Abstract
Aripiprazole is recommended for routine use in schizophrenia patients. However, the biological mechanism for the adverse drug reactions (ADRs) among schizophrenia patients with the antipsychotic drug aripiprazole is far from clear. To explore the potential genetic factors that may cause movement-related adverse antipsychotic effects in patients, we conducted an association analysis between movement-related ADRs and SNPs in schizophrenia patients receiving aripiprazole monotherapy. In this study, multiple ADRs of 384 patients were quantified within 6-week treatment, and the scores of movement-related ADRs at baseline and follow-up time points during treatment were obtained. The highest score record was used as the quantitative index in analysis, and genetic analysis at the genome-wide level was conducted. The SNP rs4149181 in SLC22A8 [P = 2.28 × 10−8] showed genome-wide significance, and rs2284223 in ADCYAP1R1 [P = 9.76 × 10−8], rs73258503 in KCNIP4 [P = 1.39 × 10−7], rs678428 in SMAD9 [P = 4.70 × 10−7], rs6421034 in NAP1L4 [P = 6.80 × 10−7], and rs1394796 in ERBB4 [P = 8.60 × 10−7] were found to be significantly associated with movement-related ADRs. The combined prediction model of these six loci showed acceptable performance in predicting adverse events [area under the curve (AUC): 0.84]. Combined with the function and network of the above genes and other candidate loci (KCNA1, CACNG1, etc.), we hypothesize that SLC22A8 and KCNIP4-Kv channel perform their respective functions as transporter or channel and participate in the in vivo metabolism or effects of aripiprazole. The above results imply the important function of ion transporters and channels in movement-related adverse antipsychotic effects in aripiprazole monotherapy schizophrenia patients.
Similar content being viewed by others
Introduction
Schizophrenia is a serious mental disorder with high inheritability1,2. It mainly manifests in clinical emotion, thinking, cognition, behavior and social functions. Epidemiological investigation shows that the prevalence rate of schizophrenia in the general population is approximately 1%1, and it usually starts slowly or onset at a young age and lasts for one’s lifetime. The process of schizophrenia is prolonged and repeated, which can lead to mental disability and bring a heavy burden to patients, family members and society. Due to the heterogeneity of the disease, the exact pathological mechanism of schizophrenia has not been clearly elucidated. It is generally believed that both genetic susceptibility and environmental factors play roles in the occurrence and development of the disease. Antipsychotic drugs can relieve patients’ clinical symptoms, but approximately 75% of patients have given up treatment due to poor efficacy or side effects of drugs3. Common adverse drug reactions to antipsychotics include extrapyramidal effects, headache, weight gain, and QTc prolongation4. Environmental factors, biological factors and therapeutic strategies may be the fickle factors behind the difference in adverse drug effects5. Serious adverse effects bring inconvenience and psychological pain to patients. Studying pharmacogenetics and identifying the genetic factors within adverse drug reactions can help us develop personalized medication guidance and cope with serious adverse symptoms. Among different adverse drug reactions, the symptoms derived from the motor and nervous systems are particularly complex. Therefore, a clear and standardized monotherapy design can help us analyze the underlying mechanisms of movement-related adverse reactions.
Compared with first-generation antipsychotics, atypical/second-generation antipsychotics have fewer extrapyramidal and asthenia-related adverse reactions. Aripiprazole is an atypical antipsychotic and is also known as a third-generation antipsychotic6,7. The pharmacological mechanism of aripiprazole is mainly due to partial agonist activity at D2 and 5-HT1A receptors and the potent antagonism of the 5HT2A receptor8,9. Its metabolism is mainly through the dehydrogenation and hydroxylation of CYP3A4 and CYP2D6 enzymes10. The possible adverse reactions caused by aripiprazole include headache, insomnia, dizziness, and restlessness4,11. Aripiprazole has less effect on weight change4. Compared with olanzapine and risperidone, aripiprazole is less likely to cause metabolic adverse drug reactions4. However, genome-wide pharmacogenomics studies on the adverse drug reactions of aripiprazole are very limited. Previous studies mainly focused on the pharmacodynamics of DRD2 and 5-HTR2A and the response to negative symptoms and cognitive performance12,13,14,15. Movement-related adverse events, including extrapyramidal side effects, motor restlessness and other abnormal movements, show individual differences. The genetic basis underlying the individual difference needs to be discovered.
Investigating the genetic mechanism of the movement-related adverse drug response to aripiprazole is an urgent research direction. Therefore, we conducted a genome-wide association analysis in schizophrenia patients treated with aripiprazole monotherapy to explore the genetic loci related to the severity of movement-related antipsychotic effects.
Subjects and methods
Study design and participants
A total of 431 patients were recruited in the discovery cohort study. According to the study protocol, we performed routine baseline assessments at the start, including the general information records, the DSM-IV-TR, and the inclusion/exclusion criteria selection. Within 2 weeks after clinical inclusion, clinicians adjusted the aripiprazole dosages based on the treatment effectiveness (10–30 mg/day). After that, the dosages remained unchanged throughout the study period. The patients received clinical evaluation at weeks 2, 4, and 6, and the assessments of adverse drug reactions were recorded. Blood samples were collected at baseline. All the clinicians and patients were blinded in the study design. The patient could decide to leave the study at any time, and then the patient will be dropped from the study. In some cases, the patient cannot continue to participate, or the clinician cannot contact the patient. If a patient did not complete the full follow-up study, the last-observation carried-forward procedure was applied, and the last recording was represented as his/her treatment response. Combined with the SNP detection and quality control results, 384 patients were included in the information and polymorphism detection results in the analysis stage. The study was approved by the research ethics committees of hospital.
The inclusion conditions of the subjects in this study were as follows: (1) diagnosed with schizophrenia based on the Structured Clinical Interview of the Diagnostic and Statistical Manual of Mental Disorders, fourth edition, Text Revision (DSM-IV-TR); (2) aged 18–45 years; (3) Han Chinese lineage; (4) total scores more than 60 on the Positive and Negative Syndrome Scale (PANSS); and (5) provided written informed consent. The exclusion criteria were as follows: (1) pregnancy or breast-feeding; (2) malignant syndrome or acute dystonia, well-documented histories of epilepsy and hyperpyretic convulsion; (3) a DSM-IV diagnosis of alcohol or drug dependence, or a history of drug-induced neuroleptic malignant syndrome; (4) had previously attempted suicide, or had experienced the symptoms of severe excitement and agitation; (5) severe or unstable physical diseases, such as abnormal liver or renal function; (6) requirement of long-acting injectable medication to maintain treatment adherence or regularly treated with clozapine for treatment over the past month; (7) had QTc prolongation, a history of congenital QTc prolongation within the past 6 months. The validation samples were from the Chinese Antipsychotics Pharmacogenetics Consortium (CAPEC), and the research protocol had been documented in the previous article16. In this set of data, serious adverse reactions such as akathisia were recorded whether they occurred at multiple timepoint. We extracted the SNPs genotype of five significant candidate gene (rs2284223 undetected in current genomic data). We used the gene’s overall risk score to predict whether patients had an adverse effect of akathisia.
Phenotype definition
In discovery cohort study, the adverse drug reaction scores for movement-related antipsychotic effects are the sum of three assessment scales, including the Barnes Akathisia Rating Scale (BARS), the Abnormal Involuntary Movement Scale (AIMS), and the Simpson-Angus Scale (SAS). At baseline, all patients were evaluated with the above assessment scales, and their psychiatric symptoms and adverse drug reactions were evaluated by clinicians at 2, 4, and 6 weeks of follow-up. The BARS is scored according to its instructions17. Objective akathisia, subjective awareness of restlessness, and subjective distress related to restlessness are rated on a 4-point scale, and the score ranges from 0 to 3. The global clinical assessment of akathisia uses a 5-point scale ranging from 0 to 4. Therefore, the summed total score ranges from 0 to 9. The AIMS is a scale designed to assess abnormal involuntary movement18, primarily tardive dyskinesia. This scale includes 12 items, and items 1–10 are graded from 0 to 4 (except items 11 and 12). The SAS is a rating scale used to assess extrapyramidal side effects with 10 items ranging from 0–419. The adverse drug reaction phenotype of validation sample was recoded as text tag, such as akathisia, insomnia, tachycardia, etc.
Genotyping
Genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The samples were genotyped with Human OmniZhongHua-8 Beadchips (Illumina, San Diego, CA, USA, http://www.illumina.com/products/human-omni-zhonghua.html), which were specifically designed for the Chinese population genome as a gene detection chip. Preliminary quality control of genome data was performed before the association analysis. Sample results in the following conditions were discarded: (1) the genotype call rate was less than 98%, (2) in the case of gender discordance, (3) samples from the individuals were first-degree or second-degree relatives, (4) samples were genetic outliers, (5) SNP minor allele frequency was less than 0.05, and (6) P-values for Hardy–Weinberg equilibrium were less than 1 × 10−6. Genotype imputation for the samples was performed with the prephasing imputation stepwise approach performed in IMPUTE2 and SHAPEIT. Haplotypes derived from phase I of the 1000 Genomes Project (release version 3) were used as references.
Statistical analyses
We hypothesized that adverse drug reactions in patients treated with aripiprazole were associated with their own genotypes and conducted association analyses at the genome-wide level. After quality control, linear regression under an additive genetic model was implemented to evaluate the associations between allele dosage and adverse drug reaction scores in PLINK (version 1.90)20,21. Gender, age, baseline adverse effect score, medication dose, and the first five principal components of population structure were used as covariates in our analysis. After that, we used a P-value less than 5 × 10−8 as the data threshold for genome-wide significance. To explore more adjacent significant sites, significance levels less than 1 × 10−6 and 1 × 10−5 were also analyzed. We used the R (version 4.2.2) CMplot package to draw the Manhattan plot. Receiver operating characteristic (ROC) analyses were performed using GraphPad Prism 6. Gene Ontology (GO) and pathway enrichment analyses for candidate genes identified by genome-wide association were performed with the R package clusterProfiler. The backup data are from the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/).
Results
ADRs scores indicate the movement-related adverse antipsychotic effects
In this study, we recruited a total of 431 schizophrenia patients, and 384 patients finally passed the clinical and genotype data quality control. The movement-related adverse response was represented by the ADRs score, which was obtained by summing the scores of the three scales, including the Barnes Akathisia Rating Scale (BARS), the Abnormal Involuntary Movement Scale (AIMS), and the Simpson-Angus Scale (SAS). Table 1 shows the age, sex ratio, average aripiprazole dose and ADRs Score. The maximum quantitative value of ADRs scores was used for analysis, and the baseline value was subtracted to eliminate the baseline effect.
Genome-wide association results of aripiprazole treatment movement-related ADRs
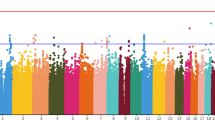
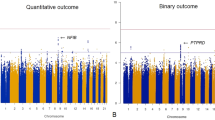
To identify genetic loci that might influence ADRs in aripiprazole treatment, genome-wide association analysis was performed. Figure 1 shows the quantile‒quantile plots, and Fig. 2 is the Manhattan plot for ADRs samples. Here, 4386312 SNPs were analyzed for movement-related adverse effects, and the linear regression analysis model was used in PLINK. Table 2 shows the 6 genes that reached (P < 1 × 10−6) significance. The genetic locus rs4149181 in SLC22A8 shows genome-wide significance [P = 2.28 × 10−8], and rs2284223 in ADCYAP1R1, rs73258503 in KCNIP4, rs678428 in SMAD9, rs6421034 in NAP1L4, and rs1394796 in ERBB4 show significance with P < 1 × 10−5. Among the 6 genes, the SNPs located in the KCNIP4 gene intron showed the best continuity in the Manhattan plot (Fig. 2, Supplementary Fig. S1). There were 120 SNPs located in KCNIP4 reaching the significance threshold (P < 1 × 10−5). The KCNIP4 gene is highly expressed in brain tissues and mainly plays a role in synaptic function. The other 5 genes had different spatiotemporal expression patterns in different brain regions (Supplementary Figs. S2–S5). Moreover, the SNPs of ADCYAP1R, KCNIP4, SMAD9, NAP1L4, and ERBB4 had eQTL effects on themselves in the brain (Supplementary Figs. S6 and S7). There are four gene loci (rs146319527, rs4747269, rs238842, and rs9605090) that are significantly associated with ADRs, but these loci currently lack gene annotation including RP11-17E2.2, 26 kb 3′ of RP11-461K13.1, CTA-481E9.4, and 14 kb 3′ of RTN4R (Supplementary Table S1).
Potential predictive effect of gene loci on movement-related ADRs
Since the six candidate genes were all significant (P < 1 × 10−6), we tried to use them to predict the movement-related adverse effects of aripiprazole. The different ADRs threshold values were applied to the receiver operating characteristic curve (ROC curve) analysis, and Fig. 3A shows the predictive effect of six SNPs to distinguish movement-related adverse responses. In the classification model, patients with ADRs scores greater than 10 points (including 10 points) were considered to have relatively serious movement-related adverse reactions. The different alleles of SNPs were weighted by the coefficient factor of the linear regression. The area under the curve (AUC) was 0.84, with 38 serious adverse response patients. Akathisia is usually seen as a more problematic adverse reaction. The six SNPs also showed a valuable potential predictive effect (AUC = 0.72), which was derived from 54 patients who had akathisia during treatment (Fig. 3B). We tested the predictive effect of our candidate risk genes in a small sample. Among 39 patients who met the pharmacogenomic analysis requirements, adverse drug reaction risk scores were calculated for five SNPs (Supplementary Fig. S8). In the clinical records of adverse drug reactions, there were four patients with severe akathisia. Compared with the ranking of risk scores, the cases in the top three of the ADRs score all had severe akathisia. This further verifies the predictive effect of our analysis results.
A ROC analysis indicated that the six most significant SNPs could effectively predict the occurrence of serious adverse reactions (ADRs), and the threshold line for serious movement-related adverse reactions in this analysis was defined as 10 or above. B The predictive effect of six SNPs in the occurrence of akathisia during treatment.
Aripiprazole is mainly metabolized by CYP2D6 and CYP3A4. As SLC22A8 has the highest expression in the kidney, we explored the possible function of SLC22A8 in drug metabolism. CPY2D6 genotyping has been suggested in personalized aripiprazole dosing10, and CYD2D6*10 has been characterized as a significantly decreased function allele22. Here, we extracted the genotype of CYD2D6*10 (rs1065852) and analyzed the adverse drug reaction score depending on the genotypes of CYD2D6*10 and SLC22A8. The G allele of rs4149181 is the higher-risk allele of SLC22A8. In the decreased function allele of CYP2D6*10 (Fig. 4), the risk genotype carriers showed higher adverse reactions, and the difference was significant (P = 0.00143; AA, 2.53 ± 0.61, n = 91; GA + GG, 11.71 ± 2.60, n = 7).
The ADRs scores of SNP (rs4149181) alleles in SLC22A8 affected by CYP2D6*10 (rs1065852). Box plot of rs4149181 (AA/GA + GG) and the ADRs scores depending on CYP2D6 polymorphism. The Kruskal‒Wallis test was used for comparisons between groups with Bonferroni correction. In the CYP2D6*10 AA genotype background, the alleles of rs4149181 were significantly different (**P = 0.00143, data are shown as the mean ± SE; AA, 2.53 ± 0.61; GA + GG, 11.71 ± 2.60).
GO and pathway analysis of genome-wide association results
To discover the molecular function and cellular pathway involved in the movement-related adverse effects of aripiprazole, GO and pathway enrichment analyses were applied with the candidate genes with a significance level less than 1 × 10−5. From gene annotation, there are several genes related to ion transporters or ion channels, including SLC22A8, KCNIP4, KCNA1 and CACNG1 (Table 2 and Supplementary Table S2). Figure 5 shows the GO terms, and detailed data on the GO terms are shown in Supplementary Table S3. Consistent with the gene function, channel or transporter functions, including voltage-gated ion channel activity (P = 0.000615) and regulation of metal ion transport (P = 0.000285), are closely related to movement-related adverse reactions induced by aripiprazole.
Previous research has reported that aripiprazole could inhibit Kv1.4 and Kv4.3 channel opening in a concentration-dependent manner23. In this study, we found a strong association between the KCNIP4 gene and adverse reactions during aripiprazole treatment in schizophrenia patients, while KCNA1/Kv1.1 also showed a significant association (rs57468930, P = 1.56 × 10−6). As KCNIP4 and KCNA1/Kv1.1 are both dominantly expressed in the brain, the protein‒protein interaction was further tested using a database (PPI website, https://string-db.org/). As shown in Supplementary Fig. S9, the interaction between KCNIP4 and KCNA1 had medium to high confidence (interaction score = 0.503).
Discussion
In this study, we explored the possible genetic loci and susceptible functional genes associated with movement-related adverse reactions to aripiprazole monotherapy in schizophrenia patients. The rs4149181 in SLC22A8 [P = 2.28 × 10−8] reached genome-wide significant associations with ADRs. The rs2284223 in ADCYAP1R1, rs73258503 in KCNIP4, rs678428 in SMAD9, rs6421034 in NAP1L4, and rs1394796 in ERBB4 showed significant associations [P < 1 × 10−6].
SLC22A8, also known as OAT3, is mainly expressed in the kidney and is also expressed in the brain, retina, and testis24. It belongs to the SLC22 family that encodes organic anion transporters, and this gene family participates in drug absorption, disposition, and/or excretion25. The substrates of SLC22A8 include anionic drugs, estrone sulfate, bile acids, flavonoids, etc. Evidence from gene knockout animals confirmed the function of SLC22 transporters in pharmacological and toxicological effects24. The genetic heterogeneity of the SLC22 family affects transporter activity26, such as rs45566039 (p.R149C), resulting in reduced transport capacity27. Through the combined analysis of the two genotypes, we hypothesized the presence of CYP2D6 and SLC22A8 risk genes might play a synergistic role in drug metabolism and clearance in vivo. The above drug metabolism and clearing are carried out in the liver and kidney, respectively. When the functions of both organs are affected, more serious adverse reactions will be produced.
KCNIP4 is a member of the Kv channel-interacting protein family, and it has a conserved EF-hand-like calcium-binding motif in the C-terminus28,29. The KCNIP4 gene is mainly expressed in the brain and plays a role in neurodevelopment and neurite outgrowth30. Previous studies have shown that two SNPs (rs876477, P = 2.69 × 10−5; rs16871892, P = 0.0109) of KCNIP4 were correlated with attention-deficit/hyperactivity disorder (ADHD) in children and adults31,32. In the genome-wide association results of schizophrenia in the CATIE study, rs1380272 (OR = 0.0522, P = 1.10 × 10−5) is an intron variant in the KCNIP4 gene locus33. In an association screen analysis of chromosome 4 for three major psychiatric disorders, including schizophrenia, bipolar and major depressive disorder, researchers identified KCNIP4 as the outstanding gene that might build a logical relationship among these disorders34. KCNIP4 is significantly associated with suicidal ideation in antidepressant treatment-related suicidal ideation35, and it also serves as a cell-type-specific module in Autism’s Pathogenesis36. In a genome-wide association study of ACE inhibitor-induced cough, KCNIP4 was significantly associated (OR = 1.3, P = 1.0 × 10−8) with ACEi-induced cough risk37. The KCNIP4 gene was originally cloned as a binding partner of Presenilin 2 (PS2), and it can co-form a complex with the voltage-gated A-type K+ channel Kv4.2 and modulate its function in the brain28,38. In a study of glutamate-induced excitotoxicity, excessive expression of KCNIP4 can produce protective effects on toxic nerves39. In conclusion, the KCNIP4 gene is deeply involved in normal brain function activities, and its gene polymorphism is generally associated with mental disorders. Moreover, the protein‒protein interaction clues between KCNIP4 and KCNA1 are highly consistent with their function in the central nervous system, as KCNA1 was identified as a pathogenic gene for epileptic ataxia and dyskinesia40,41. Combined with the evidence of the channel open blockade effect of aripiprazole on Kv channels, we hypothesized that there might be a Ca2+ signal-KCNIPs-Kv pathway involved in the movement-related adverse response in aripiprazole treatment.
Other candidate genes, including ADCYAP1R1, NAP1L4, and ERBB4, have been reported in mental disorders. The ADCYAP1R1 gene encodes a type I adenylate cyclase-activating polypeptide receptor and is highly expressed in the brain. Most clinical studies of the ADCYAP1R1 gene have focused on post-traumatic stress symptoms and children’s fear conditioning42,43,44,45,46. The results of a meta-analysis showed that the C allele of rs2267735 may increase the risk of PTSD, and the risk effect was higher in women47. Moreover, rs2267735 is associated with major depression symptoms in trauma-exposed women48, and women with lower serum estradiol and lower ADCYAP1R1 expression showed higher PTSD symptoms44. ERBB4 encodes a receptor tyrosine kinase and promotes inhibitory synapse formation in pyramidal neurons49. ERBB4 mediates amyloid β-induced neurotoxicity, which is a biomarker for Alzheimer’s disease50, and Neuregulin1-ERBB4 signaling regulates the inflammatory pain of electroacupuncture analgesia in the spinal cord51. Moreover, the function of ERBB4 in dopamine neurons is related to depression-like behaviors, and it regulates the homeostasis of extracellular dopamine and norepinephrine in catecholaminergic cells52. In animal models, ErbB4 (the homologous gene in mouse) has been shown to work with NRG1 to maintain glutaminergic activity in the amygdala, and ErbB4 is sufficient and crucial for tone-cued fear conditioning53. Loss of ErbB4 leads to dendritic spine loss in excitatory neurons, and the dendritic spine loss also occurs in many psychiatric disorders54. NAP1L4 is widely expressed in neurons and glial cells. It was reported that NAP1L4 interacts with DGKζ to attenuate hypoxic stress in the brain55. SMAD9 is involved in bone morphogenetic protein (BMP) signaling and is associated with high bone mass56. It acts as a transcriptional regulator in BMP signaling57. Its polymorphism was associated with the risk of essential hypertension in the Chinese population58.
In this study, a genome-wide association analysis was conducted for movement-related adverse antipsychotic reactions in patients treated with aripiprazole monotherapy, and candidate genes such as SLC22A8 and KCNIP4 were identified. Previous studies analyzing movement-related adverse effects in schizophrenic patients with unrestricted drug use or using different omics datasets for secondary analysis have identified some susceptible gene loci59,60. This study provides new pharmacogenomic evidence and potential signaling pathways for aripiprazole treatment and constructs an adverse reaction prediction model. Since the three rating scales (SAS, BARS, and AIMS) are also used in the clinical assessment of tardive dyskinesia (TD), our results might also have important value in TD prediction. From our perspective, the main limitation of this study is the sample size of validation cohort, and further cross-validation and calculation of genetic risk prediction models in other cohorts or disease groups.
In conclusion, we elucidated the role of ion transporter genes and their associated regulatory proteins in movement-related antipsychotic effects in aripiprazole treatment in schizophrenia patients by genetic association analyses. The important functional genes found in this study, such as SLC22A8, ADCYAP1R, and KCNIP4, will be important candidates for further research on the molecular signaling pathways of mental and nervous system diseases. These genes may become specific drug targets for future treatment of difficult clinical problems such as akathisia. The KCNIP4 was significantly associated with ADHD, autism, schizophrenia, bipolar and major depressive disorder, and this also suggested that K+ channel-related calcium regulator protein may be a common genetic basis in various mental disorders.
Data availability
The data of our study has been submitted to the Population Health Data Archive of National Population Health Data Center (NPHDC, https://www.ncmi.cn). The dataset’s accession number is 2016YFC1307000.
References
McGrath, J., Saha, S., Chant, D. & Welham, J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol. Rev. 30, 67–76 (2008).
Sullivan, P. F., Kendler, K. S. & Neale, M. C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003).
Lieberman, J. A. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia: efficacy, safety and cost outcomes of CATIE and other trials. J. Clin. Psychiatry 68, e04 (2007).
Soria-Chacartegui, P., Villapalos-Garcia, G., Zubiaur, P., Abad-Santos, F. & Koller, D. Genetic polymorphisms associated with the pharmacokinetics, pharmacodynamics and adverse effects of olanzapine, aripiprazole and risperidone. Front. Pharmacol. 12, 711940 (2021).
Zhang, X., Xiang, Q., Zhao, X., Ma, L. & Cui, Y. Association between aripiprazole pharmacokinetics and CYP2D6 phenotypes: a systematic review and meta-analysis. J. Clin. Pharm. Ther. 44, 163–173 (2019).
Preda, A. & Shapiro, B. B. A safety evaluation of aripiprazole in the treatment of schizophrenia. Expert Opin. Drug Saf. 19, 1529–1538 (2020).
Prommer, E. Aripiprazole. Am. J. Hosp. Palliat. Care 34, 180–185 (2017).
Taylor, D. M. Aripiprazole: a review of its pharmacology and clinical use. Int. J. Clin. Pract. 57, 49–54 (2003).
Shapiro, D. A. et al. Aripiprazole, a novel atypical antipsychotic drug with a unique and robust pharmacology. Neuropsychopharmacology 28, 1400–1411 (2003).
Kneller, L. A., Zubiaur, P., Koller, D., Abad-Santos, F. & Hempel, G. Influence of CYP2D6 phenotypes on the pharmacokinetics of aripiprazole and dehydro-aripiprazole using a physiologically based pharmacokinetic approach. Clin. Pharmacokinet. 60, 1569–1582 (2021).
Bernagie, C., Danckaerts, M., Wampers, M. & De Hert, M. Aripiprazole and acute extrapyramidal symptoms in children and adolescents: a meta-analysis. CNS Drugs 30, 807–818 (2016).
Chen, S.-F., Shen, Y.-C. & Chen, C.-H. HTR2A A-1438G/T102C polymorphisms predict negative symptoms performance upon aripiprazole treatment in schizophrenic patients. Psychopharmacology 205, 285–292 (2009).
Kwon, J. S. et al. Taq1A polymorphism in the dopamine D2 receptor gene as a predictor of clinical response to aripiprazole. Eur. Neuropsychopharmacol. 18, 897–907 (2008).
Ramsay, H. et al. Association between dopamine receptor D2 (DRD2) variations rs6277 and rs1800497 and cognitive performance according to risk type for psychosis: a nested case control study in a Finnish population sample. PLoS ONE 10, e0127602 (2015).
Shen, Y. C. et al. Effects of DRD2/ANKK1 gene variations and clinical factors on aripiprazole efficacy in schizophrenic patients. J. Psychiatr. Res. 43, 600–606 (2009).
Yu, H. et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry 5, 327–338.
Barnes, T. R. The Barnes Akathisia Rating Scale—revisited. J. Psychopharmacol. 17, 365–370 (2003).
Munetz, M. R. & Benjamin, S. How to examine patients using the Abnormal Involuntary Movement Scale. Hosp. Community Psychiatry 39, 1172–1177 (1988).
Janno, S., Holi, M. M., Tuisku, K. & Wahlbeck, K. Validity of Simpson-Angus Scale (SAS) in a naturalistic schizophrenia population. BMC Neurol. 5, 5 (2005).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Caudle, K. E. et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 13, 116–124 (2020).
Park, J., Cho, K. H., Lee, H. J., Choi, J. S. & Rhie, D. J. Open channel block of Kv1.4 potassium channels by aripiprazole. Korean J. Physiol. Pharmacol. 24, 545–553 (2020).
Nigam, S. K. The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Annu. Rev. Pharmacol. Toxicol. 58, 663–687 (2018).
Koepsell, H. & Endou, H. The SLC22 drug transporter family. Pflugers Arch. 447, 666–676 (2004).
Lozano, E. et al. Genetic heterogeneity of SLC22 family of transporters in drug disposition. J. Pers. Med. 8, 14 (2018).
Vávra, J. et al. Functional characterization of rare variants in OAT1/SLC22A6 and OAT3/SLC22A8 urate transporters identified in a gout and hyperuricemia cohort. Cells 11, 1063 (2022).
Morohashi, Y. et al. Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. J. Biol. Chem. 277, 14965–14975 (2002).
Liang, P., Chen, H., Cui, Y., Lei, L. & Wang, K. Functional rescue of Kv4.3 channel tetramerization mutants by KChIP4a. Biophys. J. 98, 2867–2876 (2010).
Pruunsild, P. & Timmusk, T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics 86, 581–593 (2005).
Bonvicini, C., Faraone, S. V. & Scassellati, C. Common and specific genes and peripheral biomarkers in children and adults with attention-deficit/hyperactivity disorder. World J. Biol. Psychiatry 19, 80–100 (2018).
Weißflog, L. et al. KCNIP4 as a candidate gene for personality disorders and adult ADHD. Eur. Neuropsychopharmacol. 23, 436–447 (2013).
Sullivan, P. F. et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol. Psychiatry 13, 570–584 (2008).
Tang, J., Chen, X., Cai, B. & Chen, G. A logical relationship for schizophrenia, bipolar, and major depressive disorder. Part 4: evidence from chromosome 4 high-density association screen. J. Comp. Neurol. 527, 392–405 (2019).
Perroud, N. Suicidal ideation during antidepressant treatment: do genetic predictors exist? CNS Drugs 25, 459–471 (2011).
Ji, G., Li, S., Ye, L. & Guan, J. Gene module analysis reveals cell-type specificity and potential target genes in autism’s pathogenesis. Biomedicines 9, 410 (2021).
Mosley, J. D. et al. A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J. 16, 231–237 (2016).
Norris, A. J., Foeger, N. C. & Nerbonne, J. M. Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J. Neurosci. 30, 13644–13655 (2010).
Su, Z. J., Wang, X. Y., Zhou, C. & Chai, Z. Down-regulation of miR-3068-3p enhances kcnip4-regulated A-type potassium current to protect against glutamate-induced excitotoxicity. J. Neurochem. 153, 617–630 (2020).
Verdura, E. et al. Complete loss of KCNA1 activity causes neonatal epileptic encephalopathy and dyskinesia. J. Med. Genet. 57, 132–137 (2020).
Yuan, H. et al. Two novel KCNA1 variants identified in two unrelated Chinese families affected by episodic ataxia type 1 and neurodevelopmental disorders. Mol. Genet. Genomic Med. 8, e1434 (2020).
Jovanovic, T. et al. PAC1 receptor (ADCYAP1R1) genotype is associated with dark-enhanced startle in children. Mol. Psychiatry 18, 742–743 (2013).
Jovanovic, T. et al. Impact of ADCYAP1R1 genotype on longitudinal fear conditioning in children: interaction with trauma and sex. Neuropsychopharmacology 45, 1603–1608 (2020).
Mercer, K. B. et al. Functional evaluation of a PTSD-associated genetic variant: estradiol regulation and ADCYAP1R1. Transl. Psychiatry 6, e978 (2016).
Uddin, M. et al. Adcyap1r1 genotype, posttraumatic stress disorder, and depression among women exposed to childhood maltreatment. Depress. Anxiety 30, 251–258 (2013).
Wang, L. et al. The ADCYAP1R1 gene is correlated with posttraumatic stress disorder symptoms through diverse epistases in a traumatized Chinese population. Front. Psychiatry 12, 665599 (2021).
Lind, M. J. et al. Association of posttraumatic stress disorder with rs2267735 in the ADCYAP1R1 gene: a meta-analysis. J. Traum. Stress 30, 389–398 (2017).
Lowe, S. R. et al. Gene-by-social-environment interaction (GxSE) between ADCYAP1R1 genotype and neighborhood crime predicts major depression symptoms in trauma-exposed women. J. Affect. Disord. 187, 147–150 (2015).
Luo, B. et al. ErbB4 promotes inhibitory synapse formation by cell adhesion, independent of its kinase activity. Transl. Psychiatry 11, 361 (2021).
Zhang, H., Zhang, L., Zhou, D., Li, H. & Xu, Y. ErbB4 mediates amyloid β-induced neurotoxicity through JNK/tau pathway activation: implications for Alzheimer’s disease. J. Comp. Neurol. 529, 3497–3512 (2021).
Wan, C. et al. Neuregulin1-ErbB4 signaling in spinal cord participates in electroacupuncture analgesia in inflammatory pain. Front. Neurosci. 15, 636348 (2021).
Cao, S. X. et al. ErbB4 regulate extracellular dopamine through the p38 MAPK signaling pathway. Neurosci. Lett. 751, 135830 (2021).
Lu, Y. et al. Maintenance of GABAergic Activity by Neuregulin 1-ErbB4 in Amygdala for Fear Memory. Neuron 111, 1684.
Cooper, M. A. & Koleske, A. J. Ablation of ErbB4 from excitatory neurons leads to reduced dendritic spine density in mouse prefrontal cortex. J. Comp. Neurol. 522, 3351–3362.
Takahashi, N. et al. Cellular expression and localization of DGKζ-interacting NAP1-like proteins in the brain and functional implications under hypoxic stress. Histochem. Cell Biol. 142, 461–471 (2014).
Gregson, C. L. et al. A rare mutation in SMAD9 associated with high bone mass identifies the SMAD-dependent BMP signaling pathway as a potential anabolic target for osteoporosis. J. Bone Miner. Res. 35, 92–105 (2020).
Tsukamoto, S. et al. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci. Rep. 4, 7596 (2014).
Chen, Y. et al. Association of the gene polymorphisms of BMPR2, ACVRL1, SMAD9 and their interactions with the risk of essential hypertension in the Chinese Han population. Biosci. Rep. 39, BSR20181217 (2019).
Aberg, K. et al. Genomewide association study of movement-related adverse antipsychotic effects. Biol. Psychiatry 67, 279–282 (2010).
Li, J. et al. Identification of novel proteins associated with movement-related adverse antipsychotic effects by integrating GWAS data and human brain proteomes. Psychiatry Res. 317, 114791 (2022).
Acknowledgements
We thank all subjects who participated in this study. This study was funded by the National Natural Science Foundation of China (81825009, 82101571), and National Key Technology R&D Program of China (2016YFC1307000), Academy of Medical Sciences Research Unit (2019-I2M-5-006), Chinese Institute for Brain Research at Beijing (2020-NKX-XM-12), PKUHSC-KCL Joint Medical Research (BMU2020KCL001), and the Major Science and Technology Projects of Henan Province (201300310200).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Mei, D., Lu, Z. et al. Genome-wide association study identified six loci associated with adverse drug reactions to aripiprazole in schizophrenia patients. Schizophr 9, 44 (2023). https://doi.org/10.1038/s41537-023-00369-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41537-023-00369-6