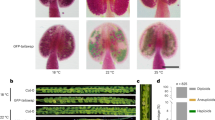
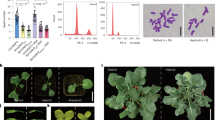
Abstract
Maize mutants of the centromeric histone H3 (CENP-A/CENH3) gene can form haploids that inherit only chromosomes of the pollinating parent but the cytoplasm from the female parent. We developed CENH3 haploid inducers carrying a dominant anthocyanin colour marker for efficient haploid identification and harbouring cytoplasmic male sterile cytoplasm, a type of cytoplasm that results in male sterility useful for efficient hybrid seed production. The resulting cytoplasmic male sterility cyto-swapping method provides a faster and cheaper way to convert commercial lines to cytoplasmic male sterile compared to conventional trait introgression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The sequences of the cenh3 edited alleles are shown on Extended Data Fig. 2. The number of embryos extracted, seedlings genotyped and confirmed haploids for both cyto-swapping experiments are provided in Extended Data Tables 1 and 2. The assays used for genotyping cenh3 alleles are in Supplementary Table 1. The genotyping results of cyto-swapped experiments are provided as Source Data. The data used for analyses of colchicine effect on HRR and seed set are in Supplementary Data.
References
Crow, J. F. 90 years ago: the beginning of hybrid maize. Genetics 148, 923–928 (1998).
Chen, L. & Liu, Y. G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606 (2014).
Kindiger, B. & Hamann, S. Generation of haploids in maize: a modification of the indeterminate gametophyte (ig) system. Crop Sci. 33, 342–344 (1993).
Evans, M. M. S. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19, 46–62 (2007).
Ravi, M. & Chan, S. W. L. Haploid plants produced by centromere-mediated genome elimination. Nature 464, 615–618 (2010).
Lv, J. et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 38, 1397–1401 (2020).
Kelliher, T. et al. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front. Plant Sci. 7, 414 (2016).
Wang, N. et al. Haploid induction by a maize cenh3 null mutant. Sci. Adv. 7, eabe2299 (2021).
Birky, C. W. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl Acad. Sci. USA 92, 11331–11338 (1995).
Whatley, F. Standard reference work on the plastids. Nature 279, 825–826 (1979).
Flood, P. J. et al. Reciprocal cybrids reveal how organellar genomes affect plant phenotypes. Nat. Plants 6, 13–21 (2020).
Dewey, R. E. et al. Chimeric mitochondrial genes expressed in the C male-sterile cytoplasm of maize. Curr. Genet. 20, 475–482 (1991).
Jaqueth, J. S. et al. Fertility restoration of maize CMS‐C altered by a single amino acid substitution within the Rf4 bHLH transcription factor. Plant J. 101, 101–111 (2020).
Hu, Y. M. et al. Identification and mapping of Rf-I an inhibitor of the Rf5 restorer gene for Cms-C in maize (Zea mays L.). Theor. Appl. Genet. 113, 357–360 (2006).
Verzegnazzi, A. L. et al. Major locus for spontaneous haploid genome doubling detected by a case–control GWAS in exotic maize germplasm. Theor. Appl. Genet. 134, 1423–1434 (2021).
Dong, S. et al. Efficient targeted mutagenesis mediated by CRISPR-Cas12a ribonucleoprotein complexes in maize. Front. Genome Ed. 3, 670529 (2021).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497 (1962).
Maluszynski, M. (ed.) Doubled Haploid Production in Crop Plants: A Manual (Springer Science & Business Media, 2003).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinf. 11, 1–9 (2010).
Acknowledgements
We thank S. Jensen for providing feedback on the manuscript, S. Rigoulot for performing protoplasts transfections and S. Bonefas for helping with maize crosses.
Author information
Authors and Affiliations
Contributions
E.B. wrote the manuscript, supervised the experiments, carried out crosses, performed the T7EI assays and evaluated CMS-converted lines. R.S. performed the CMS cyto-swapping experiments. L.T. performed the Normal B to Normal A cyto-swapping experiments. R.E. supervised the Normal B to Normal A cyto-swapping experiment and contributed to the writing. S.D. performed the protoplast transfections and supervised the maize embryo bombardment. K.B. and K.M. performed the maize embryo bombardment. J.F. contributed with statistical analyses. M.D. selected genotyping assays and created genetic distance figures. S.G. provided pictures for Fig. 1. D.M. planned and supervised greenhouse plantings. R.C.M.L. supervised and executed samplings for genotyping. K.S. contributed with plant maintenance and crossings. D.D. contributed to the design of the CMS cyto-swapping experiment and selected lines for conversions. R.K.D. provided lines carrying cenh3-KD. T.K. conceived the experiments, carried out crosses and contributed to the writing.
Corresponding author
Ethics declarations
Competing interests
E.B. and T.K. are co-inventors on patent WO23225469, Conferring cytoplasmic male sterility, filed by Syngenta relating to work in this manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Ravi Maruthachalam, Takayoshi Ishii and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Heat map of genetic distances from 2346 markers among NSS inbreds (NB-SYN-INBXXX) and their cyto-swapped Double Haploid progeny (NA-SYN-INBXXX-X).
The scale represents the percent of alleles that are different. NA-cenh3-KD-11 and −12 are the conversion lines carrying the cenh3-KD allele and Normal A cytoplasm used in this experiment.
Extended Data Fig. 2 Alleles of CENH3 used in this study.
(a) The region of CENH3 targeted by crRNA140, the location of the 1 bp deletion in cenh3-KD8 and the sequences of the alleles selected for cyto-swapping to CMS-C. (b) The deduced amino acids sequences that could potentially be encoded by the deletion alleles (Figure made in Geneious 2023.0.4).
Extended Data Fig. 3 Steps taken to produce the CMS-C conversion and Maintainer lines.
The steps shown here are simplified, some crosses have been omitted. From top to bottom: An RNP complex was bombarded into maize calli carrying the R1-scm2 allele. Plants that contained edited alleles expected to result in CENH3 loss of function were crossed as males to a CMS-C line and selfed. The resulting progeny, free of T-DNA and homozygous for R1-scm2 and the recessive restorer allele rf4, consists of a male-sterile Conversion line with a CMS-C cytoplasm and a male fertile Maintainer line with a Normal cytoplasm. The latter is used to pollinate both the CMS-C and the Maintainer lines. Both lines are propagated as segregating families (wild type 1:1 cenh3 heterozygous).
Extended Data Fig. 4 Heat map of genetic distances from 198 markers among Doubled Haploids converted to CMS-C cytoplasm and RP.
The scale represents the percent of alleles that are different.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Supplementary Data
Data for analyses of effect of colchicine on HRR and DHBC1 seed produced.
Source data
Extended Data Fig. 1
Genotypic data from 2,346 SNP markers.
Extended Data Fig. 4
Genotypic data from 198 SNP markers.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bortiri, E., Selby, R., Egger, R. et al. Cyto-swapping in maize by haploid induction with a cenh3 mutant. Nat. Plants 10, 567–571 (2024). https://doi.org/10.1038/s41477-024-01630-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-024-01630-1
This article is cited by
-
Haploids fast-track hybrid plant breeding
Nature Plants (2024)