Abstract
RuBisCO is the most abundant enzyme in nature, catalyzing the fixation of CO2 in photosynthesis. Its common form consists of eight RbcL and eight RbcS subunits, the assembly of which requires a series of chaperones that include RbcX and RuBisCO accumulation factor 1 (Raf1). To understand how these RuBisCO-specific chaperones function during cyanobacterial RbcL8RbcS8 (L8S8) holoenzyme formation, we solved a 3.3-Å cryo-electron microscopy structure of a 32-subunit RbcL8Raf18RbcX16 (L8F8X16) assembly intermediate from Anabaena sp. PCC 7120. Comparison to the previously resolved L8F8 and L8X16 structures together with biochemical assays revealed that the L8F8X16 complex forms a rather dynamic structural intermediate, favoring RbcS displacement of Raf1 and RbcX. In vitro assays further demonstrated that both Raf1 and RbcX function to regulate RuBisCO condensate formation by restricting CcmM35 binding to the stably assembled L8S8 holoenzymes. Combined with previous findings, we propose a model on how Raf1 and RbcX work in concert to facilitate, and regulate, cyanobacterial RuBisCO assembly as well as disassembly of RuBisCO condensates.
Similar content being viewed by others
Introduction
Life on earth depends on the photosynthesis pathway to convert atmospheric CO2 into organic carbon. This process is initiated by the globally most abundant enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), whose total mass in nature is ~0.7 Gt1,2. As the most common form, the form I RuBisCO in plants, eukaryotic algae, and cyanobacteria is a ~530 kDa complex consisting of eight large (RbcL, ~53 kDa) and eight small (RbcS, ~15 kDa) subunits3. Eight RbcL subunits are assembled into a tetramer of catalytic antiparallel dimers, which are capped by four RbcS subunits at the top and bottom, respectively, forming a functional holoenzyme RbcL8RbcS8 (L8S8). Remarkably, RuBisCO is a rather inefficient and error-prone enzyme due to its slow catalytic rate (~3–12 s−1) and limited specificity towards CO2 versus O24,5.
Generally, RuBisCO biogenesis is a complicated process that requires a series of molecular chaperones6,7,8. In cyanobacteria, the nascent RbcL subunits are initially folded by the chaperonin GroEL-GroES9, followed by assembly of the octameric core RbcL8, mainly assisted by individual chaperones such as RuBisCO accumulation factor Raf17,10 and RbcX11,12,13. Afterward, docking of RbcS subunits displaces Raf114 and/or RbcX13 to enable the formation of L8S8 holoenzyme. The previously solved RbcL8RbcX16 (L8X16) structure showed that RbcX is a homodimer of mostly α-helical structure with a central cleft binding to the C-terminal conserved motif of RbcL13. Our previously reported structures of Raf1 and its complex with RbcL (RbcL8Raf18, termed L8F8 for short) demonstrated that Raf1 is also a homodimer, each subunit of which consists of an N-terminal α-helical domain (Raf1α) and a C-terminal β-sheet dimerization domain (Raf1β) separated by a flexible linker10. Upon binding to RbcL, the Raf1α and Raf1β domains of Raf1 undergo rigid body rotations to embrace an RbcL dimer, forming the complex L8F8, in which Raf1β are arranged around the equator of each RbcL dimer, whereas the two Raf1α domains contact the top and bottom edges of the RbcL dimer10. Given the co-existence of Raf1 and RbcX in most cyanobacteria and plants15, the two chaperones might function in concert on RuBisCO assembly. However, how these two chaperones interplay on RuBisCO biogenesis remains elusive.
Here we solved the 3.3-Å cryo-electron microscopy (cryo-EM) structure of a ternary complex composed of RbcL, Raf1, and RbcX, representing a cyanobacterial RuBisCO assembly intermediate. Structural and biochemical analyses elucidated the mechanism underlying the concerted action of Raf1 and RbcX on the assembly of RuBisCO holoenzyme and disassembly of RuBisCO condensates. All these findings provide new insights into cyanobacterial RuBisCO assembly and potential avenues for its engineering in heterologous systems toward improving plant photosynthesis and growth16,17.
Results
Structure of the RuBisCO assembly intermediate RbcL8Raf18RbcX16
We co-expressed RbcL with the chaperones Raf1 and RbcX from Anabaena sp. PCC 7120, in the presence of the chaperonin GroEL-GroES, which facilitates the formation of a 32-subunit RbcL–Raf1–RbcX ternary complex of ~1 MDa in size (Supplementary Fig. S1). Subsequently, we purified this complex and solved its cryo-EM structure at 3.3 Å resolution (Fig. 1a; Supplementary Fig. S2a–c), representing an intermediate of RbcL8 core engaged by 24 molecules of chaperones. The overall density map, especially RbcL8 core at the center, is of high quality, whereas the surrounding regions corresponding to RbcX are relatively dispersed (Supplementary Fig. S2d). Nevertheless, thanks to the known structures of RbcL, Raf1, and RbcX10,13, we successfully fitted all protein components into the map and finally obtained the complex structure RbcL8Raf18RbcX16, termed L8F8X16 for short (Fig. 1a). This structure demonstrates the interaction patterns of a RuBisCO intermediate bound by multiple chaperones.
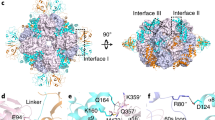
a The overall structure of L8F8X16 is shown in two orientations rotated by 90°. The RbcL octameric core is shown as the surface, with the two subunits of each dimer colored in pink and blue, respectively. The four Raf1α dimers and the eight RbcX dimers are shown as cartoons. The two subunits of each Raf1α dimer are respectively colored in marine and magenta, whereas those of each RbcX dimer are respectively colored in yellow and green. The interfaces between RbcL–Raf1α, RbcL–RbcX, and Raf1α–RbcX are indicated by dashed boxes. b–d The three interfaces of RbcL–Raf1α (b), RbcL–RbcX (c), and RbcX–Raf1α (d) in L8F8X16. The color scheme of each subunit is the same as that in the overall structure of L8F8X16. RbcL, Raf1α, and RbcX are shown as semi-transparent cartoons. The interacting residues are shown as sticks and labeled, with hydrogen bonds indicated as dashed lines. e Structural comparison of two RbcX dimers in the structures of L8F8X16 and L8X16 (PDB, 3RG6). f Structural comparison of Raf1α domains in the structures of L8F8X16 and L8F8 (PDB, 6KKM).
In the L8F8X16 structure, eight RbcL subunits form a core of four antiparallel dimers (Fig. 1a), which resembles the RbcL8 octameric core seen previously in RuBisCO-chaperone intermediary structures10,13,18. Similar to the L8F8 structure10, each Raf1α domain of Raf1 is docked onto the interface cleft between two neighboring RbcL dimers, embracing the C-terminal TIM-barrel domain of an RbcL subunit (Fig. 1b; Supplementary Table S1). Additionally, Raf1α also makes contact with the N-terminal domain of the adjacent RbcL dimer (Fig. 1b; Supplementary Table S1). However, the Raf1β domain of Raf1, which binds to the equator of RbcL8 in the structure of L8F810, could not be modeled in the final map of L8F8X16 due to the untraceable density. Moreover, similar to the chimeric structure of L8X16 composed of Synechococcus sp. PCC 6301 RbcL and Anabaena sp. CA RbcX13, two RbcX dimers are docked to one RbcL dimer via interacting with the C-terminal peptide of one RbcL subunit and the N-terminal domain of the other subunit (Fig. 1c; Supplementary Table S1). Of note, Raf1α also interacts with one subunit of RbcX dimer (RbcX2) via several hydrogen bonds, including Raf1R140–RbcXK57, Raf1L142–RbcXR102, Raf1P143–RbcXR102 bonds, to further stabilize the complex (Fig. 1d; Supplementary Table S1). In sum, the ternary complex of L8F8X16 has three different interfaces, which possess a buried interface area of ~740, 1000, and 400 Å2 for RbcL–Raf1α, RbcL–RbcX2, and Raf1–RbcX2, respectively. Sequence analyses showed that the interface residues within RbcL–Raf1 and RbcL–RbcX are highly conserved, whereas the residues at the Raf1–RbcX interface are relatively variable among Raf1 and RbcX homologs (Supplementary Fig. S3). It indicates that Raf1 and RbcX do not possess specific interactions with each other and are respectively recruited by RbcL.
Compared to L8F8 and L8X16, the L8F8X16 structure exhibits significant conformational variations, beyond similar architectures and binding patterns. In contrast to L8X16, each RbcX2 moves ~2.6 Å towards the equator of RbcL dimer in L8F8X16, forming an interface area of ~1000 Å2 between RbcL and RbcX2 in L8F8X16, compared to that of ~1400 Å2 in L8X16 (Fig. 1e). The structural comparison showed that Raf1β in L8F8 and RbcX2 in L8F8X16 share a partially overlapped binding region on RbcL (Supplementary Fig. S4). Thus Raf1β is expelled from the equator of RbcL8 upon RbcX binding, which might be flexibly tethered nearby the RbcL8 core, due to the 23-residue linker between Raf1α and Raf1β. Moreover, Raf1α in L8F8X16 moves ~3.0 Å away from the RbcL8 core compared to that in L8F8 (Fig. 1f), resulting in a dramatic decrease of RbcL–Raf1α interface from ~1000 Å2 in L8F8 to ~740 Å2 in L8F8X16. Another notable difference is the 8-residue C-terminal tail of Raf1 (C-tail), which inserts deeply into the active-site pocket of RbcL in L8F810, and is untraceable in L8F8X16. Superposition of L8F8 onto L8F8X16 revealed that Raf1 C-tail and one subunit of RbcX2 have a large steric hindrance (Supplementary Fig. S4). Notably, similar to that in L8X16, the conserved C-terminal peptide of RbcL is embedded in a narrow hydrophobic groove of RbcX2 in L8F8X16, different from that interacting with Raf1α in L8F8 (Supplementary Fig. S5). Moreover, the so-called “60s loop” of RbcL (residues 64–83) that forms a part of the catalytic pocket, is missing in both structures of L8X16 and L8F8X16, whereas it is stabilized by Raf1α in L8F8.
Generally, the L8F8X16 structure shows much looser contacts of Raf1 or RbcX with RbcL, indicating that Raf1 and RbcX partly antagonize each other upon binding to RbcL8. As a consequence, L8F8X16 adopts a rather dynamic structure with a more relaxed RbcL8 core that comprises a 25 Å central pore in diameter, which is ~5 Å larger than those in either L8F8, L8X16 or L8S8 structures (Supplementary Fig. S6).
Concerted action of Raf1 and RbcX on RuBisCO assembly
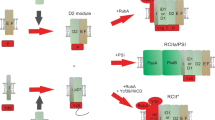
To test if this rather relaxed RbcL8 core is more favorable for RbcS recruitment, we performed in vitro RuBisCO assembly assays. Titration of RbcS at various ratios to the L8F8 complexes gradually triggered the formation of higher-molecular-mass (HMM) intermediates, corresponding to the ternary complex RbcL–Raf1–RbcS (Fig. 2a; Supplementary Fig. S7a). However, almost no RuBisCO holoenzyme could be detected, even in the presence of 10-fold RbcS in molarity to that of RbcL (Fig. 2a), indicating that excess RbcS could hardly displace Raf1 from L8F8, most likely due to the tight interaction between RbcL and Raf110. By contrast, the addition of RbcS at increasing ratios into L8F8X16 solution gradually triggers the formation of HMM intermediates that contain RbcL, RbcS, Raf1, and RbcX (Fig. 2b; Supplementary Fig. S7b). Upon the addition of RbcS up to 10-fold in molarity to that of RbcL, these HMM intermediates remain heterogeneous and finally reach a migration rate close to that of L8S8 holoenzyme (Fig. 2b), accompanied by the decrease of Raf1 in the intermediates (Supplementary Fig. S7b). Moreover, titration of RbcS at various ratios to the L8F8 solution pre-incubated with 16-fold RbcX in molarity also yielded a similar profile of HMM intermediates formation (Fig. 2c; Supplementary Fig. S7c). In contrast, pre-incubation of RbcL–Raf1 complexes with RbcXE32A&R69A mutant, in which the two RbcX residues directly interacting with RbcL13 were mutated, no longer promoted the formation of HMM intermediates that possess less Raf1 (Fig. 2d; Supplementary Fig. S7d), but led to the formation of HMM intermediates that behave quite similarly to that of RbcL–Raf1 complexes titrated with RbcS (Fig. 2a; Supplementary Fig. S7a).
a–d RbcS proteins at increasing concentrations were added to the solution of RbcL–Raf1 (a), RbcL–Raf1–RbcX (b), or RbcL–Raf1 pre-incubated with RbcX (c) or RbcXE32A&R69A (d). The concentrations of RbcS are 0, 2, 4, 8, 20, and 40 μM, with the molar ratio of 0, 0.5, 1, 2, 5, and 10 fold RbcS (as shown at the top) to RbcL. The concentrations of RbcX and RbcXE32A&R69A are 8 μM, which is 2-fold that of RbcL. The assembly intermediates are indicated by arrows on the left of the native-PAGE, in which HMM represents the complexes of high-molecular-mass intermediates. The Anabaena sp. PCC 7120 L8S8 holoenzyme was used as the positive control in lane 1 of each panel.
Furthermore, we applied the RuBisCO carboxylase activity assays to further evaluate whether the functional RuBisCO active sites are formed in these HMM intermediates. As expected, the RbcL–Raf1 complexes were almost inactive without RbcS (Supplementary Fig. S8). Addition of RbcS at increasing concentrations to RbcL–Raf1 complexes gradually augmented the carboxylase activities (Supplementary Fig. S8), which indicates that displacement of Raf1 by RbcS enables the formation of functional RuBisCO active sites. However, even 10-fold molarity of RbcS could not completely displace Raf1 from RbcL, as shown by a much lower activity compared to that of Synechococcus elongatus PCC 7942 RuBisCO holoenzyme (Supplementary Fig. S8). Moreover, compared to RbcL–Raf1 complexes, pre-incubation with RbcX2 at equal molarity in vitro led to a drastic decrease in the carboxylase activity, in the presence of RbcS at the same molarity (Supplementary Fig. S8). Apparently, the addition of extra RbcX to RbcL–Raf1 complexes facilitates the release of Raf1 from RbcL, and leads to the formation of highly dynamic HMM complexes composed of a series of RuBisCO assembly intermediates, including RbcL–RbcX and RbcL–Raf1–RbcX complexes. In fact, under the physiological conditions, the relative abundance of Raf1 and RbcX is only ~0.6% or less than that of RbcL19; thus, the higher excess RbcS in cyanobacterial cells could easily displace Raf1 and/or RbcX from RbcL. Taken together, our results suggest that RbcX acts in concert with Raf1 to maintain the homeostasis of RuBisCO assembly, which is a rather dynamic and reversible process composed of various assembly intermediates.
Notably, compared to the dynamic RbcL–Raf1 complexes that have a larger fraction of L2F2 (Fig. 2a), the bands corresponding to L2F2 sharply decreased in the samples of RbcL–Raf1–RbcX complexes (Fig. 2b), and were almost diminished in the samples of RbcL–Raf1 complexes pre-incubated with RbcX (Fig. 2c). Upon the increase of RbcX in the RbcL–Raf1 complexes, followed by the addition of RbcS (1:1 molarity to RbcL), it yielded more HMM intermediates and fewer L2F2 complexes (Supplementary Fig. S9). Once the equal molarity of RbcX2 was added to the solution, almost all RbcL proteins become the HMM intermediates, with most Raf1 released and L2F2 undetectable (Supplementary Fig. S9). These results suggested that RbcX could also facilitate the shift of equilibrium from RbcL dimer to RbcL octamer, besides promoting the release of Raf1 from RbcL.
RbcX can efficiently solubilize CcmM35-mediated RuBisCO condensates
In cyanobacteria, RuBisCO holoenzymes further form condensates that are cross-linked by the scaffold protein CcmM35, the truncated form of CcmM that harbors three RuBisCO small-subunit-like (SSUL) modules20,21. We previously found that Raf1 acts as a solubilizer that antagonizes CcmM35-mediated RuBisCO condensates formation in vitro10. When incubating Anabaena sp. PCC 7120 L8F8 complex with 8-fold RbcS in molarity, followed by the addition of 8-fold CcmM35 in solution, we prepared the RuBisCO condensates, as shown by that the turbidity finally reached the maximum absorbance (Fig. 3a). Afterward, the addition of RbcX triggers the decrease in turbidity over time (Fig. 3a), indicating the gradual disassembly of the condensates. The more RbcX that was added, the faster the condensates were solubilized. Once adding RbcX2 in equal molarity to that of RbcL, the solution became clear in ~10 min (Fig. 3a), indicating that RuBisCO condensates were almost completely solubilized. Moreover, the L8F8X16 complexes could no longer form RuBisCO condensates even in the presence of excess RbcS up to 10-fold in molarity to that of RbcL (Fig. 3b).
a Plots of solubilizing Anabaena sp. PCC 7120 RuBisCO condensates upon addition of RbcX. The RuBisCO condensates were formed by mixing 0.5 µM L8F8, 4 µM RbcS, and 4 µM CcmM35 for 5 min. RbcX2 at various concentrations (0, 0.2, 0.4, 0.8, 2 and 4 μM), corresponding to a molar ratio of 0, 0.05, 0.1, 0.2, 0.5 and 1 fold RbcX2 to RbcL, respectively, was then added to the turbid solution. b Plots of condensate formation from Anabaena sp. PCC 7120 L8F8X16 was incubated with RbcS for 30 min, and then 4 µM of CcmM35. 0.5 µM L8F8X16 was incubated with RbcS at various concentrations (0, 2, 4, 8, 20 and 40 µM), corresponding to a molar ratio of 0, 0.5, 1, 2, 5 and 10 fold RbcS to RbcL, respectively. c–e Plots of S. elongatus PCC 7942 RuBisCO condensate solubilization upon adding Raf1 (c), RbcX (d) and Raf1&RbcX (e), respectively. The RuBisCO condensates were formed by mixing 0.25 µM L8S8 and 2 µM CcmM35 for 4 min. Raf1, RbcX2 and Raf1&RbcX2 at various concentrations (0, 1, 2, 4, 10 and 20 μM), corresponding to a molar ratio of 0, 0.5, 1, 2, 5, and 10 fold Raf1 or RbcX2 to RbcL, respectively, were then added to the turbid solution. The turbidity was monitored at 340 nm over time. f–h Confocal microscopy images of the final condensates corresponding to c–e. Scale bars, 20 μm. i Structural superposition of L8F8X16 against L8S8–SSUL complex (PDB, 6HBC). The RbcL dimer is shown as surface and colored in pink/blue. The SSUL domain and 60s loop of RbcL from L8S8–SSUL complex are indicated by cyan cartoon, whereas the Raf1α domains and 60s loops of RbcL from L8F8X16 are shown as a magenta cartoon.
To further compare the effect of Raf1 and RbcX on solubilizing RuBisCO condensates, we prepared RuBisCO condensates by using S. elongatus PCC 7942 L8S8, due to that the insufficient amount of Anabaena sp. PCC 7120 L8S8 is required for multiple rounds of in vitro assays. As a previously reported solubilizer10, Raf1 could trigger the disassembly of the RuBisCO condensates, as shown by the gradual decrease in turbidity over time (Fig. 3c); however, it is of relatively lower efficiency, as even 10-fold Raf1 could only partially solubilize the condensates. In contrast, RbcX showed a much higher efficiency compared to Raf1, as the RuBisCO condensates were almost completely solubilized in ~5 min upon addition of 5-fold RbcX2 (Fig. 3d). Notably, simultaneous addition of both Raf1 and RbcX2 (at equal molarity) showed a profile of quick decrease of turbidity over time (Fig. 3e), similar to that upon addition of RbcX2 alone.
Moreover, we applied confocal fluorescence spectroscopy assays to observe the disassembly of RuBisCO condensates upon the addition of Raf1 and/or RbcX. Similar to the turbidity assays, a small fraction of RuBisCO condensates still existed even 10-fold molarity of Raf1 was added to the turbid solution, further confirming that Raf1 is a solubilizer with low efficiency (Fig. 3f). In contrast, the RuBisCO condensates were almost completely solubilized upon the addition of RbcX2 (Fig. 3g) or Raf1/RbcX2 at 5-fold molarity (Fig. 3h). These results suggested that both Raf1 and RbcX could solubilize the RuBisCO condensates in vitro under the tested conditions, and RbcX possesses a much higher efficiency.
It was previously reported that Raf1 antagonizes the RuBisCO condensation via competitively binding to the SSUL-binding site on RbcL10. Despite RbcX possessing a binding site on RbcL different from that of SSUL, the 60s loop of RbcL is disordered upon binding to RbcX, as shown in the structure of either L8X1613 or L8F8X16 (Fig. 3i). It was known that the 60s loop of RbcL directly interacts with SSUL in the structure of RuBisCO–SSUL21, and is necessary for the recruitment of RbcS to form RuBisCO holoenzyme22. Thus upon binding to RbcX, RbcL might adopt an altered conformation that is incapable of binding to RbcS and CcmM35-SSUL, which are prerequisites for the assembly of RuBisCO holoenzymes and succeeding condensation.
Discussion
Combined with previous findings of Raf17,10,15,23 and RbcX11,12,13,24, the present ternary structure of L8F8X16 intermediate together with biochemical assays provide fine molecular insights into the coordinated action of Raf1 and RbcX on RuBisCO holoenzyme assembly and CcmM35-mediated condensates disassembly (Fig. 4). Assembly of RbcL8 core could be assisted by the individual chaperone Raf1 or RbcX (Fig. 4, the upper and lower paths). However, in most cases, Raf1 and RbcX could simultaneously bind to RbcL, forming a dynamic L8F8X16 intermediate (Fig. 4, the central path), which favors RbcS displacement of the two chaperones. Notably, as an early-stage assembly chaperone7, Raf1 is a major contributor to forming the RbcL dimers in the form of L2F2, in addition to L8F8 (Fig. 2a). Given a much larger interface of RbcL–Raf1, compared to a smaller interface of RbcL–RbcX (Fig. 1b, c), it is possible that Raf1 functions as a major contributor in RuBisCO assembly since its deletion in cyanobacteria and plants precludes RuBisCO biogenesis15, whereas RuBisCO production in cyanobacteria is unaffected by RbcX deletion25. Moreover, plant RuBisCO biogenesis in Escherichia coli remains feasible upon RbcX omission, but fully reliant on Raf1 production26. Our biochemical assays showed that similar to the C-terminal tail of Raf110, RbcX also facilitates the formation of RbcL octamers from dimers in the presence of Raf1 (Fig. 2c; Supplementary Fig. S9). Thus RbcX assists and acts in concert with Raf1 to form a dynamic L8F8X16 intermediate, eventually favoring RbcS recruitment, indicating that RbcX is more likely an assistant chaperone that functions at the relatively late stage.
The previously identified paths of individual Raf1- or RbcX-assisted RuBisCO assembly are also included in the model. The proteins GroEL-GroES, RbcL, RbcS, Raf1, and RbcX are shown as surfaces and are colored in gray, pink, cyan, marine, and yellow, respectively. A putative step of condensates disassembly is indicated by the dashed line.
The ordered RuBisCO holoenzyme structure with a tightly packed RbcL8 core is a prerequisite for the proper RuBisCO lattice formation and succeeding recruitment of shell proteins of carboxysome27,28,29. Beyond functioning as a chaperone, RbcX is a more efficient solubilizer to dissolve the RuBisCO condensates in vitro under tested conditions, in contrast to Raf1 (Fig. 3). It implies that RbcX might be a key regulator that controls RuBisCO condensation, a process succeeding the maturation of RuBisCO holoenzymes in cyanobacteria. In fact, a previous report suggested that RbcX appears as one component of carboxysome and co-localizes with RuBisCO which mediates carboxysome biogenesis in vivo24.
Introducing cyanobacterial carboxysomes into plant chloroplasts has become a promising strategy for genetic engineering to improve photosynthetic performance30,31,32. However, reconstituting entire functional β-carboxysomes in heterologous hosts is still a big challenge, partly due to the sophisticated mechanisms of RuBisCO assembly and carboxysome biogenesis. Notably, the plant RuBisCO assembly requires a chloroplast-specific chaperone BSD233, which was proposed to be an end-stage assembly factor of RuBisCO26 and acts as a negative regulator of RbcL transcription34,35. It was proposed that in the chloroplast, BSD2 may have diminished the role of RbcX in RuBisCO assembly, owing to a partly overlapped binding site on RbcL26. In addition, cyanobacterial Raf1 may serve dual functions of both plant Raf1 and BSD2, where the two conserved C-terminal acidic residues of either BSD2 or cyanobacterial Raf1 insert into the RbcL catalytic pocket, contributing to RbcL octamer assembly10. Given a more complicated biogenesis pathway of plant L8S8 holoenzyme, further studies are needed to elucidate the fine mechanism by which BSD2 might provide an adaptive advantage during plant RuBisCO biogenesis and/or repair.
Despite that cyanobacteria and plants share a generally similar mechanism of RuBisCO assembly, the roles of individual chaperones differ a lot. To guarantee the proper assembly of carboxysome in the chloroplast, it is feasible to introduce cyanobacterial chaperones Raf1 and RbcX into plants, especially when cyanobacterial RuBisCO is incorporated. Notably, introducing S. elongatus PCC 7942 RuBisCO, together with RbcX and CcmM35 into tobacco chloroplast supported the autotrophic photosynthesis16, which provided an initial successful trial of replacing plant RuBisCO. However, for proper assembly and functionality of carboxysome in C3 plants, more investigations are needed to comprehend the coordinated action of cyanobacterial and chloroplast-specific chaperones on RuBisCO assembly and carboxysome biogenesis in plants.
In sum, our present findings, together with previous reports, enable us to better understand the molecular insights into RuBisCO assembly and carboxysome biogenesis in different cyanobacterial strains10,15,24,25,36,37. The chaperones Raf1 and RbcX act in concert to regulate multiple stages of cyanobacterial RuBisCO assembly and condensation. Formation of a dynamic ternary complex L8F8X16, via simultaneous binding of Raf1 and RbcX to RbcL, facilitates the release of RbcL8 core from the chaperones. Moreover, beyond functioning as a late-stage assembly factor to promote the formation of RbcL8 core, RbcX is a major contributor to regulating the dynamic balance between RuBisCO holoenzymes and condensates. These findings provide an advanced understanding of the fine functions of cyanobacterial chaperones Raf1 and RbcX in RuBisCO assembly and carboxysome formation, which will guide the design and engineering of a more efficient RuBisCO and/or carboxysome in plants for enhanced carbon fixation and agricultural productivity.
Materials and methods
Cloning and plasmids
The coding regions of RbcL, Raf1, RbcX, RbcS, GroEL-GroES, and CcmM35 were amplified from the genomic DNA of cyanobacteria Anabaena sp. PCC 7120 or S. elongatus PCC 7942, and were cloned into the pET19 and/or pCDFDuet vectors using the homologous recombination methods. The details of plasmids and protein sequences used in this study are listed in Supplementary Table S2.
Protein expression and purification
The ternary complex of Anabaena sp. PCC 7120 RbcL–Raf1–RbcX was obtained by co-expressing the plasmids of pCDFduet-GroEL-GroES-RbcX-Raf1 and pET19-His-RbcL in E. coli BL21 (DE3) strain. The transformed cells were cultured at 37 °C in 6 L Luria-Bertani medium (10 g NaCl, 10 g Bacto Tryptone, and 5 g yeast extract/L) containing ampicillin of 50 μg/mL and spectinomycin of 100 μg/mL to an A600 nm of 0.8, and then induced with 0.2 mM isopropyl-β-D-thiogalactoside for a further 20 h at 16 °C. The cells were harvested by 5 min of centrifugation at 8000× g, resuspended in 40 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 20 mM NaCl, 5 mM MgCl2), and disrupted by the Ultrasonic Cell Disruptor (SONICS). After centrifugation at 12,000× g for 30 min, the supernatant was loaded onto a Ni-NTA column (Qiagen) pre-equilibrated with the binding buffer (50 mM Tris-HCl, pH 8.0, 20 mM NaCl, 5 mM MgCl2). The target protein was eluted with the binding buffer containing 500 mM imidazole, and further purified by gel filtration (Superdex 200 Increase 10/300, GE Healthcare) in the binding buffer. The peak fractions containing the complex of RbcL–Raf1–RbcX were collected by monitoring the absorbance at 280 nm, and concentrated to 2 mg/mL for cryo-EM analysis or 10 mg/mL for biochemical assays by 100 kDa cut-off concentrators.
The Anabaena sp. PCC 7120 RbcL–Raf1 complex was obtained by co-transforming the plasmids of pET19-His-RbcL–Raf1 and pCDFduet-GroEL-GroES into E. coli (DE3) strain. The cells were cultured, overexpressed, and purified following a previously described protocol10. The target proteins were flash-frozen in liquid nitrogen and stored at −80 °C for further use.
The Anabaena sp. PCC 7120 RuBisCO holoenzyme was obtained by co-purifying the Flag-RbcS and RbcL–Raf1–RbcX complex, in which the Raf1 protein is from S. elongatus PCC 7942. The Flag-RbcS and RbcL–Raf1–RbcX complex were overexpressed in E. coli BL21 (DE3) cells with transformed plasmids of pET19-Flag-RbcS and pCDFduet-GroEL-GroES-RbcX-Raf1/pET19-His-RbcL, respectively. The harvested cells were mixed and disrupted by the Ultrasonic Cell Disruptor (SONICS). The following purification procedures were the same as that described for Anabaena sp. PCC 7120 RbcL–Raf1–RbcX complex. Notably, the purified Anabaena sp. PCC 7120 RuBisCO holoenzyme is only of little amount, which was only applicable in the native-PAGE analysis as a control marker.
The recombinant Anabaena sp. PCC 7120 RbcX, RbcXE32A&R69A, RbcS, and CcmM35 proteins were overexpressed in E. coli BL21 (DE3) strain using the plasmids of pET19-His-RbcX, pET19-His-RbcXE32A&R69A, pET19-His-RbcS, and pET19-His-CcmM35, respectively. They were purified and stored as described previously10. The recombinant Anabaena sp. PCC 7120 Flag-RbcS protein was expressed in E. coli BL21 (DE3) strain by transforming plasmid pET19-Flag-RbcS, and purified via affinity chromatography with anti-FLAG M2 gel (Sigma) and size-exclusion chromatography with Superdex 75 Increase 10/300 (GE Healthcare). These proteins were concentrated to 10 mg/mL for biochemical assays by centrifugation with 10 kDa cut-off concentrators.
The recombinant proteins of S. elongatus PCC 7942 RuBisCO holoenzyme, eGFP-RuBisCO, Raf1, RbcX, and CcmM35 were overexpressed, purified, and concentrated the same as those described in our previous report10.
The protein concentration was determined using a NanoDrop (Thermo Fisher Scientific), and the purity was assessed by SDS-PAGE.
Cryo-EM sample preparation, data collection, and processing
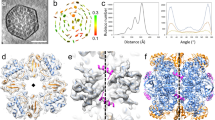
The purified ternary complex of Anabaena sp. PCC 7120 RbcL–Raf1–RbcX was concentrated to ~2 mg/mL. An aliquot of 3.5 μL of the sample was applied to glow-discharged Quantifoil R1.2/1.3 300-mesh Cu Holey Carbon Grids. The grids were blotted for 4 s with a blot force of 2 and a wait time of 20 s, and then plunged into liquid ethane using a Vitrobot Mark IV (FEI) at 4 °C and 100% humidity. The cryo-EM data sets were collected by a 300 keV Titan Krios electron microscope (FEI) at the Center for Integrative Imaging, University of Science and Technology of China. Totally, 1780 micrograph stacks (32 frames, each 0.17 s, 9 e/Å2/s, total dose ~50 e/Å2) were recorded with a K2 Summit direct electron detector (Gatan) at the super-resolution mode in a nominal magnification of 22,500× with a defocus range from −1.0 to −2.0 μm. All stacks were motion-corrected and dose weighted using MotionCor2 (version 1.3.1)38, and binned 2-fold to yield a pixel size of 1.01 Å. The defocus values were estimated using CTFFIND4 (version 4.1)39.
After manual removal of bad images, a total of 308,714 particles were automatically picked from 1538 images using RELION (version 3.0)40. Then, these particles were boxed and binned 4-fold for 2D classification. 207,936 particles from all good classes were subjected to the 3D classification with C4 symmetry, during which particles were classified into 4 classes. 54,260 particles from one class that shows clear features of the ternary complex were re-extracted without binning for the 3D auto-refinement with D4 symmetry, yielding a density map with an overall resolution of 3.3 Å after post-processing.
Model building of L8F8X16 was performed using Chimera41 by manually fitting the L8F8 structure (PDB, 6KKM) into the map. The Raf1β domains were not modeled in the final structure due to the dispersed density. Moreover, the Raf1α domains were manually adjusted to best fit the map. Then, the structure of RbcX was manually built into the map using Chimera by fitting the Anabaena sp. CA RbcX structure (PDB, 2PEO) into the extra density of the map. The model was manually refined by COOT (version 0.8.9)42, followed by the iterative positional and B-factor refinement in real space using PHENIX (version 1.14)43. The final structure showed good geometry and was further evaluated using MolProbity44 (http://molprobity.biochem.duke.edu).
The flowchart of cryo-EM data processing is shown in Supplementary Fig. S10. The parameters of cryo-EM data collection, processing, structure determination, and refinement are listed in Supplementary Table S3.
RuBisCO assembly assays
The purified L8F8, L8F8X16, RbcX, RbcXE32A&R69A, and Flag-RbcS were concentrated to 15, 15, 10, 10, and 3 mg/mL, respectively, for the RuBisCO assembly assays. All measurements were performed at 25 °C in the buffer containing 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM MgCl2, 5% glycerol. First, 0.5 µM L8F8 in the absence of RbcX2 or L8F8X16 alone was added to the solution containing RbcS at various concentrations (0, 0.5, 1, 2, 5, 10-fold to RbcL). Then, after incubation for about half an hour, 10 μL of each sample was mixed with the loading buffer and applied to native-PAGE analysis (6% Bis-Tris and boric acid). The protein compositions in the corresponding bands of the native-PAGE were further analyzed by SDS-PAGE. Moreover, 0.5 µM RbcL–Raf1 was pre-incubated with 8 µM RbcX or RbcXE32A&R69A (2 fold to RbcL) for half an hour before being added to the solution containing RbcS at various concentrations, and then applied to native-PAGE analyses. In addition, 0.5 µM RbcL–Raf1 in the presence of RbcX2 at various concentrations (0, 0.05, 0.1, 0.2, 0.5, 1 fold to RbcL) was added to the solution containing 4 µM RbcS, and also applied to native-PAGE analyses. The purified Anabaena sp. PCC 7120 RuBisCO holoenzyme was applied to the native-PAGE as a positive control, and the purified RbcL–Raf1, RbcL–Raf1-RbcX, RbcXHis and Flag-RbcS proteins were also applied to the SDS-PAGE as positive controls.
RuBisCO carboxylase activity assays
The HMM complexes analyzed by native-PAGE were also applied to the RuBisCO carboxylase activity assays. First, the RbcL–Raf1 solutions with or without RbcX2 at equal molarity were mixed with RbcS at increasing concentrations (0, 0.5, 1, 2, 5, 10-fold to RbcL). After incubation of the 100 µL reaction mixture for ~0.5 h at 25 °C, 10 μL of each sample was applied to the RuBisCO carboxylase activity assays. The amount of RbcL, corresponding to the number of active sites, is equal in all samples. The RuBisCO holoenzyme, which was used as a positive control, also contains the same amount of RbcL proteins. All the assays were tested at 25 °C for the RuBisCO carboxylase activity, using a commercial RuBisCO assay kit (BC0445, Solarbio Life Science Co., Beijing, China) according to the instruction of the manufacturer. Using a Beckman DU800 spectrophotometer, the RuBisCO carboxylase activity was measured at 340 nm in the unit of U/mg, which represents the oxidation of 1 nmol of NADH per min. The activity of each sample was repeated for three times.
Turbidimetric assays
The turbidity of the solution was measured by monitoring the absorbance at 340 nm using a Beckman DU800 spectrophotometer. The purified Anabaena sp. PCC 7120 L8F8, L8F8X16, CcmM35, RbcX2 and RbcS proteins were concentrated to 15, 10, 10, 10 and 5 mg/mL, respectively, for the turbidimetric assays. All measurements were performed at 25 °C in the buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM MgCl2. Two groups of experiments were designed to detect the effect of RbcX on RuBisCO condensate formation. First, the RuBisCO condensation was triggered by mixing 0.5 µM L8F8, 4 µM RbcS, and 4 µM CcmM35. After the condensates reached the maximum turbidity in ~5 min, RbcX2 at various concentrations (0, 0.2, 0.4, 0.8, 2, and 4 µM) was added to the solution. Second, 0.5 µM L8F8X16 was added in the solution containing RbcS at various concentrations (0, 2, 4, 8, 20 and 40 µM). After incubation for ~30 min, 4 µM CcmM35 was added to the solution. During the experiments, the absorbance at 340 nm was monitored along the time to indicate turbidity. The RbcL–Raf1 complexes without RbcS and CcmM35 proteins, which could not form turbidity, were used as a negative control.
The purified S. elongatus PCC 7942 RuBisCO holoenzyme, CcmM35, Raf1, and RbcX2 proteins were concentrated at 12, 10, 20, and 20 mg/mL, respectively, for the assays. All purified proteins were assessed by SDS-PAGE (Supplementary Fig. S11). All measurements were performed at 25 °C in the buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM MgCl2. The RuBisCO condensation was triggered by mixing 0.25 µM RuBisCO and 2 µM CcmM35. After the condensates reached the maximum turbidity in ~4 min, Raf1, RbcX2, and Raf1&RbcX2 at various concentrations (0, 1, 2, 4, 10, and 20 µM) were added to the solution, and the turbidities were monitored at 340 nm over time. The graphs were plotted using the Origin Pro software.
Laser-scanning confocal microscopy
Three groups of condensation experiments were performed in the same manner as described for the turbidimetric assays. However, the S. elongatus PCC 7942 RuBisCO was replaced with eGFP-RuBisCO, in which an eGFP tag was fused to the N-terminus of RbcL. After incubating for 20 min, the reaction mixtures were immediately imaged by a laser-scanning confocal microscope (ZEISS LSM710). The 20 μL samples were transferred to a glass-bottom cell culture dish and excited with a laser at 488 nm for green fluorescence imaging. Images were recorded by focusing on the bottom of the dish using Axio Observer Z1 microscope with a Plan-Apochromat 20×/0.8 M27 objective.
Data availability
The cryo-EM structure of L8F8X16 has been deposited at PDB under the accession code of 7XSD. The cryo-EM density map of L8F8X16 has been deposited at the Electron Microscopy Data Bank (EMD-33524).
References
Bar-On, Y. M. & Milo, R. The global mass and average rate of RuBisCO. Proc. Natl. Acad. Sci. USA 116, 4738–4743 (2019).
Ellis, R. J. The most abundant protein in the world. Trends Biochem. Sci. 4, 241–244 (1979).
Andersson, I. & Backlund, A. Structure and function of RuBisCO. Plant Physiol. Biochem. 46, 275–291 (2008).
Whitney, S. M., Houtz, R. L. & Alonso, H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, RuBisCO. Plant Physiol. 155, 27–35 (2011).
Hartman, F. C. & Harpel, M. R. Structure, function, regulation, and assembly of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63, 197–234 (1994).
Hauser, T., Popilka, L., Hartl, F. U. & Hayer-Hartl, M. Role of auxiliary proteins in RuBisCO biogenesis and function. Nat. Plants 1, 15065 (2015).
Hauser, T. et al. Structure and mechanism of the RuBisCO-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 22, 720–728 (2015).
Hayer-Hartl, M. & Hartl, F. U. Chaperone machineries of RuBisCO-the most abundant enzyme. Trends Biochem. Sci. 45, 748–763 (2020).
Goloubinoff, P., Gatenby, A. A. & Lorimer, G. H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337, 44–47 (1989).
Xia, L. Y. et al. Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1. Nat. Plants 6, 708–717 (2020).
Saschenbrecker, S. et al. Structure and function of RbcX, an assembly chaperone for hexadecameric RuBisCO. Cell 129, 1189–1200 (2007).
Liu, C. et al. Coupled chaperone action in folding and assembly of hexadecameric RuBisCO. Nature 463, 197–202 (2010).
Bracher, A., Starling-Windhof, A., Hartl, F. U. & Hayer-Hartl, M. Crystal structure of a chaperone-bound assembly intermediate of form I RuBisCO. Nat. Struct. Mol. Biol. 18, 875–880 (2011).
Kolesinski, P., Belusiak, I., Czarnocki-Cieciura, M. & Szczepaniak, A. RuBisCO accumulation factor 1 from Thermosynechococcus elongatus participates in the final stages of ribulose-1,5-bisphosphate carboxylase/oxygenase assembly in Escherichia coli cells and in vitro. FEBS J. 281, 3920–3932 (2014).
Huang, F. et al. RuBisCO accumulation factor 1 (Raf1) plays essential roles in mediating RuBisCO assembly and carboxysome biogenesis. Proc. Natl. Acad. Sci. USA 117, 17418–17428 (2020).
Lin, M. T., Occhialini, A., Andralojc, P. J., Parry, M. A. & Hanson, M. R. A faster RuBisCO with potential to increase photosynthesis in crops. Nature 513, 547–550 (2014).
Orr, D. J. et al. Hybrid cyanobacterial-tobacco RuBisCO supports autotrophic growth and procarboxysomal aggregation. Plant Physiol. 182, 807–818 (2020).
Newman, J., Branden, C. I. & Jones, T. A. Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC 6301. Acta Crystallogr. D. Biol. Crystallogr. 49, 548–560 (1993).
Wang, J. et al. The quantitative proteome atlas of a model cyanobacterium. J. Genet. Genomics 49, 96–108 (2022).
Kerfeld, C. A. & Melnicki, M. R. Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75 (2016).
Wang, H. et al. RuBisCO condensate formation by CcmM in beta-carboxysome biogenesis. Nature 566, 131–135 (2019).
Duff, A. P., Andrews, T. J. & Curmi, P. M. The transition between the open and closed states of RuBisCO is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 298, 903–916 (2000).
Feiz, L. et al. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 24, 3435–3446 (2012).
Huang, F. et al. Roles of RbcX in carboxysome biosynthesis in the cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 179, 184–194 (2019).
Emlyn-Jones, D., Woodger, F. J., Price, G. D. & Whitney, S. M. RbcX can function as a RuBisCO chaperonin, but is non-essential in Synechococcus PCC 7942. Plant Cell Physiol. 47, 1630–1640 (2006).
Aigner, H. et al. Plant RuBisCO assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278 (2017).
Faulkner, M. et al. Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9, 10662–10673 (2017).
Savage, D. F., Afonso, B., Chen, A. H. & Silver, P. A. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327, 1258–1261 (2010).
Cameron, J. C., Wilson, S. C., Bernstein, S. L. & Kerfeld, C. A. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155, 1131–1140 (2013).
Gonzalez-Esquer, C. R., Newnham, S. E. & Kerfeld, C. A. Bacterial microcompartments as metabolic modules for plant synthetic biology. Plant J. 87, 66–75 (2016).
Hanson, M. R., Lin, M. T., Carmo-Silva, A. E. & Parry, M. A. J. Towards engineering carboxysomes into C3 plants. Plant J. 87, 38–50 (2016).
Rae, B. D. et al. Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants. J. Exp. Bot. 68, 3717–3737 (2017).
Brutnell, T. P., Sawers, R. J., Mant, A. & Langdale, J. A. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849–864 (1999).
Doron, L., Segal, N., Gibori, H. & Shapira, M. The BSD2 ortholog in Chlamydomonas reinhardtii is a polysome-associated chaperone that co-migrates on sucrose gradients with the rbcL transcript encoding the RuBisCO large subunit. Plant J. 80, 345–355 (2014).
Wostrikoff, K. & Stern, D. RuBisCO large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104, 6466–6471 (2007).
Kolesinski, P., Rydzy, M. & Szczepaniak, A. Is RAF1 protein from Synechocystis sp. PCC 6803 really needed in the cyanobacterial RuBisCO assembly process? Photosyn. Res. 132, 135–148 (2017).
Onizuka, T. et al. The rbcX gene product promotes the production and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase of Synechococcus sp. PCC 7002 in Escherichia coli. Plant Cell Physiol. 45, 1390–1395 (2004).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank Dr. Yong-Xiang Gao for technical support on cryo-EM data collection at the Cryo-EM Center at the University of Science and Technology of China. We also thank Dr. Zhen-Bang Liu at the Core Facility Center for Life Sciences at the University of Science and Technology of China, for the technical assistance with confocal imaging. This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (http://www.cas.cn; XDA24020302 and XDB37020301), the National Natural Science Foundation of China (http://www.nsfc. gov.cn; 32171198), Anhui Provincial Natural Science Foundation (http://kjt.ah.gov.cn; 2108085J14) and the Key R&D Projects of Anhui Province (http://kjt.ah.gov.cn; 2022l07020034). Y.-L.J. thanks the Youth Innovation Promotion Association of Chinese Academy of Sciences for their support (Membership No. 2020452).
Author information
Authors and Affiliations
Contributions
C.-Z.Z., Y.-L.J., and Q.L. conceived, designed, and supervised the project. Y.-L.J., C.-Z.Z., Q.L., L.-Y.X., and Y.C. analyzed the data. Y.-L.J., and C.-Z.Z. wrote the manuscript. Q.L. and L.-Y.X. performed protein purification and biochemical assays. Y.-L.J. and L.-Y.X. conducted cryo-EM sample preparation, data collection, structure determination, and model refinement. All of the authors discussed the data and read the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Q., Jiang, YL., Xia, LY. et al. Structural insights into cyanobacterial RuBisCO assembly coordinated by two chaperones Raf1 and RbcX. Cell Discov 8, 93 (2022). https://doi.org/10.1038/s41421-022-00436-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-022-00436-9
This article is cited by
-
Structure and assembly of the α-carboxysome in the marine cyanobacterium Prochlorococcus
Nature Plants (2024)
-
Towards engineering a hybrid carboxysome
Photosynthesis Research (2023)