Abstract
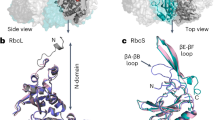
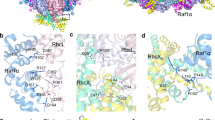
Ribulose-1,5-bisphosphate carboxylase–oxygenase (Rubisco) catalyses the first step in carbon fixation and is a strategic target for improving photosynthetic efficiency. In plants, Rubisco is composed of eight large and eight small subunits, and its biogenesis requires multiple chaperones. Here, we optimized a system to produce tobacco Rubisco in Escherichia coli by coexpressing chaperones in autoinduction medium. We successfully assembled tobacco Rubisco in E. coli with each small subunit that is normally encoded by the nuclear genome. Even though each enzyme carries only a single type of small subunit in E. coli, the enzymes exhibit carboxylation kinetics that are very similar to the carboxylation kinetics of the native Rubisco. Tobacco Rubisco assembled with a recently discovered trichome small subunit has a higher catalytic rate and a lower CO2 affinity compared with Rubisco complexes that are assembled with other small subunits. Our E. coli expression system will enable the analysis of features of both subunits of Rubisco that affect its kinetic properties.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
A list of the accession numbers of proteins expressed in this study is provided in Extended Data Fig. 4 and is publicly available at https://www.ncbi.nlm.nih.gov or https://solgenomics.net. The tobacco transcriptomic data are available at NCBI under Bioproject accession PRJNA208209. Source data are provided with this paper.
References
Whitney, S. M., Houtz, R. L. & Alonso, H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35 (2011).
Bauwe, H., Hagemann, M., Kern, R. & Timm, S. Photorespiration has a dual origin and manifold links to central metabolism. Curr. Opin. Plant Biol. 15, 269–275 (2012).
Walker, B. J., VanLoocke, A., Bernacchi, C. J. & Ort, D. R. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant. Biol. 67, 107–129 (2016).
Davidi, D. et al. Highly active rubiscos discovered by systematic interrogation of natural sequence diversity. EMBO J. https://doi.org/10.15252/embj.2019104081 (2020).
Jordan, D. B. & Ogren, W. L. A sensitive assay procedure for simultaneous determination of ribulose-1,5-bisphosphate carboxylase and oxygenase activities. Plant Physiol. 67, 237–245 (1981).
Flamholz, A. I. et al. Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry 58, 3365–3376 (2019).
Tcherkez, G. G. B., Farquhar, G. D. & Andrews, T. J. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl Acad. Sci. USA 103, 7246–7251 (2006).
Iniguez, C. et al. Evolutionary trends in Rubisco kinetics and their co-evolution with CO2 concentrating mechanisms. Plant J. 101, 897–918 (2020).
Evans, J. R. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78, 9–19 (1989).
Young, J. N. et al. Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 67, 3445–3456 (2016).
Wilson, R. H. & Hayer-Hartl, M. Complex chaperone dependence of Rubisco biogenesis. Biochemistry 57, 3210–3216 (2018).
Gatenby, A. A., van der Vies, S. M. & Bradley, D. Assembly in E. coli of a functional multi-subunit ribulose bisphosphate carboxylase from a blue-green alga. Nature 314, 617–620 (1985).
Lin, M. T., Occhialini, A., Andralojc, P. J., Parry, M. A. & Hanson, M. R. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550 (2014).
Long, B. M. et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570 (2018).
Tabita, F. R. & Small, C. L. Expression and assembly of active cyanobacterial ribulose-1,5-bisphosphate carboxylase/oxygenase in Escherichia coli containing stoichiometric amounts of large and small subunits. Proc. Natl Acad. Sci. USA 82, 6100–6103 (1985).
Whitney, S. M., Birch, R., Kelso, C., Beck, J. L. & Kapralov, M. V. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl Acad. Sci. USA 112, 3564–3569 (2015).
Sharwood, R. E., Ghannoum, O., Kapralov, M. V., Gunn, L. H. & Whitney, S. M. Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2, 16186 (2016).
Lin, M. T. & Hanson, M. R. Red algal Rubisco fails to accumulate in transplastomic tobacco expressing Griffithsia monilis RbcL and RbcS genes. Plant Direct 2, e00045 (2018).
Whitney, S. M., Baldet, P., Hudson, G. S. & Andrews, T. J. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 26, 535–547 (2001).
Brutnell, T. P., Sawers, R. J., Mant, A. & Langdale, J. A. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849–864 (1999).
Feiz, L. et al. A protein with an inactive pterin-4a-carbinolamine dehydratase domain is required for Rubisco biogenesis in plants. Plant J. 80, 862–869 (2014).
Feiz, L. et al. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 24, 3435–3446 (2012).
Fristedt, R. et al. RAF2 is a RuBisCO assembly factor in Arabidopsis thaliana. Plant J. 94, 146–156 (2018).
Aigner, H. et al. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278 (2017).
Hauser, T. et al. Structure and mechanism of the Rubisco-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 22, 720–728 (2015).
Liu, C. et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463, 197–202 (2010).
Kolesinski, P. et al. Insights into eukaryotic Rubisco assembly—crystal structures of RbcX chaperones from Arabidopsis thaliana. Biochim. Biophys. Acta 1830, 2899–2906 (2013).
Schmidt, J. A., McGrath, J. M., Hanson, M. R., Long, S. P. & Ahner, B. A. Field-grown tobacco plants maintain robust growth while accumulating large quantities of a bacterial cellulase in chloroplasts. Nat. Plants 5, 715–721 (2019).
South, P. F., Cavanagh, A. P., Liu, H. W. & Ort, D. R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077 (2019).
Laterre, R., Pottier, M., Remacle, C. & Boutry, M. Photosynthetic trichomes contain a specific Rubisco with a modified pH-dependent activity. Plant Physiol. 173, 2110–2120 (2017).
Wilson, R. H., Thieulin-Pardo, G., Hartl, F. U. & Hayer-Hartl, M. Improved recombinant expression and purification of functional plant Rubisco. FEBS Lett. 593, 611–621 (2019).
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Gong, L., Olson, M. & Wendel, J. F. Cytonuclear evolution of Rubisco in four allopolyploid lineages. Mol. Biol. Evol. 31, 2624–2636 (2014).
Sierro, N. et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5, 3833 (2014).
Morita, K., Hatanaka, T., Misoo, S. & Fukayama, H. Identification and expression analysis of non-photosynthetic Rubisco small subunit, OsRbcS1-like genes in plants. Plant Gene 8, 26–31 (2016).
Pottier, M., Gilis, D. & Boutry, M. The hidden face of rubisco. Trends Plant Sci. 23, 382–392 (2018).
Conlan, B. et al. BSD2 is a Rubisco-specific assembly chaperone, forms intermediary hetero-oligomeric complexes, and is nonlimiting to growth in tobacco. Plant Cell Environ. 42, 1287–1301 (2019).
Houtz, R. L., Magnani, R., Nayak, N. R. & Dirk, L. M. Co- and post-translational modifications in Rubisco: unanswered questions. J. Exp. Bot. 59, 1635–1645 (2008).
Morita, K., Hatanaka, T., Misoo, S. & Fukayama, H. Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of rubisco in rice. Plant Physiol. 164, 69–79 (2014).
Spreitzer, R. J. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414, 141–149 (2003).
Genkov, T. & Spreitzer, R. J. Highly conserved small subunit residues influence rubisco large subunit catalysis. J. Biol. Chem. 284, 30105–30112 (2009).
Spreitzer, R. J., Peddi, S. R. & Satagopan, S. Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl Acad. Sci. USA 102, 17225–17230 (2005).
Ishikawa, C., Hatanaka, T., Misoo, S., Miyake, C. & Fukayama, H. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol. 156, 1603–1611 (2011).
Andersson, I. & Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 46, 275–291 (2008).
Kapralov, M. V., Kubien, D. S., Andersson, I. & Filatov, D. A. Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Mol. Biol. Evol. 28, 1491–1503 (2011).
Yamada, K., Davydov, I. I., Besnard, G. & Salamin, N. Duplication history and molecular evolution of the rbcS multigene family in angiosperms. J. Exp. Bot. 70, 6127–6139 (2019).
Smith, S. A. & Tabita, F. R. Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 331, 557–569 (2003).
Mueller-Cajar, O., Morell, M. & Whitney, S. M. Directed evolution of rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry 46, 14067–14074 (2007).
Wilson, R. H., Alonso, H. & Whitney, S. M. Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 6, 22284 (2016).
Zhou, Y. & Whitney, S. Directed evolution of an improved Rubisco; in vitro analyses to decipher fact from fiction. Int. J. Mol. Sci. 20, 5019 (2019).
Hanson, M. R., Gray, B. N. & Ahner, B. A. Chloroplast transformation for engineering of photosynthesis. J. Exp. Bot. 64, 731–742 (2013).
Orr, D. J. & Carmo-Silva, E. Extraction of RuBisCO to determine catalytic constants. Methods Mol. Biol. 1770, 229–238 (2018).
Whitney, S. M. & Sharwood, R. E. Plastid transformation for Rubisco engineering and protocols for assessing expression. Methods Mol. Biol. 1132, 245–262 (2014).
Kubien, D. S., Brown, C. M. & Kane, H. J. Quantifying the amount and activity of Rubisco in leaves. Methods Mol. Biol. 684, 349–362 (2011).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Acknowledgements
We thank M. Hayer-Hartl from the Max Planck Institute of Biochemistry in Martinsried, Germany, for providing us with her laboratory’s E. coli expression vectors; and D. Orr, E. Carmo-Silva and M. Parry from Lancaster University, Lancaster, UK for providing us with purified RuBP and advice on experiments to quantify Rubisco active sites and measure RuBP carboxylation kinetics. M.T.L. and W.D.S. are supported by a grant to M.R.H. from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award no. DE-SC0020142); and M.R.H. and V.C. by the National Science Foundation (award no. MCB-1642386).
Author information
Authors and Affiliations
Contributions
M.T.L. and M.R.H. conceived the research. All of the authors designed the experiments. M.T.L., W.D.S. and V.C. performed the experiments. All of the authors analysed the data and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
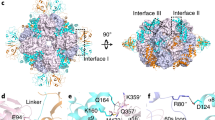
Extended Data Fig. 1 Native PAGE immunoblots of tobacco Rubisco expressed in Rosetta (DE3) E. coli with different small subunits.
Nt-Cpn60α, Nt-Cpn60β, GroES, Nt-RbcX, Nt-Raf1, At-Raf2 and Nt-Bsd2 were co-expressed. 9 µg of total soluble extract was loaded for each sample. 3 µg of total soluble extract from a young tobacco leaf (N) was also included as a control. The protein expressions were auto-induced in ZYP-5052 medium at 23 °C for 18-20 h. The top panel shows Coomassie blue staining, and the bottom panel shows the immunoblot using the antibody against Rubisco. The bands for L8S8 Rubisco (540 kDa) and chaperonin-RbcL complex (~720 kDa, indicated with asterisks) are marked next to each panel. The native PAGE was performed once for the same set of samples and multiple times for samples with RbcS-S1 and RbcS-T1 with similar results.
Extended Data Fig. 2 Comparison of tobacco Rubisco expressed in BL21 StarTM (DE3) E. coli using different culture media.
a, RuBP carboxylation rates at 42 μM [CO2] were measured from incorporation of 14C into RuBP in the absence of O2 at 25 °C. Rubisco active sites were quantified with bound 14C-CABP separated in size-exclusion chromatography. The error bars represent the mean values and standard deviations of measurements from three to six E. coli growth experiments for each condition. Data were analyzed with one-way ANOVA, and the P-value was obtained from F-statistics. Tukey’s honest significance test was then carried, and samples with P value > 0.05 are indicated with the same letter. b, The native PAGE immunoblot of the same samples in a using the antibody against wheat Rubisco. The three additional media tested are the buffered medium without additional carbon source (ZYP), the auto-induction medium without Mg (ZYP-5052ΔMg) and the auto-induction medium without trace metals (ZYP-5052Δmetals). The native PAGE was performed once for the same set of samples.
Extended Data Fig. 3 The enzyme kinetics of tobacco Rubisco expressed in E. coli with different small subunits compared to the native tobacco Rubisco.
RuBP carboxylation rates were measured from incorporation of 14C into RuBP in the absence of O2 at 25 °C. At-Cpn60α, At-Cpn60β, At-Cpn20, Nt-RBCX, Nt-RAF1, At-RAF2 and Nt-BSD2 were co-expressed in BL21 StarTM (DE3) either in LB or ZYP-5052 auto-induction medium at 23 °C for 18-20 h. [CO2] in the reaction mixtures ranged from 12.9 to 108.3 µM for the E. coli extracts and 8.3 to 103.7 µM for tobacco leaf extracts. Rubisco active sites were quantified with bound 14C-CABP separated in size-exclusion chromatography. The data were fitted to the Michaelis-Menten equation with nonlinear regression. The fitted models are shown as dotted lines for the E. coli samples and dashed line for the tobacco leaf samples.
Extended Data Fig. 4
Summary of the genes expressed in this study.
Extended Data Fig. 5
Summary of plasmids used in the expression of tobacco Rubisco in E. coli.
Supplementary information
Supplementary Information
Oligonucleotide sequences used in the construction of expression vectors.
Source data
Source Data Fig. 2
Unprocessed native PAGE and western blot.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed native PAGEs and western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed native PAGE and western blot.
Source Data Extended Data Fig. 2
Unprocessed western blot.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Rights and permissions
About this article
Cite this article
Lin, M.T., Stone, W.D., Chaudhari, V. et al. Small subunits can determine enzyme kinetics of tobacco Rubisco expressed in Escherichia coli. Nat. Plants 6, 1289–1299 (2020). https://doi.org/10.1038/s41477-020-00761-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-00761-5
This article is cited by
-
Towards engineering a hybrid carboxysome
Photosynthesis Research (2023)
-
Correlative adaptation between Rubisco and CO2-concentrating mechanisms in seagrasses
Nature Plants (2022)
-
Rubiscosome gene expression is balanced across the hexaploid wheat genome
Photosynthesis Research (2022)
-
Mix-and-match Rubisco subunits
Nature Plants (2020)