Abstract
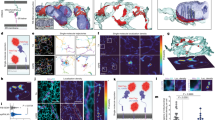
The endoplasmic reticulum (ER), a network of membranous sheets and pipes, supports functions encompassing biogenesis of secretory proteins and delivery of functional solutes throughout the cell1,2. Molecular mobility through the ER network enables these functionalities, but diffusion alone is not sufficient to explain luminal transport across supramicrometre distances. Understanding the ER structure–function relationship is critical in light of mutations in ER morphology-regulating proteins that give rise to neurodegenerative disorders3,4. Here, super-resolution microscopy and analysis of single particle trajectories of ER luminal proteins revealed that the topological organization of the ER correlates with distinct trafficking modes of its luminal content: with a dominant diffusive component in tubular junctions and a fast flow component in tubules. Particle trajectory orientations resolved over time revealed an alternating current of the ER contents, while fast ER super-resolution identified energy-dependent tubule contraction events at specific points as a plausible mechanism for generating active ER luminal flow. The discovery of active flow in the ER has implications for timely ER content distribution throughout the cell, particularly important for cells with extensive ER-containing projections such as neurons.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source image-series data for Fig. 1e and Supplementary Figs. 2 and 4e,f have been provided as Supplementary Videos 1, 2 and 8–10 respectively; and statistical information for Fig. 2 and Supplementary Fig. 2 have been provided in Supplementary Table 1. Custom code has been deposited in the Zenodo database (https://doi.org/10.5281/zenodo.1317630) with experimental raw data (https://doi.org/10.5281/zenodo.1318129). All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
References
Powers, R. E., Wang, S., Liu, T. Y. & Rapoport, T. A. Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257–260 (2017).
Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. & Rapoport, T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 (2006).
Hubner, C. A. & Kurth, I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain 137, 3109–3121 (2014).
Blackstone, C., O’Kane, C. J. & Reid, E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat. Rev. Neurosci. 12, 31–42 (2011).
Dayel, M. J., Hom, E. F. & Verkman, A. S. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 76, 2843–2851 (1999).
Nehls, S. et al. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2, 288–295 (2000).
Boersma, A. J., Zuhorn, I. S. & Poolman, B. A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 12, 227–229 (2015).
Cheezum, M. K., Walker, W. F. & Guilford, W. H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 81, 2378–2388 (2001).
Chenouard, N. et al. Objective comparison of particle tracking methods. Nat. Methods 11, 281–289 (2014).
Schuss, Z. Theory and Applications of Stochastic Processes: An Analytical Approach (Springer, New York, 2010).
Hoze, N. & Holcman, D. Residence times of receptors in dendritic spines analyzed by stochastic simulations in empirical domains. Biophys. J. 107, 3008–3017 (2014).
Hoze, N. et al. Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc. Natl Acad. Sci. USA 109, 17052–17057 (2012).
Langevin, P. The theory of Brownian movement. C. R. Hebd. Acad. Sci. 146, 530–533 (1908).
Nixon-Abell, J. et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, 433–445 (2016).
Lai, C. W., Aronson, D. E. & Snapp, E. L. BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Mol. Biol. Cell 21, 1909–1921 (2010).
Snapp, E. L., Sharma, A., Lippincott-Schwartz, J. & Hegde, R. S. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl Acad. Sci. USA 103, 6536–6541 (2006).
Tarjan, R. Depth first search and linear graph algorithms. SIAM J. Comput. 1, 146–160 (1972).
Nadeem, S. & Maraj, E. N. The mathematical analysis for peristaltic flow of nano fluid in a curved channel with compliant walls. Appl. Nanosci. 4, 85–92 (2014).
Young, L. J., Strohl, F. & Kaminski, C. F. A guide to structured illumination TIRF microscopy at high speed with multiple colors. J. Vis. Exp. 111, e53988 (2016).
Kner, P., Chhun, B. B., Griffis, E. R., Winoto, L. & Gustafsson, M. G. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6, 339–342 (2009).
Subramanian, K. & Meyer, T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell 89, 963–971 (1997).
Lo Giudice, T., Lombardi, F., Santorelli, F. M., Kawarai, T. & Orlacchio, A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp. Neurol. 261, 518–539 (2014).
Imamura, H. et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl Acad. Sci. USA 106, 15651–15656 (2009).
Frank, J. H. et al. A white light confocal microscope for spectrally resolved multidimensional imaging. J. Microsc. 227, 203–215 (2007).
Avezov, E. et al. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J. Cell Biol. 201, 337–349 (2013).
Chenouard, N., Bloch, I. & Olivo-Marin, J. C. Multiple hypothesis tracking for cluttered biological image sequences. IEEE Trans. Pattern. Anal. Mach. Intell. 35, 2736–3750 (2013).
Gustafsson, M. G. et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 (2008).
Wicker, K., Mandula, O., Best, G., Fiolka, R. & Heintzmann, R. Phase optimisation for structured illumination microscopy. Opt. Express 21, 2032–2049 (2013).
Acknowledgements
We are grateful to M. Gratian and M. Bowen (CIMR, Cambridge) and G. Strachan (Institute of Metabolic Science, Cambridge) for assistance in establishing microscopy data acquisition approaches; and S. Preissler (CIMR, Cambridge) for discussion. This study was supported by grants from the Wellcome Trust to D.R. (Wellcome 200848/Z/16/Z, WT: UNS18966); the Wellcome Trust Strategic Award for core facilities to the Cambridge Institute for Medical Research (Wellcome 100140); EPSRC (EP/L015889/1 and EP/H018301/1), MRC (MR/K015850/1 and MR/K02292X/1) and the Wellcome Trust (3-3249/Z/16/Z and 089703/Z/09/Z) to C.F.K.; an FRM team research grant to D.H.; and a DIM fellowship from Ile-de-France to P.P.; UK Dementia Research Institute grant to E.A. D.R. is a Wellcome Trust Principal Research Fellow. E.A. is a United Kingdom Dementia Research Institute Fellow.
Author information
Authors and Affiliations
Contributions
D.H. and P.P. designed, executed and interpreted the mathematic modelling and analysis of S.P.T. P.P. designed and created the custom code for the data analysis. J.E.C. executed and analysed the FLIM experiments. S.J.M. contributed to the FLIM design and analysis. M.F. and L.J.Y. developed the fast SIM system, acquired SIM images and developed custom code for image processing and analysis. C.F.K. contributed to FLIM and SIM design, analysis and interpretation. D.R. oversaw the project, contributed to the design and interpretation of the experiments, and edited the manuscript. E.A. conceived and led the project, executed analysed and interpreted cell structured illumination and single particle imaging, and contributed to the image data processing. E.A. and D.H. designed the project and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Exogenous expression levels of a fluorescent protein do not affect macromolecular crowding in the ER lumen; TMR photostability affords longer SPT trajectories.
(a) Plot of fluorescence intensity versus fluorescence lifetime of individual cells expressing an ER-localised FRET-based macromolecular crowding probe as in Fig. 1b. Fluorescence intensity serves as a measure of exogenous protein expression levels. Fluorescence lifetime correlates inversely with macromolecular crowding as described in Fig. 1b. Note, no correlation (Pearson correlation coefficient ρ = −0.2109) was observed between the broad range of exogenous protein expression levels and macromolecular crowding, n = 48 independently sampled cells (b) Distribution of trajectory lengths (central bar indicates the median, bottom and top edges of the box indicate the 25th and 75th percentiles respectively and whiskers extend to the most extreme data points), generated as in Fig. 2 from single particle signal of ER-targeted mEOS or Halo-tagged calreticulin labelled with 0.5 nM TRM-ligand. Note, the correlation between fluorophore photostability and trajectory length attest to the fidelity of the tracking procedure.
Supplementary Figure 2 Analysis of SPTs Diffusive dynamics and schematic illustration of internode and intranode motion.
(a) Distributions of diffusion coefficients computed on the individual square bins presented in Fig. 2f for the entire domain, including internode high-velocity displacements and intranode displacements. Numbers correspond to AVG+/−SD. (b) Mean Squared Displacement analysis applied on the SPTs from the experiment presented in Fig. 2. Left panel: individual MSD curves color-coded by their anomalous exponent α and right panel: distribution of the corresponding anomalous exponents (central bar indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles respectively and whiskers extend to the most extreme data points). (c) Average MSD computed on trajectory fragments contained in nodes. Left panel: average MSD curve for each node, color-coded according to its anomalous exponent and right panel: distribution of the corresponding anomalous exponents (central bar indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively and whiskers extend to the most extreme data points). (d) Averaged MSD curves computed from trajectories exiting three different nodes with anomalous exponents α>1 indicative of a super-diffusive regime. For a description of the method, refer to Suppl. Note 1 section 1.1. (e) Synchronization procedure: Left: Trajectories are synchronized on their last point spent in the indicated node (red arrow). Trajectories can then either jump from node to node or move inside a node. Right: For each time τ after escaping the node, internode (red segments) and intranode (black segments) displacements (as identified from left panel) are grouped into separate ensembles and an average instantaneous velocity is computed for each group based on formula 35 and 36 (Suppl. Note 1); analysis performed on n trajectories as denoted in individual panels.
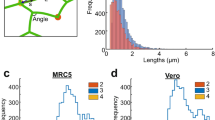
Supplementary Figure 3 Characteristics of ER luminal SPT displacements for several cell types and markers.
Statistics of SPTs motion obtained as in Fig. 2, but for a HEK-293t cell (a) and a COS-7 cell (b) expressing an inert tracer – ER-targeted HaloTag with a C-terminal ER-retention signal, KDEL. Each column is organized similarly to Fig. 2. Top: SPTs are presented color-coded according to the instantaneous velocity distributions presented below. The overlaid curves on the instantaneous velocity distributions correspond to the distributions expected for an exclusively diffusion-driven motion (solid lines, using the apparent D), or a combination of diffusion and flow (dashed lines, using D in nodes and the average instantaneous velocity between node). Inset are the corresponding cumulative distributions (color-coded as in the histogram). Middle: density and diffusion maps computed for a grid of square bins (sides of 0.2 μm) imposed on the particle displacements and flow map computed by averaging the non-Brownian velocity jumps of particles moving between pairs of neighbour nodes and color-coded according to their velocity. Bottom: distributions of apparent diffusion coefficients (left), node diffusion coefficients (middle) and average instantaneous velocity between neighbouring nodes (right). (c) SPTs recorded for a COS-7 cell expressing an ER-localised TMR labeled Halo-tagged Calreticulin (left) color-coded according to the instantaneous velocity distribution (right). Overlaid curves on the histogram are as for (a-b). Shown are characteristic images observed in three independent repeats. All values are given as AVG +/− SD, for n trajectories as denoted in individual panels.
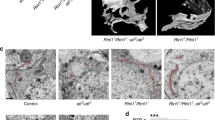
Supplementary Figure 4 Characteristics of ER structural components mobility.
(a) 3D kymograph presenting the motion of tubules and junctions across time (z-axis). (b) SPTs extracted from a stack of 50 images. Inset: magnification of an ER region showing junction trajectories. (c) Distribution of anomalous exponents obtained from SPTs (central bar indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively and whiskers extend to the most extreme data). (d) Distribution of instantaneous velocities of the SPTs. (e-f) Estimation of the characteristics of the stochastic dynamics associated to the trajectories: distribution of spring constants (e) and diffusion coefficients (f). (g) Observed and simulated junctions confinement areas. Inset: corresponding boxplots (central bar indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively and whiskers extend to the most extreme data points). A Man-Whitney U-test was used to compare the two distributions, returning a non-significant p-value (p = 0.1927), see also video S5. (a,b) images are representative of three independent repeats. Analyses performed on n tubular junctions as denoted in individual panels.
Supplementary Figure 5 Contraction points in ER tubules and ER fragmentation.
High-speed Structured Illumination Microscopy (SIM) super-resolved images of the tubular ER for a COS7 cell (a) and a HEK293 cell (b), intensity color-coded. The observed contraction is unlikely to reflect Z-dimension tubule bending at a stiff angle, as sharp bending events have not been seen along the tubules in X-Y, and given the tubule dimensions the observed phenomenon is inconsistent with Z-bending at an angle sufficient to take a small tubule-fragment out of the focal plane. Tubule contractions are visible also in 3D-SIM (Video S9). (c) Confocal image of the ER, following 20 minutes exposure to 5 µM Thapsigargin (an ER calcium uptake inhibitor), Note ER fragmentation accompanied by apparent tube-contraction, through elevation in cytoplasmic calcium21). (d) SPT velocity analysis, performed as in Fig. 2, of cell treated to induce ER fragmentation as in (c), values given as AVG +/− SD, n = 47 nodes. Ovals exemplify the nearly spherical structures of fragmented ER. (e) Fragmentation, with notable contracted tubule intermediates, was induced by intense 561 nm laser illumination of cells expressing ER-targeted HaloTag and loaded with excess TRM. (a) characteristic images observed in five independent repeats, (b-e) characteristic images observed in three independent repeats.
Supplementary information
Supplementary Information
Supplementary Figures 1–5, Supplementary Table and Supplementary Video legends.
Supplementary Note 1
Mathematical modelling and analysis.
Supplementary Table 1
Single particle motion parameters across cell types, ER tracers.
Supplementary Table 2
Plasmids list.
Supplementary Video 1
Dendra2-ER photo conversion pulse chase.
Supplementary Video 2
Raw data of ER single particle tame series.
Supplementary Video 3
Zoom in on a particle with a long trajectory.
Supplementary Video 4
Labelling control.
Supplementary Video 5
ER structural components mobility.
Supplementary Video 6
Kinetic analysis of ER tubular network remodelling.
Supplementary Video 7
Kinetic analysis of ER tubular network remodelling.
Supplementary Video 8
Transient tubule narrowing.
Supplementary Video 9
3D-SIM visualisation of tubule narrowing.
Supplementary Video 10
ATP dependence of tubular contraction frequency.
Supplementary Video 11
Light-induced reversible ER fragmentation.
Rights and permissions
About this article
Cite this article
Holcman, D., Parutto, P., Chambers, J.E. et al. Single particle trajectories reveal active endoplasmic reticulum luminal flow. Nat Cell Biol 20, 1118–1125 (2018). https://doi.org/10.1038/s41556-018-0192-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0192-2
This article is cited by
-
A local interplay between diffusion and intraflagellar transport distributes TRPV-channel OCR-2 along C. elegans chemosensory cilia
Communications Biology (2022)
-
Stress-induced protein disaggregation in the endoplasmic reticulum catalysed by BiP
Nature Communications (2022)
-
Single-particle tracking photoactivated localization microscopy of membrane proteins in living plant tissues
Nature Protocols (2021)
-
Network organisation and the dynamics of tubules in the endoplasmic reticulum
Scientific Reports (2021)
-
Impact of global structure on diffusive exploration of organelle networks
Scientific Reports (2020)