute (HHMI) Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryn and Gene Bishop Distinguished Chair in Pediatric Research, the director of the Hamon Laboratory for Stem Cells and Cancer and a Cancer Prevention and Research Institute of Texas Scholar. This work was supported by the National Institutes of Health (DK118745), the Moody Medical Research Institute, the Josephine Hughes Sterling Foundation and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (all to S.J.M.). C.E.M. was supported by a Postdoctoral Fellowship from the American Cancer Society (PF-13-245-01-LIB). E.C.J. was supported by a Postdoctoral Fellowship from the Damon Runyon Cancer Research Foundation (2278-16). R.J.B. was supported by Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowship from the National Heart, Lung, and Blood Institute (F32HL122095-01). Funding for the UT Dallas/UT Southwestern Green Fellows Program (C.D.S.) was supported, in part, by the Cecil and Ida Green Foundation. We thank G. Karsenty for providing the Adipor1fl/fl and Adipor2fl/fl mice, N. Loof, C. Cantu, T. Shih, G. Wilson and the Moody Foundation Flow Cytometry Facility, M. Mulkey for mouse colony management, and the BioHPC high-performance computing cloud at the University of Texas Southwestern Medical Center for providing computational resources.
Author information
Authors and Affiliations
Contributions
C.E.M. and S.J.M. conceived the project, and designed and interpreted experiments. C.E.M. performed most of the experiments. E.C.J. and R.J.B. discussed experiments and helped interpret data. E.C.J. performed the stromal cell analysis. A.M.S. processed samples for flow cytometric analysis in Fig. 2. E.M.C. processed samples for flow cytometric analysis in Fig. 4. C.D.S. and M.A.A. performed ELISAs in Fig. 3 and processed samples for flow cytometric analysis in Fig. 6. A.M.S., E.M.C. and M.A.A. analysed blood samples from transplanted mice. G.M.C. performed histopathological analysis of spleen sections. Z.Z. performed bioinformatic and statistical analyses. C.E.M. and S.J.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Maria Carolina Florian, and the other, anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Adiponectin receptor deficient mice are born in normal numbers and are normal in size (related to Fig. 1).
a-c. Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice were born at the expected mendelian frequencies (a) and were grossly normal in size (b) and appearance (c) (n = 4 female and n = 8 male Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice and n = 10 female and n = 3 male control mice in one experiment (b)). d. The number of bone marrow cells in one femur and one tibia or in the spleen of 8-10-week-old Vav1-Cre and control mice (five mice per genotype in one experiment). e. White blood cell, red blood cell, and platelet counts in the blood of 8–10-week-old Vav1-Cre and control mice (five mice per genotype in one experiment). f-i. The frequencies of HSCs (f), MPPs (g), HPCs (h), CMPs, MEPs, and GMPs (i) in bone marrow from one femur and one tibia of Vav1-Cre and control mice (five mice per genotype in one experiment). j-m. The frequencies of HSCs (j), MPPs (k), HPCs (l), CMPs, MEPs, and GMPs (m) in splenocytes from Vav1-Cre and control mice (five mice per genotype in one experiment). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using Mann-Whitney tests followed by Holm-Sidak’s multiple comparisons adjustments (b and d-e) or Student’s t-tests followed by Holm-Sidak’s multiple comparisons adjustments (f-m). All statistical tests were two-sided. Source numerical data are available in the source data files.
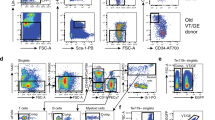
Extended Data Fig. 2 Flow cytometry gating strategy for the isolation of hematopoietic stem cells, progenitor cells, and differentiated cells (related to Figs. 1–6).
a-c. Representative flow cytometry gates used to identify hematopoietic stem and progenitor cells (a-b), NK cells, CD4 + T cells, and CD8 + T cells (c) in the bone marrow. The markers used to identify each of the cell populations characterized in this study are listed in Supplementary Table 1. d. Hematoxylin and Eosin stained sections from the spleens of Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice. In Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice the spleens were enlarged (Fig. 1d) and the red pulp was expanded throughout the spleen with increased numbers of myeloid, erythroid, and megakaryocytic cells. In control spleens there was limited extramedullary hematopoiesis confined to the subcapsular region. Representative images from three mice per genotype in one experiment are shown.
Extended Data Fig. 3 Adiponectin receptors are required to sustain HSC function (related to Fig. 2).
a-b. The percentages of HSCs that formed colonies (n = 6 mice per genotype in two independent experiments) (a) and colony size (n = 3 mice per genotype in one experiment) (b) from Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice. c. The percentage of whole bone marrow cells that formed CFU-GEMM, CFU-GM, CFU-G, CFU-M, and BFU-E colonies (n = 3 mice per genotype in one experiment). d. Donor CD45 + cells, T, B, and myeloid cells from the blood of mice competitively transplanted with donor bone marrow cells from Mx1-Cre; Adipor1fl/fl; Adipor2fl/fl (n = 27 recipients) or control (n = 25 recipients) mice (five donors per genotype in five independent experiments). e. Secondary recipients of bone marrow cells from the mice in (d) (n = 25 recipients from six Mx1-Cre; Adipor1fl/fl; Adipor2fl/fl donors and n = 24 recipients from five control donors in five independent experiments). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using Mann-Whitney tests (d-e), a Student’s t-test (a), a multinomial logistic regression (b), and Student’s t-tests followed by Holm-Sidak’s multiple comparisons adjustments (c), or Mann-Whitney tests (d-e). All statistical tests were two-sided. Source numerical data are available in the source data files.
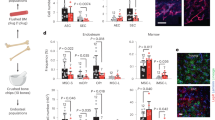
Extended Data Fig. 4 Adiponectin receptors suppress the production of inflammatory factors (related to Fig. 3).
a. ELISA analysis of inflammatory cytokines in spleen lysates from Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice (spleen lysates from 16 (IFNγ, TNF, IL6, and Il1β) or 13 (IFNα and IFNβ) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice and 19 (IFNγ, TNF, IL6, and Il1β) or 14 (IFNα and IFNβ) control mice in two independent ELISAs per cytokine). b. qRT-PCR analysis of Tnf transcript levels in sorted bone marrow cells. Data are normalized to transcript levels in wildtype whole bone marrow (WBM) cells (cells sorted from six (WBM), three (HSC, HPC, CMP, MEP, GMP, T-cells, B-cells, and erythroid progenitors), or five (myeloid cells) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice and from seven (WBM), three (HSC, HPC, CMP, MEP, GMP, and erythroid progenitors), or four (T-cells, B-cells, and myeloid cells) control mice in one experiment). c. qRT-PCR analysis of Tnf transcripts in sorted CD4 + T cells, CD8 + T cells, or NK cells from the bone marrow (cells sorted from three mice per genotype in one experiment). d. qRT-PCR analysis of Tnf transcripts in sorted macrophages, inflammatory monocytes, and neutrophils from the bone marrow. Data are normalized to transcript levels in wildtype WBM (cells sorted from seven mice per genotype in two independent experiments). e. The frequency of LepR+ stromal cells in bone marrow from 8-10 week old Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice (n = 7 mice per genotype in two independent experiments). f. RNAseq analysis of transcripts for Ifng and Tnf in LepR+ cells from 8-10 week old Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice (n = 3 mice per genotype one experiment). g. The frequencies of donor and competitor HSCs in the spleens of recipient mice co-transplanted with Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl or control donor cells and wildtype competitor cells 16 weeks after transplantation (n = 19 recipients from Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl donors and n = 20 recipients from control donors in four independent experiments). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using Student’s t-tests followed by Holm-Sidak’s multiple comparisons adjustments (a-b and d), a matched samples two-way ANOVA followed by Sidak’s multiple comparisons adjustment (c), Student’s t-tests (e), or Mann-Whitney tests followed by Holm-Sidak’s multiple comparisons adjustments (a and g). All statistical tests were two-sided. Source numerical data are available in the source data files.
Extended Data Fig. 5 Adiponectin receptors promote HSC function by reducing IFNγ receptor signaling (related to Fig. 4).
a. Bone marrow (one femur and one tibia) and spleen cellularity in Vav1-Cre; Ifngr1fl/fl and control mice (n = 7 Vav1-Cre; Ifngrfl/fl mice and n = 6 control mice in three independent experiments). b-d. The frequencies of HSCs, MPPs, HPCs (b), CMPs, MEPs, GMPs (c), and differentiated T, B, myeloid, and erythroid cells (d) in the bone marrow of Vav1-Cre; Ifngr1fl/fl and control mice (n = 7 Vav1-Cre; Ifngrfl/fl mice and n = 6 control mice in three independent experiments). e−g. The frequencies of HSCs, MPPs, HPCs (e), CMPs, MEPs, GMPs (f), and differentiated cells (g) in the spleens of Vav1-Cre; Ifngr1fl/fl and control mice (n = 7 Vav1-Cre; Ifngrfl/fl mice and n = 6 control mice in three independent experiments). h. The percentage of HSCs that incorporated a 72 hour pulse of BrdU (n = 3 Vav1-Cre; Ifngrfl/fl mice and n = 4 control mice one experiment). i-o. The frequencies of HSCs (i), MPPs (j), HPCs (k), CMPs (l), MEPs (m), GMPs (n) and differentiated cells (o) in the bone marrow of control, Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl, and Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Ifngrfl/fl mice (n = 15 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice, n = 13 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Ifngrfl/f mice and n = 21 control mice in five independent experiments). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using a matched samples two-way ANOVA followed by Sidak’s multiple comparisons adjustment (a), Mann-Whitney tests followed by Holm-Sidak’s multiple comparisons adjustments (b-g), a Student’s t-test (h), or one−way ANOVAs followed by Tukey’s multiple comparisons adjustments (i-o). All statistical tests were two-sided. Source numerical data are available in the source data files.
Extended Data Fig. 6 Adiponectin receptors promote HSC function by reducing TNF levels (related to Fig. 5).
a. Bone marrow (one femur and one tibia) and spleen cellularity in Tnf deficient and control mice (n = 5 Tnf-/- mice and n = 7 control mice in four independent experiments). b-d. The frequencies of HSCs, MPPs, HPCs (b), CMPs, MEPs, GMPs (c) and differentiated T, B, myeloid, and erythroid cells (d), in the bone marrow of Tnf deficient and control mice (n = 5 Tnf-/- mice and n = 7 control mice in four independent experiments). e-g. The frequencies of HSCs, MPPs, HPCs (e), CMPs, MEPs, GMPs (f) and differentiated cells (g), in the spleens of Tnf deficient and control mice (n = 5 (e and g) or n = 4 (f) Tnf-/- mice and n = 7 control mice in four independent experiments). h. The percentage of HSCs that incorporated a 72 hour pulse of BrdU (n = 4 Tnf-/- mice and n = 9 control mice in two independent experiments) i-o. The frequencies of HSCs (i), MPPs (j), HPCs (k), CMPs (l), MEPs (m), GMPs (n) and differentiated cells (o) in the bone marrow of control, Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl, and Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Tnf-/- mice (n = 12 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice, n = 11 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Tnf-/- mice and n = 12 control mice in five independent experiments). p-q. ELISA of TNF (p) or INFg (q) in blood plasma from Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Ifngrfl/fl (p), Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Tnf-/- (q), Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl and control mice (n = 6 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice, n = 10 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Ifngrfl/f mice and n = 17 control mice run in one ELISA per cytokine (p) and n = 13 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice, n = 6 Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl; Tnf-/- mice and n = 12 control mice in one ELISA per cytokine (q)). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using a matched samples two-way ANOVA followed by Sidak’s multiple comparisons adjustment (a), Student’s t-tests (b-h) followed by Holm-Sidak’s multiple comparisons adjustments (b-g), one-way ANOVAs followed by Tukey’s multiple comparisons adjustments (i-o and q), a Welch’s one-way ANOVA followed by Dunnett’s T3 multiple comparisons adjustment (o), or a Kruskal-Wallis test followed by Dunn’s multiple comparisons adjustment (o and p). All statistical tests were two-sided. Source numerical data are available in the source data files.
Extended Data Fig. 7 Adiponectin receptor function reduces the frequencies of splenic myeloid cells in 5-6 and 19-24 month old mice (related to Fig. 6).
a-d. The frequencies of CMPs, MEPs, GMPs (a, c), differentiated T, B, myeloid, and erythroid cells (b, d) in the bone marrow from 5-6 month old (a-b) or 19-24 month old (c-d) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl or control mice (n = 7 (a-b) or n = 5 (c-d) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice and n = 10 (a-b) or n = 6 (c-d) control mice in two independent experiments per age group). e-n. The frequencies of HSCs (e, j), MPPs (f, k), HPCs (g, l), CMPs, MEPs, GMPs (h, m), and differentiated T, B, myeloid, and erythroid cells (i, n) in the spleens of 5–6-month-old (e-i) and 19-24 month-old (j-n) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl or control mice (n = 7 (e-i) or n = 5 (j-n) Vav1-Cre; Adipor1fl/fl; Adipor2fl/fl mice and n = 10 (e-i) or n = 6 (j-n) control mice in two independent experiments per age group). All data represent mean ± standard deviation and each dot reflects a different mouse. Statistical significance was assessed using Mann-Whitney tests followed by Holm-Sidak’s multiple comparisons adjustments (a-n). All statistical tests were two-sided. Source numerical data are available in the source data files.
Supplementary information
Supplementary Table 1
Cell populations analysed by flow cytometry in this study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Meacham, C.E., Jeffery, E.C., Burgess, R.J. et al. Adiponectin receptors sustain haematopoietic stem cells throughout adulthood by protecting them from inflammation. Nat Cell Biol 24, 697–707 (2022). https://doi.org/10.1038/s41556-022-00909-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00909-9