Abstract
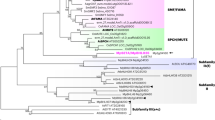
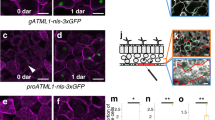
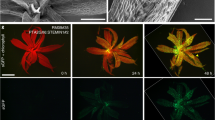
The leaf epidermis represents a multifunctional tissue consisting of trichomes, pavement cells and stomata, the specialized cellular pores of the leaf. Pavement cells and stomata both originate from regulated divisions of stomatal lineage ground cells (SLGCs), but whereas the ontogeny of the stomata is well characterized, the genetic pathways activating pavement cell differentiation remain relatively unexplored. Here, we reveal that the cell cycle inhibitor SIAMESE-RELATED1 (SMR1) is essential for timely differentiation of SLGCs into pavement cells by terminating SLGC self-renewal potency, which depends on CYCLIN A proteins and CYCLIN-DEPENDENT KINASE B1. By controlling SLGC-to-pavement cell differentiation, SMR1 determines the ratio of pavement cells to stomata and adjusts epidermal development to suit environmental conditions. We therefore propose SMR1 as an attractive target for engineering climate-resilient plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data are available in the main text or the supplementary materials. Source data are provided with this paper.
References
Liu, S., Jobert, F., Rahneshan, Z., Doyle, S. M. & Robert, S. Solving the puzzle of shape regulation in plant epidermal pavement cells. Annu. Rev. Plant Biol. 72, 525–550 (2021).
Glover, B. J. Differentiation in plant epidermal cells. J. Exp. Bot. 51, 497–505 (2000).
Jacques, E., Verbelen, J.-P. & Vissenberg, K. Review on shape formation in epidermal pavement cells of the Arabidopsis leaf. Funct. Plant Biol. 41, 914–921 (2014).
Javelle, M., Vernoud, V., Rogowsky, P. M. & Ingram, G. C. Epidermis: the formation and functions of a fundamental plant tissue. N. Phytol. 189, 17–39 (2011).
Zuch, D. T. et al. Cell biology of the leaf epidermis: fate specification, morphogenesis and coordination. Plant Cell 34, 209–227 (2022).
Geisler, M., Nadeau, J. & Sack, F. D. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086 (2000).
Pillitteri, L. J. & Dong, J. Stomatal development in Arabidopsis. Arabidopsis Book 11, e0162 (2013).
Shpak, E. D., McAbee, J. M., Pillitteri, L. J. & Torii, K. U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293 (2005).
Ho, C.-M. K., Bringmann, M., Oshima, Y., Mitsuda, N. & Bergmann, D. C. Transcriptional profiling reveals signatures of latent developmental potential in Arabidopsis stomatal lineage ground cells. Proc. Natl Acad. Sci. USA 118, e2021682118 (2021).
De Veylder, L., Larkin, J. C. & Schnittger, A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634 (2011).
Lammens, T. et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl Acad. Sci. USA 105, 14721–14726 (2008).
Churchman, M. L. et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18, 3145–3157 (2006).
Roeder, A. H. et al. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 8, e1000367 (2010).
Han, S.-K. et al. Deceleration of the cell cycle underpins a switch from proliferative to terminal divisions in plant stomatal lineage. Dev. Cell 57, 569–582 (2022).
Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E. & Dengler, N. G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419 (1999).
Andriankaja, M. et al. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78 (2012).
MacAlister, C. A., Ohashi-Ito, K. & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007).
Lampard, G. R., Macalister, C. A. & Bergmann, D. C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008).
Nadeau, J. A. & Sack, F. D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 (2002).
Horst, R. J. et al. Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genet. 11, e1005374 (2015).
Van Leene, J. et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397 (2010).
Boudolf, V. et al. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955 (2004).
Xie, Z. et al. Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22, 2306–2321 (2010).
Vanneste, S. et al. Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30, 3430–3441 (2011).
Dubois, M. et al. SIAMESE-RELATED1 is regulated posttranslationally and participates in repression of leaf growth under moderate drought. Plant Physiol. 176, 2834–2850 (2018).
Bhave, N. S. et al. TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta 229, 357–367 (2009).
Kumar, R. et al. Development of an efficient and reproducible regeneration system in wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 23, 945–954 (2017).
Bhosale, R., Maere, S. & De Veylder, L. Endoreplication as a potential driver of cell wall modifications. Curr. Opin. Plant Biol. 51, 58–65 (2019).
Han, S.-K., Kwak, J. M. & Qi, X. Stomatal lineage control by developmental program and environmental cues. Front. Plant Sci. 12, 751852 (2021).
Skirycz, A. et al. Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29, 212–214 (2011).
Doheny-Adams, T., Hunt, L., Franks, P. J., Beerling, D. J. & Gray, J. E. Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Phil. Trans. R. Soc. B 367, 547–555 (2012).
Bramsiepe, J. et al. Endoreplication controls cell fate maintenance. PLoS Genet. 6, e1000996 (2010).
Bhosale, R. et al. A spatiotemporal DNA endoploidy map of the Arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell 30, 2330–2351 (2018).
Boudolf, V. et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16, 2683–2692 (2004).
Wang, K. et al. The CDK inhibitor SIAMESE targets both CDKA;1 and CDKB1 complexes to establish endoreplication in trichomes. Plant Physiol. 184, 165–175 (2020).
Kumar, N. et al. Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell 27, 3065–3080 (2015).
Yang, K.-Z. et al. Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in Arabidopsis. Mol. Plant 8, 783–795 (2015).
Claeys, H. & Inze, D. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779 (2013).
Nowack, M. K. et al. Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22, 1030–1040 (2012).
Vlieghe, K. et al. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15, 59–63 (2005).
Fendrych, M. et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 24, 931–940 (2014).
Karimi, M., Inzé, D. & Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Decaestecker, W. et al. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887 (2019).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
De Veylder, L. et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667 (2001).
Acknowledgements
We thank A. Bleys for critical reading and helping in preparing the manuscript and M. Vermeersch for expertise on the drought phenotyping platform. This work was supported by grants from the Research Foundation Flanders (nos. G011420N and G010820N). I.A. and M.D. acknowledge support from a ‘Don Carlos Antonio Lopez’ by El Programa Nacional de Becas from Paraguay predoc (BECAL no. 164/2017) and a Research Foundation Flanders postdoc grant (no. 12Q7923N), respectively.
Author information
Authors and Affiliations
Contributions
L.D.V. supervised the project. M.D., I.A., D.I., J.E.G. and L.D.V. designed the experiments. M.D., I.A., S.P., R.T.B., R.A.B. and I.V. performed the experiments. M.D., I.A. and L.D.V. wrote and modified the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1 and 2.
Supplementary Data 1
Source data for Supplementary Figs. 1–5.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dubois, M., Achon, I., Brench, R.A. et al. SIAMESE-RELATED1 imposes differentiation of stomatal lineage ground cells into pavement cells. Nat. Plants 9, 1143–1153 (2023). https://doi.org/10.1038/s41477-023-01452-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01452-7
This article is cited by
-
Leaf form diversity and evolution: a never-ending story in plant biology
Journal of Plant Research (2024)