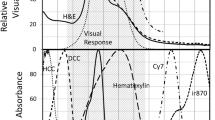
Abstract
Histological examination of tissues is central to the diagnosis and management of neoplasms and many other diseases and is a foundational technique for preclinical and basic research. However, commonly used bright-field microscopy requires prior preparation of micrometre-thick tissue sections mounted on glass slides—a process that can require hours or days, contributes to cost and delays access to critical information. Here, we introduce a simple, non-destructive slide-free technique that, within minutes, provides high-resolution diagnostic histological images resembling those obtained from conventional haematoxylin and eosin histology. The approach, which we named microscopy with ultraviolet surface excitation (MUSE), can also generate shape and colour-contrast information. MUSE relies on ~280 nm ultraviolet light to restrict the excitation of conventional fluorescent stains to tissue surfaces and it has no significant effects on downstream molecular assays (including fluorescence in situ hybridization and RNA sequencing). MUSE promises to improve the speed and efficiency of patient care in both state-of-the-art and low-resource settings and to provide opportunities for rapid histology in research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rastogi, V. et al. Artefacts: a diagnostic dilemma—a review. J. Clin. Diagn. Res.7, 2408–2413 (2013).
Neil, M. A., Juskaitis, R. & Wilson, T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett.22, 1905–1907 (1997).
Dobbs, J. et al. Confocal fluorescence microscopy for rapid evaluation of invasive tumor cellularity of inflammatory breast carcinoma core needle biopsies. Breast Cancer Res. Treat.149, 303–310 (2015).
Tao, Y. K. et al. Assessment of breast pathologies using nonlinear microscopy. Proc. Natl Acad. Sci. USA111, 15304–15309 (2014).
Kang, D. et al. Endoscopic probe optics for spectrally encoded confocal microscopy. Biomed. Opt. Express4, 1925–1936 (2013).
Orringer, D. A. et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng.1, 0027 (2017).
Liu, J. T. et al. Efficient rejection of scattered light enables deep optical sectioning in turbid media with low-numerical-aperture optics in a dual-axis confocal architecture. J. Biomed. Opt.13, 034020 (2008).
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat. Photon.9, 113–119 (2015).
Gabriele, M. L. et al. Optical coherence tomography: history, current status, and laboratory work. Invest. Ophthalmol. Vis. Sci.52, 2425–2436 (2011).
Lin, B., Urayama, S., Saroufeem, R. M., Matthews, D. L. & Demos, S. G. Real-time microscopic imaging of esophageal epithelial disease with autofluorescence under ultraviolet excitation. Opt. Express17, 12502–12509 (2009).
Lin, B., Urayama, S., Saroufeem, R. M., Matthews, D. L. & Demos, S. G. Characterizing the origin of autofluorescence in human esophageal epithelium under ultraviolet excitation. Opt. Express18, 21074–21082 (2010).
Kather, J. N. et al. New colours for histology: optimized bivariate colour maps increase perceptual contrast in histological images. PLoS ONE10, e0145572 (2015).
Fereidouni, F., Datta-Mitra, A., Demos, S. & Levenson, R. Microscopy with UV surface excitation (MUSE) for slide-free histology and pathology imaging. In Progress in Biomedical Optics and Imaging - Proc. SPIE9318, 93180F (SPIE, 2015).
Ho, D., Fereidouni, F., Levenson, R. M. & Jagdeo, J. Real-time, high-resolution, in vivo characterization of superficial skin with microscopy using ultraviolet surface excitation (MUSE). J. Drugs Dermatol.15, 1344–1346 (2016).
Zeskind, B. J. et al. Nucleic acid and protein mass mapping by live-cell deep-ultraviolet microscopy. Nat. Methods4, 567–569 (2007).
Zaak, D. et al. Ultraviolet-excited (308 nm) autofluorescence for bladder cancer detection. Urology60, 1029–1033 (2002).
Sharma, A. & Schulman, S. G. Introduction to Fluorescence Spectroscopy (John Wiley & Sons, New York, 1999).
Kenny, K. B. System and methods for generating a brightfield image using fluorescent images. US patent US8639013 B2 (2011).
Garsha, K. et al. Polyfocal interferometric image acquisition. Google patents CA2849985 C (2016).
Hui, P. & Buza, N. Atlas of Intraoperative Frozen Section Diagnosis in Gynecologic Pathology (Springer, Switzerland, 2015).
Foschini, M. P., Baldovini, C., Ishikawa, Y. & Eusebi, V. The value of large sections in surgical pathology. Int. J. Breast Cancer2012, 785947 (2012).
Abels, E. & Pantanowitz, L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J. Pathol. Inform.8, 23–23 (2017).
Adeyi, O. A. Pathology services in developing countries—the West African experience. Arch. Pathol. Lab. Med.135, 183–186 (2011).
Benediktsson, H., Whitelaw, J. & Roy, I. Pathology services in developing countries: a challenge. Arch. Pathol. Lab. Med.131, 1636–1639 (2007).
Srinivasan, M., Sedmak, D. & Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol.161, 1961–1971 (2002).
Kapp, J. R. et al. Variation in pre-PCR processing of FFPE samples leads to discrepancies in BRAF and EGFR mutation detection: a diagnostic RING trial. J. Clin. Pathol.68, 111–118 (2015).
Penland, S. K. et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab. Invest.87, 383–391 (2007).
Maes, E. et al. Analysis of the formalin-fixed paraffin-embedded tissue proteome: pitfalls, challenges, and future prospectives. Amino Acids45, 205–218 (2013).
Gnanapragasam, V. J. Unlocking the molecular archive: the emerging use of formalin-fixed paraffin-embedded tissue for biomarker research in urological cancer. BJU Int.105, 274–278 (2010).
Ragan, T. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods9, 255–258 (2012).
Hapke, M. R. & Dehner, L. P. The optically clear nucleus. A reliable sign of papillary carcinoma of the thyroid? Am. J. Surg. Pathol.3, 31–38 (1979).
Lakowicz, J. R. Principles of Fluorescence Spectroscopy (Springer Science + Business Media, Berlin, 2013).
Lechpammer, M. et al. Pathology of inherited manganese transporter deficiency. Annals Neurol.75, 608–612 (2014).
Woehrer, A. et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice—a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin. Neuropathol.30, 47–55 (2010).
Borodina, T., Adjaye, J. & Sultan, M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol.500, 79–98 (2011).
Levin, J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods7, 709–715 (2010).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature456, 53–59 (2008).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-Seq experiments. Bioinformatics28, 2184–2185 (2012).
Wang, L. et al. Measure transcript integrity using RNA-Seq data. BMC Bioinformatics17, 58 (2016).
Acknowledgements
We acknowledge L. Brandi, A. Datta-Mitra, T. McBroom, A. Adelaja, A. Krueger and L. Martinez for helping with the sample preparation and imaging, J. Wilson and D. Peabody for assisting with the tissue procurement, and E. Hillman for providing critical feedback. This work was partially supported by UC Davis Department of Pathology and Laboratory Medicine start-up funds, a UC Davis Science Translation and Innovative Research grant and an unrestricted gift from Agilent Technologies. This work was performed in part under the auspices of the US Department of Energy by the Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344.
Author information
Authors and Affiliations
Contributions
R.L. and S.G.D. developed the original observations of tissue imaging using fluorescing stains under UV LED excitation. F.F., S.G.D. and R.L. designed and fabricated the MUSE microscope based on the original design by S.G.D. and conducted the wavelength-dependent depth measurements. Z.T.H. was responsible for the colour-mapping software and maintaining the github repository. R.L., F.F., M.T., A.T. and J.A.K. performed the experiments on sample preparation, staining and comparisons with traditional histology. R.L., M.T., J.A.K. and M.L. designed and performed the comparison of MUSE and traditional histology in the central nervous system cases. R.L. and A.T. designed and performed the comparison of MUSE tissues with traditional histology, which were viewed and interpreted by J.B. and A.D.B. R.L., F.F., M.L. and J.D.M. designed and performed the FISH and RNA-Seq experiments. All authors discussed and interpreted the results and wrote and edited the paper.
Corresponding author
Ethics declarations
Competing interests
R.L. and S.G.D. are co-founders of a start-up company, Muse Microscopy, which is involved in commercializing MUSE technology. The other authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary figures, tables, notes and references
Rights and permissions
About this article
Cite this article
Fereidouni, F., Harmany, Z.T., Tian, M. et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Eng 1, 957–966 (2017). https://doi.org/10.1038/s41551-017-0165-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-017-0165-y
This article is cited by
-
DeepDOF-SE: affordable deep-learning microscopy platform for slide-free histology
Nature Communications (2024)
-
Artificial intelligence in diagnostic pathology
Diagnostic Pathology (2023)
-
Deep ultraviolet fluorescence microscopy of three-dimensional structures in the mouse brain
Scientific Reports (2023)
-
Multifractal analysis of cellular ATR-FTIR spectrum as a method for identifying and quantifying cancer cell metastatic levels
Scientific Reports (2023)
-
Artificial intelligence for digital and computational pathology
Nature Reviews Bioengineering (2023)