Abstract
Reactive force fields have enabled an atomic level description of a wide range of phenomena, from chemistry at extreme conditions to the operation of electrochemical devices and catalysis. While significant insight and semi-quantitative understanding have been drawn from such work, the accuracy of reactive force fields limits quantitative predictions. We developed a neural network reactive force field (NNRF) for CHNO systems to describe the decomposition and reaction of the high-energy nitramine 1,3,5-trinitroperhydro-1,3,5-triazine (RDX). NNRF was trained using energies and forces of a total of 3100 molecules (11,941 geometries) and 15 condensed matter systems (32,973 geometries) obtained from density functional theory calculations with semi-empirical corrections to dispersion interactions. The training set is generated via a semi-automated iterative procedure that enables refinement of the NNRF until a desired accuracy is attained. The root mean square (RMS) error of NNRF on a testing set of configurations describing the reaction of RDX is one order of magnitude lower than current state of the art potentials.
Similar content being viewed by others
Introduction
The development of general-purpose reactive force fields, most notably ReaxFF, since the early 2000s enabled an atomistic description of complex, emergent phenomena that are beyond the reach of electronic structure calculations. Examples include physical and chemical processes in high-energy-density (HE) materials at extreme conditions1,2,3,4,5, oxidation of hydrocarbons6, and the operation of electrochemical metallization cells7. Molecular dynamics (MD) simulations with reactive force fields are reaching the micron scale and play a central role in multiscale modeling8 connecting first principles, electronic structure, calculations, and the continuum scales. The ability to describe the complex mechanical, chemical, thermal processes at a relatively low computational cost has enabled significant scientific progress and, in some cases, provided semi-quantitative predictions. Reactive potentials9 like ReaxFF6,10, COMB11,12, and REBO13,14 are designed using chemical and physical intuition about atomic bonding, but unavoidable limitations of the underlying functional forms limit their accuracy. For example, even after nearly two decades of development15, and despite its numerous contributions to the field, the state-of-art ReaxFFs for HE materials have well-documented deficiencies5. Capitalizing on the recent progress in the use of data science tools in the physical sciences, a new class of data-driven interatomic potentials has emerged16,17. These potentials use physics-agnostic models to relate the local atomic environment to energies and forces and have been shown to provide accurate descriptions for simple molecules18,19, metals20,21, alloys22, and other materials23,24,25 over a wide range of atomic environments26,27. In this paper we present the first data-driven force field using physics-agnostic models capable of describing four-element systems and complex, multi-step, chemical reactions of the nitramine RDX (cyclic [CH2-NNO2-]3).
Reactive MD simulations of HE materials are critical to develop a predictive understanding of these materials. This is particularly true regarding their chemical initiation under thermal and mechanical insults, where thermochemical models and electronic structure calculations are insufficient. The initiation of a detonation involves complex, coupled, processes involving mechanical, thermal, and chemical processes. Reactive MD simulations can provide a description of these unit processes making few, well-understood, approximations. The largest sources of uncertainties in such calculations are the interatomic potentials used and the use of classical (as opposed to quantum) dynamics28. To address the first of these limitations, reactive force fields have been continuously improved and quantum-mechanics based methods, like total energy tight-binding potentials, have been developed29,30,31,32. Despite progress, the uncertainties in these methods remain poorly quantified and so is their effect on the quantities of interest (that are invariably complex emergent phenomena). In this paper we use an extensive training set of electronic structure calculations to train a reactive force field that combines descriptors of local atomic environments using two and three-body symmetry functions with deep neural networks. Our neural network reactive force field (NNRF) is capable of describing the complex chemistry of RDX under a wide range of temperatures, resulting in product molecules and overall activation energy for decomposition in agreement with experiments. Importantly, the root mean squared (RMS) errors with respect to electronic structure calculations in atomic forces during the decomposition and reaction of RDX from the crystal to final products is one order of magnitude smaller than the current state of the art reactive force fields.
Results and discussion
Iterative training procedure of NNRF
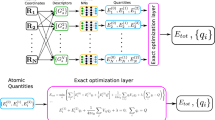
The selection of training, validation, and testing sets is important for the development of any force field, but it is particularly critical in the case of data-driven force field where the models are agnostic to the underlying physics. Thus, we designed an iterative approach, schematically represented in Fig. 1, to generate progressively more relevant configurations and an automated procedure that can be easily generalized to other systems.
Two different feedback loops enhance the training set: automatic generation of configurations during decomposition and reaction (left red box) and extraction of chemical species and intermediates (right blue box). The list of entries and their numbers are all tabulated in Supplementary Table 4.
We start from an isothermal, isochoric ReaxFF MD simulation of the decomposition of crystalline RDX at the experimental density and at 2500 K with a total duration of 400 ps (using a simulations cell with 8 molecules and periodic boundary conditions). This simulation samples the original crystal, the initiation of chemistry, and the formation of intermediates as well as final products. We extracted 4000 frames (one every 0.1 ps) from the ReaxFF trajectory and performed single point density functional theory (DFT) calculations using Perdew–Burke–Ernzerhof (PBE) exchange-correlation functional33 and the empirical D2 correction for London dispersion interactions34 to obtain energies and atomic forces to be used as training data. This level of theory will be denoted DFT PBE-D2. In addition, the initial training set includes bond dissociation of all possible di-atomic molecules and snapshots from a T = 300 K MD simulation of isolated molecules (including RDX, intermediates, and products) to provide additional information about the key species involved in the decomposition5. Intermediate and product species were obtained from prior theoretical studies of thermal decomposition of RDX35,36,37 and other nitramine3. In addition, we included the equations of state (with ~10% volume compression and expansion) for RDX, graphite, and diamond crystals to describe the reference states and pressure effects.
All energies used in the training are referenced to O2, N2, and H2 molecules and graphite. The data were randomly divided into a training set (with 90% of the data) used to optimize the neural network and a validation set (10%) used to assess convergence and avoid overfitting during the training. The accuracy of each force field generation was monitored using independent testing data, obtained from five sets of data which include ReaxFF MD simulation of the decomposition of RDX at 2000 K with a total duration of 400 ps, and MD simulations using generation 1.5 (Gen1.5) and generation 1.6 (Gen1.6) NNRF (see below) under two conditions (2000 K for 20 ps and 2500 K for 200 ps). These test sets were never used in training.
With the initial data discussed above, including 10,462 structures and 2,807,361 force components in total, we obtained an initial NNRF (Gen 1.1). NNRF generations will be denoted with two integers, the first marking significant additions to the training set and the second one the iteration number. Following NNRF Gen1.1 we marched along the above process to increase the training data both in size and relevance and generated a family of increasingly accurate models. Parity plots comparing NNRF and DFT energies and forces and histograms with deviations over the test set are shown in Fig. 2 for Gens 1.1, 1.6, and 1.9 NNRF. The evolution of RMS errors in energy and force over the training, validation, and testing sets vs. generation number is shown in Fig. 3 and tabulated in Supplementary Table 3. Trained with a ReaxFF-2014 thermal decomposition trajectory, monomer data, and equations of state, NNRF Gen1.1 is clearly biased towards the configurations explored by ReaxFF. Since it is certain that the ReaxFF dynamics will explore unlikely configurations and might miss important decomposition paths, reproducing the training data do not guarantee an accurate force field. This is the motivation behind the addition of an iterative loop to our training procedure (left in Fig. 1), where each generation NNRF is used to simulate the decomposition of RDX and the resulting configurations are added to the training of subsequent generations. This is also the reason why our testing set contains reactive simulations using Gen1.5 and Gen1.6 NNRF, in addition to ReaxFF, as they explore different decomposition paths. In addition to reactive MD simulations (left loop in Fig. 1), the training set is continuously enhanced with intermediate and final molecules identified during these reactive simulations to improve the description of individual chemical reactions (right loop). These configurations are obtained using the following steps. (i) Extract molecules from the NNRF MD trajectories (every 1 ps) based on ReaxFF bond orders, (ii) identify molecules not present in the training set based on the connectivity from bond order analysis, (iii) run DFT MD simulations at 500 K for 10 ps for each molecule, (iv) add the energy and forces of the initial and final structure of DFT MD simulation. Supplementary Table 4 provides a detailed description of the data used in the training of each generation NNRF and Supplementary Fig. 1 represents the progression in the number of data points with generation. This approach can be thought of as active learning in sampling the training space38.
The energy RMS error (ERMSE) in (a) and the force RMS error (FRMSE) in (b) from Gen1.1 to Gen1.9 NNRF for three datasets (Training set, Validation set, and Testing set). The ERMSE and FRMSE for a single iteration (Gen1.7 NNRF) can be found in Supplementary Fig. 5.
The results in Figs. 2 and 3 (and Supplementary Figs. 2–4) show that the accuracy of NNRF improves from Gen1.1 to Gen1.9, validating our iterative approach. As expected, NNRF Gen1.1 is accurate only for the portion of the test set generated using ReaxFF-2014 (2000 K/400 ps data set). Figure 3 shows the increase in accuracy as a function of generation and convergence in Gen 1.7–1.9. It might be surprising that the NNRFs describe the test set more accurately than the training and validation sets in terms of energies. This is because, early NNRFs generate trajectories that explore unlikely, distorted clusters; these are included in the training and validation sets but not in the testing set. We consider the Gen1.9 NNRF to be appropriately converged and performed a series of validation tests. Before comparing NNRF with four widely used ReaxFF parameterizations for HE materials in the verification and validation section, we assess its accuracy in the description of important quantities that govern the decomposition and reaction of RDX.
Accuracy of Gen 1.9 NNRF
The formation energies of intermediate and product species observed during the thermal decomposition of nitramines are critical values for an accurate description of chemistry. Figure 4 compares the formation energy of key molecules obtained from NNRF Gen1.9, various parameterizations of ReaxFF for HE materials, and more accurate quantum chemistry calculations and experimental results. The RMS error in formation energy with respect to PBE-D2, see inset of Fig. 4, indicates that NNRF provides an accurate description of these key species. This demonstrates that NNRF learns the formation energies of molecules even though these quantities represent a small fraction of the training set. The largest deviations of the formation energy of Gen1.9 NNRF was observed only for HNO3 and NO2, because these molecules were sampled less frequently in the MD simulations using NNRF. It should be noted that the PBE-D2 data deviates from the experimental and higher-accuracy quantum chemistry calculation39 results, see for example HON and O2, due to intrinsic limitations of DFT; the formation energies of NNRF are close to DFT data. Supplementary Table 5 shows the RMS error of the various methods with respect to experiments. We find that the RMS errors with respect to experiments for NNRF are slightly larger than ReaxFF-iw, but comparable to DFT PBE-D2 and smaller than ReaxFFs-2014, ReaxFF-2018, and ReaxFF-lg. (Supplementary Table 5).
We note that ReaxFFs provides a remarkably accurate description of the formation energies of intermediate and final products. We believe this success is behind its popularity to investigate HE materials and their contribution to the field. However, the NNRF not only results in a more accurate description of these gas-phase formation energies compared to ReaxFF-2014, ReaxFF-2018, and ReaxFF-lg but, more importantly, the description of energetics and forces along the MD simulations (even along ReaxFF trajectories) are one order of magnitude better, as shown in Supplementary Fig. 6. While ReaxFF has not been parameterized to match energies and forces during decomposition, the comparison is important as it quantifies the accuracy of the description throughout the process and conditions of interest.
Figure 5 shows the dissociation curves of ten dimers predicted by the Gen1.9 NNRF and DFT. All dissociation curves were calculated using spin-polarized DFT for different spin states and the lowest-energy spin configurations were selected for each bond distance. The NNRF accuracy for all these bond dissociation curves is quite striking; visual inspection clearly shows significant improvements with respect to ReaxFF, see, for example, Fig. 3 in ref. 40. We note that NNRF learns the difference between single, double, and triple C–C bonds, see Fig. 6.
One limitation of the current force field is its inability to capture the high barrier associated with torsions around a double bond, see Supplementary Fig. 7. We note that we do not include four body terms that could be important to describe torsions. Torsional potentials for various chemical environments could be addressed via such four-body terms or combinations of two and three-body terms, see ref. 41, just like van der Waals repulsion controls the torsional potential around a single C–C bond. Future work should explore these options.
As a final assessment of the ability of NNRF Gen1.9 to describe the chemistry of RDX, we investigated four unimolecular reaction paths proposed from ab initio simulations35,36. Supplementary Fig. 8 shows the energies of transition states and intermediates the NO2 dissociation, HONO, NO2 insertion, and concerted decomposition paths. Even though these decomposition paths were not used during training, NNRF Gen 1.9 captures the main differences in these paths. The deviations between NNRF and high-accuracy quantum chemistry calculations, using B3LYP hybrid functionals42,43,44,45, are similar to those observed in DFT-PBE. Additional discussion is included in the Supplementary Information (SI).
Verification and validation
We now perform verification and validation tests on Gen1.9 NNRF, designed to assess their description of the decomposition of RDX under isothermal (NVT ensemble) and adiabatic (NVE) conditions. The results are then compared to experiments and ReaxFF. The calculation of vibration density of state and crystalline structures were conducted using the 3 × 3 × 3 supercell of RDX unit-cell structure obtained from the Cambridge Crystallographic Data Centre46 with the experimental density of 1.86 g cm−3. The system containing 4536 atoms or 216 RDX molecules were minimized for energy and forces and thermalized under constant volume, and constant temperature (NVT ensemble) at 300 K for 20 ps.
In order to verify the implementation of NNRF in LAMMPS, we quantified the energy conservation of Gen1.9 NNRF during an adiabatic MD simulation (NVE ensemble). We simulated five different initial temperatures with integration timesteps (in parenthesis) appropriate to each value: T0 = 1500 K (0.1 fs), 1750 K (0.05 fs), 2000 K (0.05 fs), 2250 K (0.025 fs), and 2500 K (0.025 fs). These samples were initialized by setting the velocities to twice the target temperature so that after thermal equilibration they would reach approximately the desired value. We tracked the reaction and decomposition until the system temperature achieved steady state and measured the total energy drifted due to the numerical integration. Interestingly, Gen1.9 NNRF conserves energy significantly better than the ReaxFF parameterizations for the same initial temperature and timestep, see Supplementary Fig. 10. For example, the 1500 K simulations with a timestep of 0.1 fs Gen 1.9 NNRF exhibited a total energy drift of −0.4 kcal mol−1 in 400 ps; this can be compared to 11.7 kcal mol−1 using ReaxFF-2014, 18.1 kcal mol−1 using ReaxFF-2018, 103.3 kcal mol−1 using ReaxFF-lg, and 94.3 kcal mol−1 using ReaxFF-iw. Using the most conservative timestep of 0.025 fs in our T = 2500 K simulations, Gen1.9 NNRF had a total energy drift of −0.3 kcal mol−1 in ~100 ps, compared to ReaxFF-2014 with 9.7 kcal mol−1, ReaxFF-2018 with 6.4 kcal mol−1, ReaxFF-lg with 37.2 kcal mol−1, and ReaxFF-iw with 32.9 kcal mol−1. We note that while NNRF enables longer integration timesteps as compared to ReaxFF, energy and force calculations take, approximately, a factor of four times longer; scaling with system size and parallel performance are similar between ReaxFF and NNRF.
The crystal structure and the equilibrium volume of α-RDX is also described accurately with NNRF, as shown in Fig. 7a and b. Energy-volume curves were used to predict both the equilibrium volume and the bulk modulus of the RDX crystal. The bulk modulus predicted by Gen1.9 NNRF (13.7 GPa), Supplementary Table 7, is in good agreement with experimental results (11.8–13.9 GPa)47,48,49. The predicted equilibrium was very close to the DFT PBE-D2 result and slightly higher than the experimental equilibrium volume. This can be attributed to the discrepancy between PBE-D2 and experiments. We note that Gen1.9 provides an inaccurate description when the crystal is expanded by more than 20% of volume. Generation 2 NNRFs address this shortcoming as will be discussed in the Transferability to other nitramines. For completeness, Fig. 7 also shows the equation of state of Gen2.3 NNRF. Besides the lattice parameters, Fig. 7b shows excellent agreement between the NNRF predicted X-ray diffraction (XRD) for α-RDX crystal and experiments. We note that ReaxFF-lg is also in good agreement with experiments.
a Equation of state for RDX crystal with experimental volume (Exp vol indicated as a vertical dashed line, corresponding to ρ = 1.86 g cm−3) and b The X-ray diffraction peaks of RDX crystal from Experiment and available force fields (Gen1.9 NNRF, ReaxFFs). aExperimental RDX crystal structure from ref. 78.
We computed the vibrational density of states and infrared spectrum of RDX from 20 ps-long MD trajectories at 300 K from the Fourier transformation of the velocity50 and dipole51 autocorrelation functions, respectively. Figure 8 compares both quantities between the four parameterizations of ReaxFF and Gen1.9 NNRF and experimental results from refs. 52 and 53. Experiments show major infrared active peaks around 1200, 1600, and 3100 cm−1, corresponding to N-NO2 symmetric, N-NO2 antisymmetric, and C–H antisymmetric vibrational modes, respectively52,53,54. Interestingly, NNRF matches all three features more closely than ReaxFF. For example, all four versions of ReaxFF overestimate the frequencies of the hydrogenic modes at 3100 cm−1.
We now focus on the decomposition and reaction of RDX. Figure 9 tracks key intermediates and final products during an isothermal decomposition simulation at 2000 and 2500 K. Gen1.9 NNRF at 2500 K predicts N2 to be the most abundant final product, followed by similar amounts of H2O and CO2, and trace amounts of HNCO, NH3, and NH4; at 2000 K the populations appear to be evolving towards similar proportions. This matches bomb calorimetry experimental observations55 where products of detonation were approximately broken down into 37% N2, 31% H2O, 18% CO2, and 14% CO. The lack on CO is our predictions can be attributed to PBE-D2 underestimating its stability, see Fig. 4. ReaxFF-2014 and ReaxFF-2018 predicted N2 as the dominant product followed by H2O and CO2. On the other hand, ReaxFF-lg and ReaxFF-iw predicted comparable amounts of H2O and N2 with less CO2. Consistent with NNRF ReaxFF does not predict appreciable amounts of CO which was observed experimentally55. It is worth mentioning that NNRF predicts NO2 and NO as the key intermediates. Using B3LYP flavor of DFT, Chakraborty et al.35 calculated, in the gas phase, that the first bond-breaking event should be N-NO2, as compared with HONO elimination and concerted ring break, because it has the lowest dissociation energy. NNRF is consistent with this picture. As an aside, the amount of HONO and H2CNHNO2 was negligible so both species are not shown in Fig. 9. The presence of NO2 is consistent with the ReaxFF-lg and ReaxFF-iw results. However, the ReaxFF-2014 and ReaxFF-2018 versions underestimate the NO bond strength and predict the formation of O radicals4.
Finally, we assess the overall kinetics of the decomposition and reaction processes. In order to obtain an overall decomposition timescale, we defined the half-exothermicity time (t1/2) as that required for the potential energy to reach the mid-point between the initial and final values. Figure 10 shows t1/2 the as a function of inverse temperature. Consistent with prior results, the overall kinetics of reaction follows, approximately, Arrhenius behavior for the range of temperatures we can simulate4, from which we can obtain effective activation energies. This is an admittedly simplistic description of the chemical kinetics for a material like RDX, which requires multiple steps to capture their complex chemistry56,57, but useful to assess overall behavior. Figure 10 shows that NNRF kinetics are slower than the ReaxFF-2014 and 2018. Gen1.9 NNRF results in the largest activation energy at 25.7 kcal mol−1, as compared to ReaxFF-2014 with 23.5 kcal mol−1, ReaxFF-2018 with 18.7 kcal mol−1, ReaxFF-lg with 11.9 kcal mol−1, and ReaxFF-iw yielding 12.0 kcal mol−1. The NNRF Gen1.9 value is close to the experimental value by Rogers and Smith58 using Kissinger’s method59 over multiple differential thermal analysis experiments, and is on the lower end of the 25–50 kcal mol−1 range covering many experiments compiled by Brill et al.60. An important assumption in applying Kissinger’s method is that the decomposition reaction of the sample must have a reaction order close to 1. Rogers and Smith showed that Kissinger’s equation will underapproximate the activation energy when the reaction order deviates away from unity, or when the decomposition scheme is assumed to be more complex. The assumption underlying Kissinger’s method is similar to our one-step approximation. Activation energies and pre-factors are provided in Supplementary Table 8 in the SI; we highlight again the approximate nature of these models.
Transferability to other nitramines (HMX, NM, PETN): Gen2.X
The transferability of a force field is critical for its widespread use and ReaxFF has excelled in this aspect. We conducted thermal decomposition dynamic simulations (2500 K/400 ps) using Gen1.9 NNRF for NM, HMX, PETN crystal systems. Single point DFT calculations of each dynamic trajectories were conducted and the corresponding parity plots for energy and forces are shown in Supplementary Fig. 11a and b. Not surprisingly, for a force field trained only on RDX, we found relatively large deviations in energies and forces. We note that these deviations are comparable to those of ReaxFF.
To improve the ability of the NNRF formulation to describe equations of state and its description of other HE materials, we added the following data to the training set: (i) NPT ensemble dynamics of RDX and PETN (pentaerythritol tetranitrate) crystals, (ii) PETN MD simulations at various temperatures (300 K/20 ps, 1000 K/20 ps, 1500 K/20 ps, 2000 K/20 ps, 2500 K/20 ps, 2500 K/200 ps, 3000 K/20 ps), and (iii) monomers of nitromethane (NM) and HMX (1,3,5,7-Tetranitro-1,3,5,7-tetrazoctane). We name this series Gen 2.X and with three iterations, Gen2.3 NNRF provides an improved description of the equation of state of all HEs tested for volume expansion beyond 130%, shown in Fig. 7a, and transferability for NM, HMX, PETN crystal systems. As shown in Supplementary Fig. 11c and d, NNRF Gen 2.3 provides an accurate description of the decomposition of these HEs.
In summary, we developed a neural network reactive force field (NNRF) for CHNO systems based on large sets of electronic structure data (we used DFT at the PBE level with D2 correction). The NNRF utilizes a high dimensional neural network to capture the short-range interatomic interactions (<5 Å) with the radial and angular descriptors and it employs the van der Waals and Coulomb interaction parameterization of ReaxFFs for the long-range interatomic interactions (<10 Å). We developed a semi-automated procedure to generate and refine training sets that uses the best current model to efficiently generate new configurations. The training set consists of a relatively small system, <200 atoms, which can be performed easily at the level of theory chosen since only single-point energy calculations are required.
Prediction of dissociation curves, formation energies of monomers, energies, and forces of test set clearly indicates that the NNRF learned the underlying bonding. We further tested Gen1.9 NNRF with larger-scale dynamic simulations (4536 atoms or 216 RDX molecules) predicting the crystal structure, bulk modulus, vibrational spectra, chemical reactions, and apparent activation energy of the process. More importantly, the high-temperature decomposition of RDX shows good agreement with experiments in terms of final products and overall activation energy. In addition, NNRF was easily extendable to other HE materials including nitromethane (CH3NO2), HMX (1,3,5,7-Tetranitro-1,3,5,7-tetrazoctane, C4H8N8O8), and PETN (pentaerythritol tetranitrate, C5H8N4O12). These results indicate that the iterative training could enable the study complex chemistry with DFT-level accuracy.
Methods
Neural network force field
Machine learning potentials relate a set of descriptors of local atomic environments to atomic energies and forces via a physics-agnostic mapping. This mapping can be performed using a neural network61, linear regression17, polynomials25, Gaussian process regression16, or using other regression techniques. Physics is embedded in these approaches via the local environment descriptors that should be invariant to rigid translations and rotations and have the flexibility to discriminate subtle differences in environments17,61. In this work, we adopted neural network potentials based on symmetry functions as descriptors, which have shown promise in describing multicomponent systems24,62. We hypothesize that this description is flexible enough to capture the chemistry of RDX.
We use atom-centered, weighted-Gaussian, symmetry functions developed by Gastegger et al.63 to build the input layer of NNRF. Two types of Gaussian symmetry functions, radial and angular in Eqs. (1) and (2), are used to extract unique and invariant fingerprints from molecular geometries. The equations for the radial \((wG_i^{\mathrm{rad}})\) and angular \((wG_i^{\mathrm{ang}})\) symmetry functions for each atom (i) are given by:
In Eqs. (1) and (2), Zj is the atomic weight included to differentiate different elements. rs, η, λ, and ξ are hyperparameters to be specified, and fc(r) is a cutoff function, which decays to zero smoothly beyond the cutoff radius of 5 Å, see the SI. We constructed 18 radial symmetry functions and 24 angular symmetry functions with a set of hyperparameters described in the SI.
With the descriptors at hand, independent neural networks are used to describe the energy and force for each element. Each neural network consists of 42 nodes as the input layer, two hidden layers with 50 nodes each, and one output node (the atomic energy); the atomic forces are obtained as the negative gradient of the energy. The hyperbolic tangent activation functions are used with the given neural network architecture (see the SI).
While such a description can be expected to describe covalent interactions, its short-range limits its applicability to describe non-bonded interactions, including van der Waals and Coulomb. Fortunately, we have rather accurate descriptions for such terms. Thus, the total energy of NNRF is the sum of the neural network described above and pre-parameterized van der Waals and electrostatics taken from the ReaxFF force field64,65. van der Waals interactions are described using shielded Morse potentials and electrostatics using environment-dependent charges with the electronegativity equalization method (EEM)66 and tapered electrostatics energy67,68. These choices are motivated by the availability of these terms in CHNO ReaxFF and were taken from the ReaxFF-2014 parametrization65. Thus, the neural network is trained to describe the difference between DFT results and the reference potential.
Density functional theory
The training data were obtained from DFT calculations performed with the Vienna ab initio simulation package (VASP, version 5.4.4)69,70,71,72,73, using PBE exchange and correlation functional33 and the semi-empirical D2 method of Grimme34 to improve the description of dispersion forces. The convergence criteria for the wave function optimization was set to a total energy difference between subsequent steps of 10−5 eV. The kinetic energy cutoff is set to 500 eV. Given the relatively large simulation cells integrals in reciprocal space using only the Γ point. The interactions between ions and electrons were described by the projector augmented wave (PAW)74 method with the 1s of H and 2s2p of C, N and O treated as valence electrons.
Data availability
DFT training data and all generations of NNRF are available in the repository at https://github.rcac.purdue.edu/StrachanGroup/nnrf_nitramines.
Code availability
In order to facilitate the assessment and adoption of NNRF we created an open, online, tool in nanoHUB75 (www.nanohub.org/tools/nnrf). Using online Jupyter notebooks, users can assess the accuracy of the various generation force fields against the various training sets. A separate notebook can be used to perform MD simulations on various HE materials using the various NNRFs. The tool page also points to the repository containing training data and the NNRF development code. We plan to continuously update the tool as we improve NNRF and new generations become available. NNRF parameters for all generations can be downloaded to perform offline simulations or used for online simulations in nanoHUB.
References
Shan, T.-R. & Thompson, A. P. Shock-induced hotspot formation and chemical reaction initiation in PETN containing a spherical void. J. Phys. Conf. Ser. 500, 172009 (2014).
Wood, M. A., Cherukara, M. J., Kober, E. M. & Strachan, A. Ultrafast chemistry under nonequilibrium conditions and the shock to deflagration transition at the nanoscale. J. Phys. Chem. C 119, 22008–22015 (2015).
Islam, M. M. & Strachan, A. Decomposition and reaction of polyvinyl nitrate under shock and thermal loading: a ReaxFF reactive molecular dynamics study. J. Phys. Chem. C 121, 22452–22464 (2017).
Sakano, M., Hamilton, B., Islam, M. M. & Strachan, A. Role of molecular disorder on the reactivity of RDX. J. Phys. Chem. C 122, 27032–27043 (2018).
Islam, M. M. & Strachan, A. Reactive molecular dynamics simulations to investigate the shock response of liquid nitromethane. J. Phys. Chem. C 123, 2613–2626 (2019).
Chenoweth, K., van Duin, A. C. T. & Goddard, W. A. ReaxFF reactive force field for molecular dynamics simulations of hydrocarbon oxidation. J. Phys. Chem. A 112, 1040–1053 (2008).
Onofrio, N., Guzman, D. & Strachan, A. Atomic origin of ultrafast resistance switching in nanoscale electrometallization cells. Nat. Mater. 14, 440–446 (2015).
van der Giessen, E. et al. Roadmap on multiscale materials modeling. Model. Simul. Mater. Sci. Eng. 28, 043001 (2020).
Liang, T. et al. Reactive potentials for advanced atomistic simulations. Annu. Rev. Mater. Res. 43, 109–129 (2013).
Senftle, T. P. et al. The ReaxFF reactive force-field: development, applications and future directions. Npj Comput. Mater. 2, 1–14 (2016).
Yu, J., Sinnott, S. B. & Phillpot, S. R. Charge optimized many-body potential for the Si/SiO2 system. Phys. Rev. B 75, 085311 (2007).
Shan, T.-R. et al. Second-generation charge-optimized many-body potential for Si/SiO2 and amorphous silica. Phys. Rev. B 82, 235302 (2010).
Brenner, D. W. et al. A second-generation reactive empirical bond order (REBO) potential energy expression for hydrocarbons. J. Phys. Condens. Matter 14, 783 (2002).
O’Connor, T. C., Andzelm, J. & Robbins, M. O. AIREBO-M: a reactive model for hydrocarbons at extreme pressures. J. Chem. Phys. 142, 024903 (2015).
Strachan, A., van Duin, A. C., Chakraborty, D., Dasgupta, S. & Goddard, W. A. III Shock waves in high-energy materials: the initial chemical events in nitramine RDX. Phys. Rev. Lett. 91, 098301 (2003).
Bartók, A. P., Payne, M. C., Kondor, R. & Csányi, G. Gaussian approximation potentials: the accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010).
Thompson, A. P., Swiler, L. P., Trott, C. R., Foiles, S. M. & Tucker, G. J. Spectral neighbor analysis method for automated generation of quantum-accurate interatomic potentials. J. Comput. Phys. 285, 316–330 (2015).
Schütt, K. T., Sauceda, H. E., Kindermans, P.-J., Tkatchenko, A. & Müller, K.-R. SchNet – A deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Morawietz, T., Singraber, A., Dellago, C. & Behler, J. How van der Waals interactions determine the unique properties of water. Proc. Natl Acad. Sci. 113, 8368–8373 (2016).
Zong, H., Pilania, G., Ding, X., Ackland, G. J. & Lookman, T. Developing an interatomic potential for martensitic phase transformations in zirconium by machine learning. Npj Comput. Mater. 4, 1–8 (2018).
Dragoni, D., Daff, T. D., Csányi, G. & Marzari, N. Achieving DFT accuracy with a machine-learning interatomic potential: thermomechanics and defects in bcc ferromagnetic iron. Phys. Rev. Mater. 2, 013808 (2018).
Zhang, L. et al. End-to-end Symmetry Preserving Inter-atomic Potential Energy Model for Finite and Extended Systems. in Advances in Neural Information Processing Systems 31 (eds. Bengio, S. et al.) 4436–4446 (Curran Associates, Inc., 2018).
Sosso, G. C., Miceli, G., Caravati, S., Behler, J. & Bernasconi, M. Neural network interatomic potential for the phase change material GeTe. Phys. Rev. B 85, 174103 (2012).
Artrith, N., Morawietz, T. & Behler, J. High-dimensional neural-network potentials for multicomponent systems: applications to zinc oxide. Phys. Rev. B 83, 153101 (2011).
Lindsey, R. K., Fried, L. E. & Goldman, N. ChIMES: a force matched potential with explicit three-body interactions for molten carbon. J. Chem. Theory Comput. 13, 6222–6229 (2017).
Eshet, H., Khaliullin, R. Z., Kühne, T. D., Behler, J. & Parrinello, M. Ab initio quality neural-network potential for sodium. Phys. Rev. B 81, 184107 (2010).
Behler, J., Martoňák, R., Donadio, D. & Parrinello, M. Metadynamics simulations of the high-pressure phases of silicon employing a high-dimensional neural network potential. Phys. Rev. Lett. 100, 185501 (2008).
Hamilton, B. W., Kroonblawd, M. P., Islam, M. M. & Strachan, A. Sensitivity of the Shock Initiation Threshold of 1,3,5-Triamino-2,4,6-trinitrobenzene (TATB) to Nuclear Quantum Effects. J. Phys. Chem. C 123, 21969–21981 (2019).
Niklasson, A. M. & Cawkwell, M. J. Fast method for quantum mechanical molecular dynamics. Phys. Rev. B 86, 174308 (2012).
Wang, C. et al. Environment-dependent tight-binding potential model. MRS Online Proc. Libr. Arch. 491, 211 (1997).
Elstner, M. et al. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 58, 7260 (1998).
Frauenheim, T. et al. A self‐consistent charge density‐functional based tight‐binding method for predictive materials simulations in physics, chemistry and biology. Phys. Status Solidi B 217, 41–62 (2000).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865 (1996).
Grimme, S. Semiempirical GGA‐type density functional constructed with a long‐range dispersion correction. J. Comput. Chem. 27, 1787–1799 (2006).
Chakraborty, D., Muller, R. P., Dasgupta, S. & Goddard, W. A. The Mechanism for Unimolecular Decomposition of RDX (1,3,5-Trinitro-1,3,5-triazine), an ab Initio Study. J. Phys. Chem. A 104, 2261–2272 (2000).
Strachan, A., Kober, E. M., van Duin, A. C., Oxgaard, J. & Goddard, W. A. III Thermal decomposition of RDX from reactive molecular dynamics. J. Chem. Phys. 122, 054502 (2005).
Schweigert, I. V. Ab initio molecular dynamics of high-temperature unimolecular dissociation of gas-phase RDX and its dissociation products. J. Phys. Chem. A 119, 2747–2759 (2015).
Smith, J. S., Nebgen, B., Lubbers, N., Isayev, O. & Roitberg, A. E. Less is more: Sampling chemical space with active learning. J. Chem. Phys. 148, 241733 (2018).
Johnson III, R. D. NIST 101. Computational chemistry comparison and benchmark database (1999).
Ashraf, C. & Van Duin, A. C. Extension of the ReaxFF combustion force field toward syngas combustion and initial oxidation kinetics. J. Phys. Chem. A 121, 1051–1068 (2017).
Wen, M. & Tadmor, E. B. Hybrid neural network potential for multilayer graphene. Phys. Rev. B 100, 195419 (2019).
Becke, A. D. Density‐functional thermochemistry. I. The effect of the exchange‐only gradient correction. J. Chem. Phys. 96, 2155–2160 (1992).
Becke, A. D. Density‐functional thermochemistry. II. The effect of the Perdew–Wang generalized‐gradient correlation correction. J. Chem. Phys. 97, 9173–9177 (1992).
Becke, A. D. Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Lee, C., Yang, W. & Parr, R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785 (1988).
Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. The Cambridge Structural Database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 72, 171–179 (2016).
Olinger, B., Roof, B. & Cady, H. Symposium international sur le comportement des milieux denses sous hautes pressions dynamiques. Paris Fr. 3 (1978).
Schwarz, R., Hooks, D., Dick, J., Archuleta, J. & Martinez, A. Resonant ultrasound spectroscopy measurement of the elastic constants of cyclotrimethylene trinitramine. J. Appl. Phys. 98, 056106 (2005).
Yoo, C.-S., Cynn, H., Howard, M. & Holmes, N. Equation of State of Unreacted High Explosives at High Pressures. 951–957 (Office of Naval Research, 1999).
Boulard, B., Kieffer, J., Phifer, C. C. & Angell, C. A. Vibrational spectra in fluoride crystals and glasses at normal and high pressures by computer simulation. J. Non-Cryst. Solids 140, 350–358 (1992).
Berens, P. H., Mackay, D. H. J., White, G. M. & Wilson, K. R. Thermodynamics and quantum corrections from molecular dynamics for liquid water. J. Chem. Phys. 79, 2375–2389 (1983).
Infante-Castillo, R., Pacheco-Londoño, L. & Hernández-Rivera, S. P. Vibrational spectra and structure of RDX and its 13C- and 15N-labeled derivatives: a theoretical and experimental study. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 76, 137–141 (2010).
Elbasuney, S. & El-Sherif, A. F. Complete spectroscopic picture of concealed explosives: laser induced Raman versus infrared. TrAC Trends Anal. Chem. 85, 34–41 (2016).
Li, G. et al. Synthesis and properties of RDX/GAP nano-composite energetic materials. Colloid Polym. Sci. 293, 2269–2279 (2015).
Ornellas, D. L. Calorimetric determinations of the heat and products of detonation for explosives: October 1961 to April 1982. https://apps.dtic.mil/docs/citations/ADA409329 (1982).
Tarver, C. M., Chidester, S. K. & Nichols, A. L. Critical conditions for impact-and shock-induced hot spots in solid explosives. J. Phys. Chem. 100, 5794–5799 (1996).
Sakano, M. N. et al. Unsupervised Learning-Based Multiscale Model of Thermochemistry in 1, 3, 5-Trinitro-1, 3, 5-triazinane (RDX). J. Phys. Chem. A 124, 9141–9155 (2020).
Rogers, R. N. & Smith, L. C. Application of scanning calorimetry to the study of chemical kinetics. Thermochim. Acta 1, 1–9 (1970).
Kissinger, H. E. Reaction kinetics in differential thermal analysis. Anal. Chem. 29, 1702–1706 (1957).
Brill, T. B., Gongwer, P. E. & Williams, G. K. Thermal decomposition of energetic materials. 66. Kinetic compensation effects in HMX, RDX, and NTO. J. Phys. Chem. 98, 12242–12247 (1994).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Artrith, N. & Urban, A. An implementation of artificial neural-network potentials for atomistic materials simulations: performance for TiO2. Comput. Mater. Sci. 114, 135–150 (2016).
Gastegger, M., Schwiedrzik, L., Bittermann, M., Berzsenyi, F. & Marquetand, P. wACSF—Weighted atom-centered symmetry functions as descriptors in machine learning potentials. J. Chem. Phys. 148, 241709 (2018).
van Duin, A. C. T., Dasgupta, S., Lorant, F. & Goddard, W. A. ReaxFF: a reactive force field for hydrocarbons. J. Phys. Chem. A 105, 9396–9409 (2001).
Wood, M. A., van Duin, A. C. T. & Strachan, A. Coupled thermal and electromagnetic induced decomposition in the molecular explosive αHMX; a reactive molecular dynamics study. J. Phys. Chem. A 118, 885–895 (2014).
Islam, M. M., Kolesov, G., Verstraelen, T., Kaxiras, E. & van Duin, A. C. eReaxFF: a pseudoclassical treatment of explicit electrons within reactive force field simulations. J. Chem. Theory Comput. 12, 3463–3472 (2016).
Mortier, W. J., Ghosh, S. K. & Shankar, S. Electronegativity-equalization method for the calculation of atomic charges in molecules. J. Am. Chem. Soc. 108, 4315–4320 (1986).
Janssens, G. O., Baekelandt, B. G., Toufar, H., Mortier, W. J. & Schoonheydt, R. A. Comparison of cluster and infinite crystal calculations on zeolites with the electronegativity equalization method (EEM). J. Phys. Chem. 99, 3251–3258 (1995).
Kresse, G. & Hafner, J. Ab initio molecular-dynamics simulation of the liquid-metal–amorphous-semiconductor transition in germanium. Phys. Rev. B 49, 14251 (1994).
Kresse, G. & Hafner, J. Ab initio molecular dynamics for liquid metals. Phys. Rev. B 47, 558 (1993).
Kresse, G. & Furthmüller, J. Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys. Rev. B 54, 11169 (1996).
Kresse, G. & Furthmüller, J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput. Mater. Sci. 6, 15–50 (1996).
Kresse, G. & Joubert, D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys. Rev. B 59, 1758 (1999).
Blöchl, P. E. Projector augmented-wave method. Phys. Rev. B 50, 17953 (1994).
Yoo, P. et al. Tool for Neural network reactive force field for CHNO systems. https://nanohub.org/tools/nnrf (2020).
Burgess, D. R., Jr. In NIST Chemistry WebBook, NIST Standard Reference Database Number 69 (eds. Linstrom, P. J. & Mallard, W. G.). https://doi.org/10.18434/T4D303 (2018).
Ruscic, B. et al. Introduction to active thermochemical tables: Several “key” enthalpies of formation revisited. J. Phys. Chem. A 108, 9979–9997 (2004).
Yan, Q.-L. et al. High density assembly of energetic molecules under the constraint of defected 2D materials. J. Mater. Chem. A 7, 17806–17814 (2019).
Acknowledgements
This work was support by the US Office of Naval Research, Multidisciplinary University Research Initiatives (MURI) Program, Contract: N00014-16-1-2557. Program managers: Chad Stoltz and Kenny Lipkowitz. P.Y. and P.L. acknowledge startup support from the School of Materials Engineering at Purdue University and the Donors of the American Chemical Society Petroleum Research Fund for partial support of this research. The computational research was supported in part through computational resources provided by the Information Technology department at Purdue University, West Lafayette, Indiana.
Author information
Authors and Affiliations
Contributions
P.Y., P.L., and A.S. conceived the idea; P.Y. developed the training algorithm, performed DFT computations, and trained ML models; M.S. and M.I. performed validations and molecular dynamic simulations; P.Y. and S.D. developed the nanoHUB tool; A.S. and P.L. are the corresponding authors; All authors contributed to the discussion and writing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoo, P., Sakano, M., Desai, S. et al. Neural network reactive force field for C, H, N, and O systems. npj Comput Mater 7, 9 (2021). https://doi.org/10.1038/s41524-020-00484-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-020-00484-3
This article is cited by
-
Exploring the frontiers of condensed-phase chemistry with a general reactive machine learning potential
Nature Chemistry (2024)
-
A deep learning framework to emulate density functional theory
npj Computational Materials (2023)
-
Mapping microstructure to shock-induced temperature fields using deep learning
npj Computational Materials (2023)
-
Development of the reactive force field and silicon dry/wet oxidation process modeling
npj Computational Materials (2023)
-
The transformational role of GPU computing and deep learning in drug discovery
Nature Machine Intelligence (2022)