Abstract
The adsorption energies of molecular adsorbates on catalyst surfaces are key descriptors in computational catalysis research. For the relatively large reaction intermediates frequently encountered, e.g., in syngas conversion, a multitude of possible binding motifs leads to complex potential energy surfaces (PES), however. This implies that finding the optimal structure is a difficult global optimization problem, which leads to significant uncertainty about the stability of many intermediates. To tackle this issue, we present a global optimization protocol for surface adsorbate geometries which trains a surrogate machine learning potential on-the-fly. The approach is applicable to arbitrary surface models and adsorbates and minimizes both human intervention and the number of required DFT calculations by iteratively updating the training set with configurations explored by the algorithm. We demonstrate the efficiency of this approach for a diverse set of adsorbates on the Rh(111) and (211) surfaces.
Similar content being viewed by others
Introduction
The adsorption energies of key reactants are the most commonly used descriptors for the activity of heterogeneous catalysts, and thus play an important role in understanding and designing catalysts1,2. In the case of small adsorbates (OHx, CHx, NHx) and ideal metal surfaces, adsorption energies are relatively straightforward to calculate with first-principles methods. In this case, the adsorbates feature simple monodentate binding motives and the number of symmetry inequivalent surface sites on low-index crystalline facets is small. Indeed, one can even benefit from simple linear scaling relations to estimate adsorption energies across different surfaces3.
The situation is different when larger reactants are involved in the process. A prime example of this is synthesis gas (syngas) conversion, which is an important industrial process that can be used to produce valuable chemicals and fuels such as ethanol and higher hydrocarbons. Here, the reaction network features a wide range of differently sized intermediates4,5,6,7. Understanding the selectivity of syngas conversion on a given catalyst therefore inevitably requires at least an approximate notion of the adsorption energies of these larger molecules.
The main problem with this is that there are typically many local minima on the adsorbate binding energy surface for larger adsorbates. This is due to the fact that the possible binding motives on a catalyst surface are numerous in this case, including multidentate geometries. Furthermore, the adsorbates may display significant internal flexibility, so that the convergence of a local geometry optimization will strongly depend on the details of the initial geometry used to set up the calculation. These issues are exacerbated when working with nontrivial surface models (e.g., high-index, defected or amorphous surfaces). In combination, these factors presently hinder the robust and reproducible prediction of the adsorption energies of large, flexible molecules on catalyst surfaces.
The most common way of addressing this issue is so-called “brute-intuition”8. This basically means constructing a number of reasonable starting geometries by hand and carrying out local relaxation for each in order to find the most stable adsorbate geometry. However, this approach is clearly biased by the intuition of the user. To avoid this, a full “brute-force” search can be performed, which amounts to the high-throughput screening of all viable candidate geometries9,10. This requires an unbiased and efficient strategy for enumerating the viable candidates, e.g., via a graph-based approach (as in CatKit)11 or by sampling in internal coordinate ranges (as in DockOnSurf)12. Unfortunately, the brute-force strategy quickly becomes computationally prohibitive due to the large number of candidates that are typically obtained. Furthermore, both CatKit and DockOnSurf still require the definition of molecular conformers, surface binding sites and anchor-points on the molecule, all of which are themselves potential sources of bias.
Ultimately, these issues are common to all global optimization problems. We can therefore draw from the wealth of algorithms developed for this purpose, such as simulated annealing13, basin hopping14 or minima hopping (MH)15. These approaches can in principle be used to perform an unbiased search of the potential energy surface (PES) without enumerating candidates, but they are typically prohibitively expensive in conjunction with first-principles methods like density functional theory (DFT). To overcome this, semiempirical methods or empirical potentials have been used5,8,16, but their availability and quality across the periodic table is often lacking, particularly for the description of molecule/surface interactions. This makes the use of global optimization algorithms like MH still rather uncommon in computational catalysis research.
With the development of machine learning (ML) techniques over the past decades, there is promise that the computational bottleneck towards the unbiased search for ground-state adsorbate geometries can be overcome with a data-driven approach. Specifically, ML surrogate models can replace DFT calculations with orders of magnitude lower computational cost. Importantly, this can be achieved with little sacrifice of accuracy, provided that the training configurations are sufficiently representative of the PES. Indeed, there are already ML models which accurately predict adsorption energies, mostly focusing on simple adsorbates on different surface models11,17,18,19. Recently, these efforts have been expanded toward larger adsorbates with promising results4,20,21,22, though these approaches still rely on a static representation of the adsorbate/surface systems and avoid a full global optimization of the geometric structure. Since ML surrogate models have already shown great potential in local23,24,25,26,27,28 and global29,30,31,32,33,34,35,36,37,38 geometry optimization problems, we herein want to expand this approach to obtain an efficient and universal algorithm for finding the global minimum geometries of adsorbate/surface systems.
Specifically, we present an active learning workflow based on Gaussian Approximation Potentials (GAP)39, which constructs its own training set on-the-fly during constrained MH simulations. Only a small number of single-point DFT calculations are used for training, while the GAP PES is used to thoroughly explore the configuration space, yielding a diverse set of promising candidates. These can be further refined with local DFT relaxations. Importantly, the necessary hyperparameters of the ML model are automatically determined by robust heuristics, leading to a method with high data-efficiency and requiring minimal human intervention.
Results
Global optimization workflow
The idea of iteratively generating training data for ML potentials goes back at least to the work of Behler and Parinello40. De Vita and coworkers subsequently showed that, in the context of molecular dynamics simulations, training sets can actually be constructed completely on-the-fly, using Gaussian Process models with uncertainty estimation41. This concept was recently further developed by Bokdam and co-workers42 and Kozinsky43,44 and co-workers. For the task of exploring adsorbate binding motives on catalyst surfaces, plain molecular dynamics (MD) simulations are typically insufficient, however, as they easily get trapped in local basins. Furthermore, an uncertainty-driven training scheme would likely overemphasize high energy configurations at the expense of local and global minima.
The global optimization protocol presented herein follows a similar philosophy as the GAP-driven random structure search (RSS) proposed by Deringer, Pickard and Csányi45 in two aspects. First, it requires no assumptions about the adsorbate geometry or site a priori. Secondly, the global structure search and GAP fitting are performed simultaneously, since a pool of candidate structures is iteratively generated by MH and used to select new training samples. Nonetheless, the specific setting of searching for surface-adsorbate geometries requires a specialized workflow, since RSS is ill-suited for maintaining the molecular identity of the adsorbates.
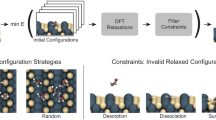
The full protocol is composed of three parts. In the first part, iterative GAP fitting is performed, using a series of MH searches to generate new training samples. Once this training process is converged, an extensive MH production run is performed, using parallel simulations that contribute to a shared pool of minima. From this pool, a diverse set of promising structures is selected, using Kernel Principal Component Analysis (PCA) and k-means clustering. These structures are subsequently locally relaxed at the DFT level. The full workflow is illustrated in Fig. 1 and the individual steps will be described in detail in the following.
a Schematic diagram of the global optimization workflow, with colors representing the use of different geometric constraints. See text for the heuristics behind the sampling of 5 structures involved in this workflow. b During minima hopping, Hookean constraints are imposed on the adsorbate bonds and the metal slab is fixed (blue). During DFT relaxations, only the two bottom layers of the slab are fixed (orange). c Exemplary trajectory of a minima hopping run, illustrating the sampling strategy of selecting the global minimum and two random local minima, as well as two random Molecular Dynamics snapshots.
Iterative training
The workflow is initiated by providing a SMILES string46 representing the adsorbate and the relaxed geometry of the clean surface as input. A rough gas-phase geometry of the adsorbate is obtained using the Merck Molecular Force Field (MMFF)47 as implemented in RDkit48. It should be noted that empirical force fields like MMFF are not well suited for describing (poly-)radical adsorbates that frequently occur in heterogeneous catalysis. Nevertheless, the obtained geometries are sufficient for our purpose, as they avoid unphysical contacts, preserve the bonding topology of the adsorbate and yield reasonable bond lengths, which are in turn used for defining so-called Hookean constraints8. These ensure that the molecular identity of the molecule of interest is maintained throughout the simulation (see Methods section). Additionally, the metal surface atoms are constrained for all GAP simulations, whereas only the lower layers of the surface are constrained in the final DFT relaxation (see Fig. 1b).
The initial geometry is then randomly placed onto the catalyst surface and the energy and forces of the full system are evaluated with DFT. This single configuration represents the initial training set of the GAP. Not surprisingly, the quality of the corresponding potential is low and results in rather unphysical structures for the first MH run (see Fig. 2). While these structures are therefore not very useful from a global optimization perspective, they nevertheless help to substantially improve the GAP, marking high energy regions of the PES. Note that subsequent iterations begin with newly randomized initial structures to further aid the MH runs in exploring as different region of the PES as possible.
Based on these structures, a small number of DFT calculations is performed. These serve the dual purpose of validating the quality of the GAP model and generating new training data for the next iteration. This leads to an important design choice for the workflow, namely how to select which structures are added to the training set. Our primary goal is to find the global minimum geometry for a given combination of adsorbate and surface. To this end, the GAP should provide accurate geometries of all local minima and reliably rank their relative stability. Clearly, minimum structures will thus form an important part of the training set. However, training only on putative minima would preclude any infomation about energetic barriers on the PES and potentially lead to numerical instabilities during the MD runs required for MH (see Fig. 1c).
To obtain a robust data selection procedure, several options were considered (assuming a fixed budget of five DFT calculations per iteration). As a baseline, five random systems were drawn from all configurations generated by a MH run. This scheme is termed full random in the following. Since there are typically many more MD snapshots than relaxed geometries, full random selection is intrinsically biased towards the former. To overcome this imbalance a stratified random approach was tested, where two configurations each were randomly drawn from the MD snapshots and local minima, while the global minimum at each iteration was always added to the training set. This stratification reflects the expected importance of the respective configurations in the training set.
While random selection tends to be a strong baseline, many active learning schemes use more sophisticated selection criteria, e.g., based on estimated uncertainties43,44,49,50 or farthest-point heuristics7,51,52. We therefore also tested selection schemes using the Farthest Point Sampling (FPS) method51. In analogy to the random approaches, these schemes are termed full FPS and stratified FPS.
All four selection approaches were tested by performing global searches for eighteen surface/adsorbate combinations (see SI for details). This reveals that stratification is essential for converging the energy and force errors on the minima in a reasonable number of iterations (less than 30). Meanwhile, there is only a small difference between FPS and random selection, with the random schemes slightly outperforming FPS overall. This is in line with previous reports that FPS tends to only be beneficial when starting from reasonably large datasets, since its focus on outliers can be problematic for very small training sets7,51. Based on this comparison, we therefore use the stratified random approach in the following.
Model convergence
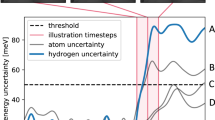
In addition to providing training data for the next GAP model, the DFT calculations are also used to estimate the out-of-sample error of the current GAP. In principle, the iterative training procedure can be considered as converged once the predicted energies and forces for the minima are sufficiently accurate (while the accuracy for the high temperature MD structures is less important). However, due to the small number of calculations performed at each iteration, the Root Mean Square Deviation (RMSD) with respect to the DFT reference only provides a noisy estimate of the true out-of-sample error. This is shown in Fig. 3 for the specific case of CH2CO on Rh(211), where the RMSD oscillates significantly across iterations. More importantly, the RMSD can be very low for some iterations and subsequently spike, meaning that it is not a robust measure of convergence.
To circumvent this issue, we use the Exponential Moving Average (EMA) of the RMSD to estimate the convergence of the training procedure:
Here, the hyperparameter α determines how quickly the weights of previous RMSDs decay in the average, with α = 1.0 recovering the current RMSD at each iteration i. As shown in Fig. 3, the EMA displays a slower and smoother decay than the RMSD. We use 0.3 for α throughout the workflow as it exhibits a reasonable balance between providing a conservative error estimate without unduly inflating the number of iterations. We consider the GAP to be converged when the EMA falls below 8 meV/atom for the energy and 0.15 eV Å−1 for the forces. As usual, these criteria are somewhat arbitrary and have been found empirically to yield a good balance between data-efficiency and accuracy for the systems investigated here.
Parallel minima hopping
Upon convergence, the training process has already yielded an extensive set of putative minimum structures. However, the quality of these structures is rather low for the initial iterations. The converged GAP model is therefore used in an additional extensive MH production run. To this end, we use a parallel MH scheme where a number of independent MH simulations are spawned and explore distinct regions of the PES simultaneously, sharing the same history of visited minima8. This avoids the danger of a single MH run spending a significant amount of time trapped in a PES region far from the global minimum due to high barriers53. Initial structures are diversified by random rotations of the rigid molecule with respect to its center of mass and random translations along the metal surface. Forty parallel processes are executed.
Since the global minimum structure is generally not known a priori, deciding when to terminate the parallel MH run is non-trivial. In previous work, the temperature of the MD simulations has been used as a stopping criterion, since the algorithm increases the temperature by a certain factor whenever it revisits known minima54. We also adopt this strategy herein, with each process performing an independent simulation with its own temperature and terminating when the temperature reaches twice the initial temperature (i.e., 4000 K) or a maximum number of iterations is exceeded (80). Note that convergence is strongly accelerated in the parallel MH approach, as several processes commonly fall into overlapping regions of the PES, rediscovering minima previously found by a nearby walker.
Candidate selection and DFT refinement
The parallel MH production run attempts an exhaustive exploration of the adsorbate conformer and binding site space, which typically results in a large ensemble of minimum adsorption structures. However, these are minima on the GAP PES (and additionally subject to the Hookean constraints on the adsorbate geometry), whereas our goal is to ultimately obtain the unconstrained minima on the DFT PES. To this end, a subset of the filtered minima (ten structures in the current work) is further refined at the DFT level (using the Bayesian Error Estimation Functional with a non-local van-der-Waals correction (BEEF-vdW), see Methods section)55.
Naively, one could select the ten lowest energy structures from the GAP ensemble, given that we are looking for the global minimum. However, this turns out to be a poor sampling strategy, because it often yields a set of minima with very similar geometries. Furthermore, molecular dissociation is sometimes observed during DFT relaxation, e.g., when the candidate structure is only a minimum on the constrained PES or because of inconsistencies between the GAP and DFT PES.
In light of these issues, it is helpful to investigate the structural diversity within the candidate ensemble prior to selecting configurations to refine. To this end, we map the structures from the conformer ensemble into a 2D space using Kernel PCA, with the averaged SOAP vector as a representation of each system (see Fig. 4)56,57. This allows the visualization of how the candidate structures are distributed in terms of their structural similarity. As we are assuming a fixed computational budget of ten DFT relaxations, we apply k-means clustering to partition the Kernel PCA space into k = 10 distinct regions. Finally, the structure with the lowest formation energy, Eform, (according to GAP) from each cluster is selected as a representative candidate for DFT relaxation (for the definition of Eform, see Methods section).
Kernel PCA representation of the adsorbate ensemble for CH3CHOH on Rh(211), highlighting the clusters obtained via k-means and the respective lowest Eform structures in each group (top, left). Five of the selected geometries are shown in the bottom frame. Kernel PCA representations colored by Eform (right).
Application to molecular adsorbates on rhodium
To demonstrate the applicability of the proposed workflow in the context of heterogeneous catalysis, we considered a set of thirteen small to mid-sized adsorbates on Rh(111) and five on Rh(211). These were previously studied in detail by Yang et al.5, while investigating the selectivity of CO hydrogenation on Rh. Importantly, these authors also used MH simulations to determine putative global minima, using approximate energies and forces from a custom density functional tight binding model (DFTB) implemented in Hotbit58. This allows us to benchmark the accuracy of the present workflow for a set of complex adsorbates. To this end, the same computational settings as in Yang et al. are used (see computational details).
The formation energies obtained from the present workflow are compared to the ones of Yang et al. in Fig. 5. Here, the global minima suggested by the proposed workflow mostly display comparable or lower formation energies than the previously reported ones, except in two cases where global minima from Yang et al. are marginally lower lying. This indicates that both the quality of the GAP potential and the MH search are sufficient to predict useful starting points for DFT relaxation. The largest difference was observed for CH3CHOH on Rh(211), where we find a configuration that lies 0.26 eV lower in energy than the previously reported global minimum. Clearly, such energy differences can have significant implications in catalysis.
(Upper panel) Comparison of global minima from ref. 5 (blue) and this work (red) for Rh(111) (left) and Rh(211) (right). Faint red bars indicate additional local minima discovered by the present workflow. (Lower panel) Comparison of adsorption structures of three example cases with the largest ΔEform compared to ref. 5, where ΔEform is the difference in formation energies between the putative global minima from ref. 5 and the current work in eV.
Given these discrepancies, it is instructive to explore the differences between the new and previously reported structures in more detail, as shown on the bottom of Fig. 5. This reveals that in some cases, large energy differences can be attributed to different adsorption sites, as for CHCO on Rh(211) and CH2CO on Rh(111). In other cases, however, the decisive factor appears to be the orientation and conformation of the adsorbate on the site, as can be seen for CH3CHOH on Rh(211). Here, both configurations are attached at equivalent sites but display different orientations. The proposed GAP/MH workflow is potentially advantageous compared to discrete graph-based algorithms in this situation, since it simultaneously explores binding sites and molecular conformations.
Notably, both the current workflow and the work of Yang et al. combines MH on an approximate PES with local DFT relaxations. Nonetheless, our workflow yields a more comprehensive set of minima which consistently includes low lying minima and even finds significantly more stable configurations in some cases. The observed differences likely stem from the use of a DFTB model (fitted to BEEF+vdW energies) for the MH search in ref. 5. Indeed, the authors of ref. 5 report that the energetics of their DFTB model are generally poor and therefore only use it as a structure generator. However, the current results indicate that reasonable agreement between the approximate PES used for MH and the target PES is beneficial for reliably predicting low-energy adsorbate geometries, even if the candidate structures are ultimately refined with the target method.
Computational cost
To put the computational benefit of the current workflow into perspective, we briefly discuss the timings for a representative system (CH2CO on Rh(211), see SI). In terms of core-hours, the dominating factors are the DFT single-point calculations used for generating the training set (15%) and the final DFT relaxations (81%), whereas the cost of fitting the GAP models and running the MH simulations is overall almost negligible. Importantly, the cost for single-point calculations depends on the complexity of the PES and is thus somewhat system-dependent. In this context, CH2CO on Rh(211) represents the worst-case scenario among the adsorbates we studied, requiring 26 iterations to achieve convergence. In contrast, simpler adsorbates like H2O and CH3 only required 15-16 iterations. Nevertheless, even for a complex adsorbate like CH2CO, the full workflow is executed in less than 8000 core-hours (on a 40 core Intel Skylake 6148 node). Given that the bulk of this time is taken up by the local DFT relaxations, our workflow thus provides a full global optimization at a cost on the order of a local optimization. For comparison, performing the full parallel MH run at the DFT level would entail approximately the 155-fold cost.
Discussion
In this manuscript, a global optimization workflow for surface adsorbates was presented and tested on important reaction intermediates for ethanol synthesis on Rh(111) and (211) surfaces. The workflow is applicable to any kind of adsorbate-surface system, since no assumption about surface sites or binding motifs are made. To achieve high computational efficiency, a GAP surrogate model is used to explore the potential energy surface. Importantly, this model is iteratively trained during the MH simulations, achieving high data-efficiency. Furthermore, the fitting procedure is fully automated, requiring minimum human intervention thanks to the use of robust heuristics for training set and hyperparameter selection.
The current work is mainly motivated by surface catalysis, where the formation energies of reaction intermediates are key ingredients of microkinetic models. Here, the global minimum configuration is of particular importance. The exploration of different adsorbate binding motifs also has other applications, however, e.g., for simulating spectroscopic properties of surface adsorbates. In this case, competing conformations may have different properties, so that good agreement with experiment can only be obtained by using Boltzmann-weighted averages of the individual spectra. In principle, the proposed method is also well suited for these applications, with the caveat that obtaining accurate conformer ensembles requires a greater number of DFT relaxations. Further discussion on competing local minima is included in the SI.
It should furthermore be noted that we have focused on the low-coverage regime herein by considering single adsorbates in supercells. Extension to larger coverage and/or multiple adsorbates is relatively straightforward. Since the cost of generating the training data is one of the main contributions to the overall effort, it would be desirable to further increase the data efficiency of the models in the future. Here, the use of well-parameterized baselines models is a promising route23,59. Finally, in order to be universally applicable, this workflow builds the surrogate model for each adsorbate from scratch. Clearly, reusing data and/or models for different adsorbates would be more efficient in many use cases. This is the subject of ongoing work.
As an alternative to the presented approach, it would in principle also be possible to fit a single interatomic potential for all adsorbates and surface types60. In our view, this would be significantly more challenging, however. While the presented method does not yield transferable interatomic potentials, the algorithm itself is transferable to a wide range of adsorbates and surface types. This means that we can easily generate bespoke interatomic potentials for a limited but important task (constrained MH for a given surface/adsorbate combination) using a small number of DFT reference calculations and no human intervention. In practice this has several advantages: On one hand, these interatomic potentials can directly be tested and validated for the task at hand, so that extrapolative capabilities are not required. On the other hand, we do not need to define the range of adsorbates and surfaces that a transferable interatomic potential should be applied to at the outset, in order to define a training set with sufficient coverage. Indeed, reaction network exploration is usually an iterative process and it is often unclear which reaction intermediates are important for a given process61. Overall, the robust and efficient global optimization workflow presented herein is thus ideally suited for application to complex processes in heterogeneous catalysis.
Methods
Gaussian approximation potentials
Gaussian approximation potentials are ML-based interatomic potentials that provide accurate representations of high-dimensional PESs39. Since the GAP methodology has recently been extensively reviewed62, we only provide a brief overview of the aspects which are most pertinent to the current paper.
In this work, we use GAPs that contain a two-body term and a many-body term based on the SOAP representation63. In order to improve the stability of the potentials in the initial training iterations (when very little data is available), an additional purely repulsive two-body baseline potential is added, which prevents unphysically close contacts between atoms and is zero otherwise (see SI). Apart from these basic design choices, there are a number of hyperparameters that need to be set in order to fit a GAP. These pertain to the basis expansions used in the two- and many-body representations, element dependent cutoff radii and switching functions, regularization of the fit and the prior weightings of two- and many-body terms.
Generally, the hyperparameters in ML models can simply be optimized, e.g., with respect to the cross-validation error of the potential. However, cross-validation and similar approaches require a sufficiently large dataset, in order to obtain a robust estimate of the generalization error. In the present work, we aim to develop an active learning workflow which starts from scratch, so that this is clearly not the case. We therefore use a mix of robust defaults and simple heuristics to set and update the hyperparameters, as detailed in the SI.
Briefly, physically motivated lengthscale parameters for the SOAP representation are available for all elements and used without modification57. Additionally, robust defaults are used for the remaining hyperparameters necessary for defining the representations and kernels. This leaves a small number of system-dependent hyperparameters, namely the regularization strength for energies and forces (σE and σF) and the prior weights of the two and many-body terms (δ2B and δMB). These are determined according to the heuristics discussed in ref. 62, see SI for details.
Constrained minima hopping
Minima hopping is a global optimization algorithm, which has been extensively applied to surface and bulk structure searching problems15,53. The basic idea is to use short, high-temperature MD runs to escape a given minimum on the PES, followed by local relaxations into the next local minimum. Importantly, MH keeps track of previously visited minima making it more efficient in finding new structures. Moreover, the algorithm drives towards lower energy structures by adaptively adjusting the temperature and energy threshold parameters, which determine the intensity of the hopping moves and the acceptance criteria for new minima. For a detailed description, we refer readers to the original minima hopping publication15.
In the context of adsobate optimization, a common problem with MH is that the high temperature MD often leads to the dissociation of the adsorbate. To address this issue, Peterson proposed using so-called Hookean constraints on bond distances, which add a harmonic energy penalty to the total energy when covalent bonds in the adsorbate are stretched beyond a certain length, thus preserving the molecular identity of the adsorbate8. Throughout this work, a spring constant (k) of 20 eV Å−2 is used. The threshold distances were set individually for each bond, using 1.05 times the bond distance in the gas-phase molecular geometry as optimized with the MMFF. In some cases the adsorbate detaches and remains floating above the metal surface owing to the high temperature in the MD. To avoid this, an additional Hookean constraint can be imposed, which pushes the adsorbate back towards the surface when it moves too far away from it.
Computational details
As representative catalytic surface models, surface slabs were constructed from (3 × 3) Rhodium (Rh) surface supercells with a thickness of four metal layers. Both the low-index (111) and stepped (211) facets were used. Due to their relevance in catalysis modeling, we report formation energies (Eform) with respect to gas-phase H2O, CO, and H2:
where Eslab+mol is the energy of the combined surface and adsorbate system, Eslab is the energy of the clean slab, εi is the gas-phase reference atomic energy of element i, and ni denotes the number of occurrences of element i in the adsorbate molecule.
All DFT calculations were carried out using the plane-wave QuantumEspresso code64 with the BEEF-vdW functional55. Ultrasoft pseudopotentials were used for the description of core electrons and a kinetic energy cutoff of 500 eV and a charge density cutoff of 5000 eV were used. The Brilloun zone was sampled via the Monkhorst-Pack scheme with a 4 × 4 × 1 grid. A dipole correction was added to compensate spurious polarization within periodic boundary conditions along z-axis. For DFT geometry optimization, the Broyden–Fletcher–Goldfarb–Shanno algorithm was used as implemented in the Atomic Simulation Environment65.
Data availability
All reported local/global minimum geometries are available on Figshare with the identifier https://doi.org/10.6084/m9.figshare.23285156.
Code availability
The code for the global optimization workflow is provided at https://gitlab.mpcdf.mpg.de/hjung/gap_workflow_surface.
References
Bruix, A., Margraf, J. T., Andersen, M. & Reuter, K. First-principles-based multiscale modelling of heterogeneous catalysis. Nat. Catal. 2, 659–670 (2019).
Nørskov, J. K., Bligaard, T., Rossmeisl, J. & Christensen, C. H. Towards the computational design of solid catalysts. Nat. Chem. 1, 37–46 (2009).
Abild-Pedersen, F. et al. Scaling properties of adsorption energies for hydrogen-containing molecules on transition-metal surfaces. Phys. Rev. Lett. 99, 016105 (2007).
Ulissi, Z. W., Medford, A. J., Bligaard, T. & Nørskov, J. K. To address surface reaction network complexity using scaling relations machine learning and dft calculations. Nat. Commun. 8, 1–7 (2017).
Yang, N. et al. Intrinsic selectivity and structure sensitivity of rhodium catalysts for C2+ oxygenate production. J. Am. Chem. Soc. 138, 3705–3714 (2016).
Margraf, J. T. & Reuter, K. Systematic enumeration of elementary reaction steps in surface catalysis. ACS Omega 4, 3370 (2019).
Stocker, S., Csányi, G., Reuter, K. & Margraf, J. T. Machine learning in chemical reaction space. Nat. Commun. 11, 227 (2020).
Peterson, A. A. Global optimization of adsorbate-surface structures while preserving molecular identity. Top. Catal. 57, 40–53 (2014).
Montoya, J. H. & Persson, K. A. A high-throughput framework for determining adsorption energies on solid surfaces. NPJ Comput. Mater. 3, 1–4 (2017).
Andriuc, O., Siron, M., Montoya, J. H., Horton, M. & Persson, K. A. Automated adsorption workflow for semiconductor surfaces and the application to Zinc Telluride. J. Chem. Inf. Model. 61, 3908 (2021).
Boes, J. R., Mamun, O., Winther, K. & Bligaard, T. Graph theory approach to high-throughput surface adsorption structure generation. J. Phys. Chem. A 123, 2281–2285 (2019).
Martí, C. et al. Dockonsurf: a python code for the high-throughput screening of flexible molecules adsorbed on surfaces. J. Chem. Inform. Model. 61, 3386–3396 (2021).
Kirkpatrick, S., Gelatt Jr, C. D. & Vecchi, M. P. Optimization by simulated annealing. Science 220, 671–680 (1983).
Wales, D. J. & Doye, J. P. Global optimization by basin-hopping and the lowest energy structures of lennard-jones clusters containing up to 110 atoms. J. Phys. Chem. A 101, 5111–5116 (1997).
Goedecker, S. Minima hopping: an efficient search method for the global minimum of the potential energy surface of complex molecular systems. J. Chem. Phys. 120, 9911–9917 (2004).
Panosetti, C., Krautgasser, K., Palagin, D., Reuter, K. & Maurer, R. J. Global materials structure search with chemically motivated coordinates. Nano Lett. 15, 8044–8048 (2015).
Andersen, M., Levchenko, S. V., Scheffler, M. & Reuter, K. Beyond scaling relations for the description of catalytic materials. ACS Catal. 9, 2752–2759 (2019).
Xu, W., Andersen, M. & Reuter, K. Data-driven descriptor engineering and refined scaling relations for predicting transition metal oxide reactivity. ACS Catal. 11, 734–742 (2020).
Andersen, M. & Reuter, K. Adsorption enthalpies for catalysis modeling through machine-learned descriptors. Acc. Chem. Res. 54, 2741–2749 (2021).
Xu, W., Reuter, K. & Andersen, M. Predicting binding motifs of complex adsorbates using machine learning with a physics-inspired graph representation. Nat. Comput. Sci. 2, 443–450 (2022).
Deshpande, S., Maxson, T. & Greeley, J. Graph theory approach to determine configurations of multidentate and high coverage adsorbates for heterogeneous catalysis. NPJ Comput. Mater. 6, 1–6 (2020).
Chanussot, L. et al. Open catalyst 2020 (oc20) dataset and community challenges. ACS Catal. 11, 6059–6072 (2021).
Musielewicz, J., Wang, X., Tian, T. & Ulissi, Z. W. Finetuna: fine-tuning accelerated molecular simulations. Mach. Learn. Sci. Technol. 3, 03LT01 (2022).
del Río, E. G., Mortensen, J. J. & Jacobsen, K. W. Local bayesian optimizer for atomic structures. Phys. Rev. B 100, 104103 (2019).
Garijo del Río, E., Kaappa, S., Garrido Torres, J. A., Bligaard, T. & Jacobsen, K. W. Machine learning with bond information for local structure optimizations in surface science. J. Chem. Phys. 153, 234116 (2020).
Denzel, A. & Kästner, J. Gaussian process regression for geometry optimization. J. Chem. Phys. 148, 094114 (2018).
Schmitz, G. & Christiansen, O. Gaussian process regression to accelerate geometry optimizations relying on numerical differentiation. J. Chem. Phys. 148, 241704 (2018).
Meyer, R. & Hauser, A. W. Geometry optimization using gaussian process regression in internal coordinate systems. J. Chem. Phys. 152, 084112 (2020).
Bisbo, M. K. & Hammer, B. Efficient global structure optimization with a machine-learned surrogate model. Phys. Rev. Lett. 124, 086102 (2020).
Westermayr, J., Chaudhuri, S., Jeindl, A., Hofmann, O. & Maurer, R. Long-range dispersion-inclusive machine learning potentials for structure search and optimization of hybrid organic-inorganic interfaces. Digit. Discov. 1, 463–475 (2022).
Jørgensen, M. S., Larsen, U. F., Jacobsen, K. W. & Hammer, B. Exploration versus exploitation in global atomistic structure optimization. J. Phys. Chem. A 122, 1504–1509 (2018).
Yamashita, T. et al. Crystal structure prediction accelerated by bayesian optimization. Phys. Rev. Mater. 2, 013803 (2018).
Todorović, M., Gutmann, M. U., Corander, J. & Rinke, P. Bayesian inference of atomistic structure in functional materials. NPJ Comput. Mater. 5, 1–7 (2019).
Järvi, J., Todorović, M. & Rinke, P. Efficient modeling of organic adsorbates on oxygen-intercalated graphene on Ir(111). Phys. Rev. B 105, 195304 (2022).
Mortensen, H. L., Meldgaard, S. A., Bisbo, M. K., Christiansen, M.-P. V. & Hammer, B. Atomistic structure learning algorithm with surrogate energy model relaxation. Phys. Rev. B 102, 075427 (2020).
Timmermann, J. et al. Data-efficient iterative training of Gaussian approximation potentials: application to surface structure determination of rutile IrO2 and RuO2. J. Chem. Phys. 155, 244107 (2021).
Kaappa, S., del Río, E. G. & Jacobsen, K. W. Global optimization of atomic structures with gradient-enhanced Gaussian process regression. Phys. Rev. B 103, 174114 (2021).
Sauerland, L. Machine-Learned Interatomic Potentials for the Syngas Conversion on Rhodium. Master’s thesis, Ludwig-Maximilians-Universität München (2021).
Bartók, A. P., Payne, M. C., Kondor, R. & Csányi, G. Gaussian approximation potentials: the accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 583 (2007).
Li, Z., Kermode, J. R. & De Vita, A. Molecular dynamics with on-the-fly machine learning of quantum-mechanical forces. Phys. Rev. Lett. 114, 096405 (2015).
Jinnouchi, R., Lahnsteiner, J., Karsai, F., Kresse, G. & Bokdam, M. Phase transitions of hybrid perovskites simulated by machine-learning force fields trained on the fly with bayesian inference. Phys. Rev. Lett. 122, 225701 (2019).
Vandermause, J. et al. On-the-fly active learning of interpretable bayesian force fields for atomistic rare events. NPJ Comput. Mater. 6, 104108 (2020).
Vandermause, J., Xie, Y., Lim, J. S., Owen, C. J. & Kozinsky, B. Active learning of reactive bayesian force fields applied to heterogeneous catalysis dynamics of H/Pt. Nat. Commun. 13, 307 (2022).
Deringer, V. L., Pickard, C. J. & Csányi, G. Data-driven learning of total and local energies in elemental boron. Phys. Rev. Lett. 120, 156001 (2018).
Weininger, D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inform. Comput. Sci. 28, 31–36 (1988).
Halgren, T. A. Merck molecular force field. i. basis, form, scope, parameterization, and performance of mmff94. J. Comput. Chem. 17, 490–519 (1996).
Landrum, G. et al. Rdkit: a software suite for cheminformatics, computational chemistry, and predictive modeling (2013).
Jinnouchi, R., Miwa, K., Karsai, F., Kresse, G. & Asahi, R. On-the-fly active learning of interatomic potentials for large-scale atomistic simulations. J. Phys. Chem. Lett. 11, 6946–6955 (2020).
Tran, K. et al. Methods for comparing uncertainty quantifications for material property predictions. Mach. Learn. Sci. Technol. 1, 025006 (2020).
Bartók, A. P. et al. Machine learning unifies the modeling of materials and molecules. Sci. Adv. 3, e1701816 (2017).
Bernstein, N., Csányi, G. & Deringer, V. L. De novo exploration and self-guided learning of potential-energy surfaces. NPJ Comput. Mater. 5, 99 (2019).
Amsler, M. Minima Hopping Method for Predicting Complex Structures and Chemical Reaction Pathways (eds Andreoni, W. & Yip, S.) Handbook of Materials Modeling 2791–2810 (Springer International Publishing, Cham, 2020).
Kim, S. K., Qiu, Y., Zhang, Y.-J., Hurt, R. & Peterson, A. Nanocomposites of transition-metal carbides on reduced graphite oxide as catalysts for the hydrogen evolution reaction. Appl. Catal. B 235, 36–44 (2018).
Wellendorff, J. et al. Density functionals for surface science: exchange-correlation model development with bayesian error estimation. Phys. Rev. B 85, 235149 (2012).
Schölkopf, B., Smola, A. & Müller, K.-R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 10, 1299–1319 (1998).
Cheng, B. et al. Mapping Materials and Molecules. Acc. Chem. Res. 53, 1981–1991 (2020).
Koskinen, P. & Mäkinen, V. Density-functional tight-binding for beginners. Comput. Mater. Sci. 47, 237–253 (2009).
Wengert, S., Csányi, G., Reuter, K. & Margraf, J. T. Data-efficient machine learning for molecular crystal structure prediction. Chem. Sci. 12, 4536–4546 (2021).
Lan, J. et al. Adsorbml: accelerating adsorption energy calculations with machine learning. Preprint at https://doi.org/10.48550/arXiv.2211.16486 (2022).
Margraf, J. T., Jung, H., Scheurer, C. & Reuter, K. Exploring catalytic reaction networks with machine learning. Nat. Catal. 6, 112–121 (2023).
Deringer, V. L. et al. Gaussian process regression for materials and molecules. Chem. Rev. 121, 10073–10141 (2021).
Bartók, A. P., Kondor, R. & Csányi, G. On representing chemical environments. Phys. Rev. B 87, 184115 (2013).
Giannozzi, P. et al. Advanced capabilities for materials modelling with quantum espresso. J. Phys. Condens. Matter 29, 465901 (2017).
Larsen, A. H. et al. The atomic simulation environment-a python library for working with atoms. J. Phys. Condens. Matter 29, 273002 (2017).
Acknowledgements
H.J. gratefully acknowledges support from the Alexander-von-Humboldt (AvH) Foundation.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
H.J. performed DFT calculations and implemented the workflow in code. H.J., S.S., L.S., and J.T.M. developed the methodology. K.R. and J.T.M. conceived and supervised the project. All authors contributed to analyzing the data and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jung, H., Sauerland, L., Stocker, S. et al. Machine-learning driven global optimization of surface adsorbate geometries. npj Comput Mater 9, 114 (2023). https://doi.org/10.1038/s41524-023-01065-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-023-01065-w
This article is cited by
-
AdsorbML: a leap in efficiency for adsorption energy calculations using generalizable machine learning potentials
npj Computational Materials (2023)