Abstract
Aerobic methanotrophic bacteria are considered strict aerobes but are often highly abundant in hypoxic and even anoxic environments. Despite possessing denitrification genes, it remains to be verified whether denitrification contributes to their growth. Here, we show that acidophilic methanotrophs can respire nitrous oxide (N2O) and grow anaerobically on diverse non-methane substrates, including methanol, C-C substrates, and hydrogen. We study two strains that possess N2O reductase genes: Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6. We show that N2O respiration supports growth of Methylacidiphilum caldifontis at an extremely acidic pH of 2.0, exceeding the known physiological pH limits for microbial N2O consumption. Methylocella tundrae simultaneously consumes N2O and CH4 in suboxic conditions, indicating robustness of its N2O reductase activity in the presence of O2. Furthermore, in O2-limiting conditions, the amount of CH4 oxidized per O2 reduced increases when N2O is added, indicating that Methylocella tundrae can direct more O2 towards methane monooxygenase. Thus, our results demonstrate that some methanotrophs can respire N2O independently or simultaneously with O2, which may facilitate their growth and survival in dynamic environments. Such metabolic capability enables these bacteria to simultaneously reduce the release of the key greenhouse gases CO2, CH4, and N2O.
Similar content being viewed by others
Introduction
Anthropogenic emissions of greenhouse gases (GHGs)—primarily carbon dioxide (CO2), methane (CH4), and nitrous oxide (N2O)—are responsible for a historically rapid increase in Earth’s average annual temperature of more than 0.2 °C per decade1,2. In addition to achieving net-zero CO2 emissions by 2050, significant reductions in the emissions of other GHGs including CH4 and N2O are now critically needed. Compared to CO2, the warming effect of CH4 is around 28 to 34 times greater3,4. However, its much shorter mean lifetime of approximately 12–13 years5 provides an additional opportunity to mitigate future climate change. Like CO2, N2O—the third most important GHG—has a long half-life (roughly 120 years) in the atmosphere6, and its warming potential is about 300 times greater than CO2 over a 100-year time scale1. In addition, N2O is a major cause of ozone depletion in the stratosphere7,8.
Although human activities are by far the most important reason for the unprecedented rise in atmospheric GHGs9, microbial activities also play a direct role in this rise10,11. GHG net accumulation is regulated by the biogeochemical source-sink dynamics of GHGs exchanged between terrestrial, marine, and atmospheric reservoirs9. GHG production and consumption in both natural and anthropogenic ecosystems are driven primarily by microbes10,12. Methane fluxes in natural environments are controlled by activities of methane-producing (methanogenic) and methane-consuming (methanotrophic) microorganisms. It is estimated that 69% of the atmospheric CH4 budget originates from microbial activities (methanogenesis) while about 50−90% of the produced CH4 is oxidized by methanotrophs before reaching the atmosphere13,14.
Microbes can oxidize methane under aerobic and anaerobic conditions. Aerobic methanotrophs oxidize methane to methanol by employing either particulate methane monooxygenases (pMMO) or soluble methane monooxygenases (sMMO)15. There are two ways in which aerobic methanotrophs use molecular oxygen (O2): as the terminal electron acceptor of aerobic respiration and for methane activation via the methane monooxygenase15. Under strictly anoxic conditions, anaerobic methanotrophic microorganisms mitigate CH4 emissions by oxidizing methane with alternative terminal electron acceptors including NO3−, Fe3+, Mn4+, SO42-, and humic acid using reverse methanogenesis pathways16,17,18,19. Furthermore, intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium ‘Candidatus Methylomirabilis oxyfera’ using pMMO was reported20.
Interestingly, the genomes of some aerobic methanotrophs encode denitrification enzymes including nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), and N2O reductases21,22,23,24. Surprisingly, however, none of the methanotroph genomes or MAGs known to date encode a complete set of denitrification genes (Supplementary Dataset 1). Kits and colleagues21,22 demonstrated that some aerobic methanotrophs can couple NO3− and NO2− reduction to the oxidation of methane and other electron donors, including methanol, formaldehyde, formate, ethane, ethanol, and ammonia in suboxic conditions. However, whether these aerobic methanotrophs are capable of anaerobic growth with NO3− and NO2− as terminal electron acceptors remain to be seen.
More than two-thirds of N2O emissions arise from bacterial and fungal denitrification and nitrification processes in soils25,26. N2O emissions are a major concern in acidic environments due to the high production of N2O via abiotic reactions and the inhibition of biological N2O reduction27,28. Although multiple sources of N2O exist25, there is only one known sink for N2O in the biosphere—the microbial reduction of N2O to N2, catalyzed by a copper-dependent enzyme, N2O reductase (N2OR) encoded by nosZ29. The NosZ enzymes found in prokaryotes are phylogenetically classified into two clades: the canonical NosZ (clade I NosZ), found mostly in denitrifiers30, and the recently described cNosZ (clade II NosZ)31, which has an additional c-type heme domain at the C terminus, found commonly in non-denitrifiers31,32. Thus, bacteria and archaea harboring the nosZ-type genes, in particular those classified as incomplete- or non-denitrifiers because they do not encode the full denitrification pathway, are receiving increasing attention in the search for technologies to combat N2O emissions32. Previous studies have reported the presence of the nosZ gene in the aerobic methanotrophs, Methylocystis sp. SC2 (ref. 23) and Methylocella tundrae24. Further genomic analysis from this study suggests that this enzyme is present in some other aerobic methanotrophs, too (Supplementary Dataset 1). Pure culture studies have unequivocally shown that denitrifiers can grow by respiring N2O (refs. 33,34). Moreover, an electron sink/spill role for N2OR has been proposed for Gemmatimonas aurantiaca T-27 (ref. 35) without biomass production (i.e., growth). Despite the presence of N2OR in Methylocystis sp. SC2, its ability to grow in anoxia under N2O-reducing conditions is unverified36. Thus, the ability to grow by converting N2O to N2 has not yet been reported for any of the known aerobic or anaerobic methanotrophs, even with non-methane substrates such as methanol.
Methanotrophs using MMO enzymes are considered to be obligate aerobes. Paradoxically, however, they are often detected at high relative abundance in extremely hypoxic and even anoxic zones of peat bogs, wetlands, rice paddies, forest soils, and geothermal habitats37,38. It is therefore critical to investigate the ability of aerobic methanotrophs to use N2O as the sole terminal electron acceptor for energy conservation and biomass production, a metabolic trait that could allow them to thrive in these anoxic ecosystems. Here, we used a multi-faceted approach to investigate the role of N2O respiration in defining the physiology and ecology of selected aerobic methanotrophs. Growth experiments demonstrated that the presence of N2OR in an acidophilic proteobacterial methanotroph, Methylocella tundrae T4, and an extremely acidophilic verrucomicrobial methanotroph, Methylacidiphilum caldifontis IT6, enables these organisms to respire N2O and to produce biomass while oxidizing a wide variety of electron donors, including methanol, acetol, pyruvate, and hydrogen. In contrast to N2O, respiration of NO3− and NO2− did not support anaerobic growth of these methanotrophs on C1 substrates. We also demonstrate that Methylocella tundrae T4 can reduce both O2 and N2O simultaneously, allowing it to oxidize more CH4 and generate more biomass under O2-limiting conditions. Our findings significantly expand the potential ecological niche of aerobic methanotrophs and reveal that some methanotrophic microbial strains could be used to mitigate multiple GHG emissions.
Results and discussion
N2OR-encoding genes in aerobic methanotrophs
To identify methanotrophs capable of using N2O as an alternative electron acceptor, publicly available genomes and metagenome-assembled genomes (MAGs) of methanotrophs were screened for nosZ genes. We found genes encoding N2OR in genomes and MAGs of methanotrophs from three bacterial phyla: Pseudomonadota, Verrucomicrobiota, and Gemmatimonadota (Supplementary Dataset 1). They were confined to the alphaproteobacterial methanotrophs and absent in gammaproteobacterial methanotrophs in the case of the phylum Pseudomonadota and represented by only two genera, Methylocella and Methylocystis, which also accounted overall for the majority of the methanotroph genomes encoding nosZ. Similarly, nosZ genes were exclusively found in one representative genome in each of the phyla Verrucomicrobiota (represented by the genus Methylacidiphilum) and Gemmatimonadota (represented by the candidate genus ‘Methylotropicum’), respectively. Phylogenetic analysis of predicted NosZ protein sequences revealed that those found in Methylocella and Methylocystis are from the clade I NosZ lineage, while those found in Methylacidiphilum and ‘Ca. Methylotropicum’ are from the clade II NosZ lineage (Fig. 1, Supplementary Fig. 1).
The phylogenetic tree was constructed with IQ-TREE (IQ-TREE options: -B 1000 -m LG + F + R5) using aligned NosZ (details in Materials and Methods) and rooted at the mid-point. Bootstrap values ≥ 70% based on 1000 replications are indicated. The scale bar represents a 0.5 change per amino acid position. Organization of the nos operon in methanotrophic strains (labeled in blue text) and closely related non-methanotrophic bacteria are shown. The genes, represented by arrows, are drawn to scale. Homologs are depicted in identical colors. The NosZ amino acid sequences and gene arrangement information were retrieved using the following genome accessions: GCF_017310505.1, Methylacidiphilum caldifontis IT6; GCF_000010785.1, Hydrogenobacter thermophilus TK-6; GCF_011006175.1, Hydrogenobacter sp. T-8; GCF_900215655.1, Hydrogenobacter hydrogenophilus DSM 2913; GCF_000619805.1, Sulfurihydrogenibium subterraneum DSM 15120; GCF_000021565.1, Persephonella marina EX-H1; GCF_000022145.1, Anaeromyxobacter dehalogenans 2CP1; GCF_000013385.1, Anaeromyxobacter dehalogenans 2CP-C; GCF_003054705.1, Opitutus sp. ER46; GCF_000019965.1, Opitutus terrae PB90-1; GCF_901905185.1, Methylocella tundrae PC4; GCA_901905175.1, Methylocella tundrae PC1; CP139089.1, Methylocella tundrae T4; FO000002.1, Methylocystis sp. SC2; GCF_000025965.1, Aromatoleum aromaticum EbN1; GCF_022760775.1, ‘Candidatus Rhodoblastus alkanivorans’ PC3; GCF_000143145.1, Hyphomicrobium denitrificans ATCC 51888; GCF_000344805.1, Bradyrhizobium oligotrophicum S58; GCF_027923385.1, Methylocystis echinoides LMG27198; GCA_003963405.1, Methylocystis sp. AWTPI-1. * indicates that the nosZ genes are truncated due to genome fragmentation. Source Data contains genome annotation information for Methylocella tundrae T4, Methylacidiphilum caldifontis IT6, Methylocystis spp. (strains IM2, IM3, and IM4), and ‘Ca. Methylotropicum kingii’.
Three Methylocella tundrae strains: T4 (re-sequenced genome), PC1 (ref. 39), and PC4 (ref. 39), have nos gene clusters (NGC) (Fig. 1). These are incorporated into nosRZDFYLX operons in strains PC4 and T4 and a nosZDFYLX operon in strain PC1 (Fig. 1). Strain PC1 has truncated nosZ and missing nosR genes. This is most likely due to its genome being highly fragmented into several small contigs containing missing and truncated genes. The NGC composition and operon arrangement, nosRZDFYLX, were largely similar in the genomes of the six N2OR-containing Methylocystis species (Fig. 1), including Methylocystis sp. SC2 (ref. 23), Methylocystis echinoides LMG27198, three in-house Methylocystis echinoides-like isolates (strains IM2, IM3, and IM4), and a metagenome-assembled genome (MAG) of a Methylocystis sp. AWTPI-1 recovered from a water treatment facility40. A notable feature in their NGC organization was the absence of the gene encoding the membrane-anchored copper chaperon, NosL, which is primarily involved in Cu(I) delivery to apo-NosZ41. Methanotrophs with pMMO usually possess multiple copper chaperones42 that may complement NosL, making it non-essential for NosZ maturation. Altogether, the NGC in these alphaproteobacterial methanotrophs has a similar organization to those of clade I N2O-reducers (Fig. 1). BLAST results further revealed that the individual nos genes in the Methylocella and Methylocystis strains shared a high degree of similarity to each other and other non-methanotrophic Alphaproteobacteria (Supplementary Dataset 2). Also, their NosZ proteins share high homology with proteins annotated as twin-arginine translocation (Tat)-dependent N2OR (35–89%) and also possess the Tat signal peptide with a characteristic SRRx[F | L] motif43 found in clade I NosZ32.
The NGC in the genome of Methylacidiphilum caldifontis IT6 (ref. 44), comprises a nosCZBLDFYC operon (Fig. 1) but lacks the typical nosX and nosR found in clade I N2O-reducers31,32, involved in NosR maturation45 and electron transfer to NosZ46, respectively. Notably, the NGC (IT6_00904–11) was found within the cluster of genes (IT6_00903, IT6_00912–7) encoding alternative complex III (refer to Source Data for annotation information). Both the aa3-type and cbb3-type cytochrome c oxidase-encoding genes are also located next to these genes. Genes encoding two c-type cytochromes (nosC) within the nos operon (Fig. 1) could serve electron transport functions47. Interestingly, BLAST and synteny analyses of the NGC show that the individual genes are most closely related to genes found in genomes of extremely thermophilic Hydrogenobacter species of the phylum Aquificota (amino acid identities of 72.41–91.96%) (Supplementary Dataset 2) with a similar genetic organization (Fig. 1). Strain IT6 NosZ shares high similarities to proteins annotated as Sec-dependent N2OR (35–89%) with an N-terminal Sec-type signal peptide found in clade II NosZ31,32; the highest identities (79–89%) were with NosZ proteins from other Hydrogenobacter species. Hydrogenobacter thermophilus TK-6, a hydrogen-oxidizing bacterium, can completely denitrify NO3- to N2 gas48, indicating the presence of a functional N2OR. As a result, Methylacidiphilum caldifontis IT6 may also have a functional N2OR due to the high similarity of its NGC to those of Hydrogenobacter species. Although genomes of other Methylacidiphilum species, including Methylacidiphilum fumariolicum, lacked the gene encoding the N2OR catalytic subunit, NosZ, some genes encoding Nos accessory proteins were found (Supplementary Dataset 2). Interestingly, the N2OR genes for Methylacidiphilum caldifontis IT6 were found in a genomic island (Supplementary Dataset 3) and were most likely acquired through horizontal gene transfer, which is consistent with its NosZ phylogeny (Fig. 1, Supplementary Fig. 1). This is not surprising since many key metabolic genes in verrucomicrobial methanotrophs, including those encoding the MMO, are believed to have been acquired through horizontal gene transfer49. As a result, Methylacidiphilum fumariolicum strains might have acquired the NGC before losing the key functional genes but retaining some of the accessory genes. Finally, we found a nosZBDF operon in the MAG of the uncultured methanotrophic bacterium ‘Ca. Methylotropicum kingii’50 that resembles clade II NGC, with a truncated nosZ and multiple missing genes like nosY, nosL, and nosC (Fig. 1). These are also likely the result of multiple MAG fragmentations. Multiple sequence alignments of the predicted NosZ proteins of methanotrophs and other microorganisms (clade I and II) were constructed. All the expected metal-binding residues present in N2OR were mostly conserved in the methanotroph NosZ sequences (Supplementary Fig. 2, Supplementary Note 1).
N2O-dependent anaerobic growth of methanotrophs
The presence of genes predicted to encode N2OR in the genomes of Methylocella tundrae strains, Methylacidiphilum caldifontis IT6, and Methylocystis strains (SC2, IM2, IM3, and IM4) (Supplementary Datasets 1, 2) led us to investigate whether this enzyme can support the anaerobic growth of these aerobic methanotrophs when N2O is supplied as their sole electron acceptor. Physiological studies on N2O reduction by methanotrophs focused on Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 since preliminary experiments showed that the N2OR-containing Methylocystis strains, including Methylocystis sp. SC2 and the in-house Methylocystis strains (IM2, IM3, and IM4) failed to reduce N2O under various anoxic growth conditions. We set up anoxic batch cultures of Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 using methanol as a sole electron donor with or without N2O as the sole electron acceptor. For these incubations, 2 mM ammonium (NH4+) was used as the nitrogen source instead of NO3− to avoid the involvement of dissimilatory nitrate reduction particularly in the Methylocella strains with nitrate-reducing potential. As a negative control, closely related methanotrophs lacking a predicted N2OR (Methylocella silvestris BL2 and Methylacidiphilum infernorum IT5, respectively) were included in the study design. The growth experiments were conducted in LSM medium at pH 5.5 for Methylocella species (strains T4 and BL2) and at pH 2.0 for Methylacidiphilum species (strains IT5 and IT6). As expected, in control incubations provided with O2 as the terminal electron acceptor, all four strains grew on CH3OH (Fig. 2A, D, G, J). In these controls, the maximum specific growth rates (µmax) of the Methylocella strains (strain T4: µmax = 2.83 ± 0.03 d−1; strain BL2: µmax = 1.79 ± 0.05 d−1) were higher than those of the Methylacidiphilum strains (strain IT6: µmax = 1.57 ± 0.04 d−1; strain IT5: µmax = 1.49 ± 0.01 d−1).
Methylocella tundrae T4, Methylocella silvestris BL2, Methylacidiphilum caldifontis IT6, and Methylacidiphilum infernorum IT5 cells were grown in LSM medium supplemented with 30 mM methanol as the electron donor and NH4+ as the N-source. Aerobic growth of the 4 strains with O2 (A, D, G, J), anaerobic growth with N2O (B, E, H, K), and anaerobic growth without N2O (C, F, I, L) as the sole terminal electron acceptor were determined by optical density measurements at 600 nm, followed by measurements of O2 and N2O consumption in the headspaces of the culture bottles. Note that the trace O2 present at the start of the incubation in the anaerobic cultures without N2O did not contribute to obvious growth (C, F, I, L). All experiments were performed in triplicates. Data are presented as mean ± 1 standard deviation (SD), and the error bars are hidden when they are smaller than the width of the symbols. Source data are provided as Source Data file.
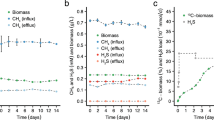
Under N2O-containing anoxic conditions, Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 reduced N2O and grew on methanol (Fig. 2B, H). When N2O was depleted, the growth of strains T4 and IT6 ceased. To verify that OD600 measurements indicated anaerobic cell growth rather than an artifact such as exopolysaccharide production, we demonstrated that cell counts and counts of 16S rRNA genes increased in parallel with OD600 during anaerobic growth (Supplementary Fig. 3). No growth was observed in N2O-free anoxic conditions used as negative controls (Fig. 2C, I). These results demonstrate that the anaerobic growth of these methanotrophs was dependent on N2O as the sole electron acceptor. The observed N2O reduction was catalyzed by a functional respiratory N2OR, as the N2OR-lacking relatives (Methylacidiphilum infernorum IT5 and Methylocella silvestris BL2) used as negative controls did not grow or reduce N2O under anoxic conditions (Fig. 2E, F, K, L). In addition, other known electron donors of Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6, which support their aerobic growth44,51,52, also supported their growth under anoxic N2O-reducing conditions (Supplementary Dataset 4). Methylocella tundrae T4 grew on pyruvate and acetol, while Methylacidiphilum caldifontis IT6 grew on acetol under anoxic N2O-reducing conditions. Further, molecular hydrogen supported the chemolithoautotrophic growth of Methylacidiphilum caldifontis IT6 as the sole electron donor under anoxic N2O-reducing conditions (Supplementary Fig. 4). The transcriptomic analysis (see below) suggests that the group 1d [NiFe] hydrogenase encoded in the genome of Methylacidiphilum caldifontis IT6 could be involved in chemolithoautotrophic growth under anoxic N2O respiring conditions.
Methylocella tundrae T4 exhibited a higher growth rate (µmax = 0.47 ± 0.02 d−1) than Methylacidiphilum caldifontis IT6 (µmax = 0.18 ± 0.01 d−1) on methanol and N2O. However, these values are approximately 6 and 9 times, respectively, lower than the growth rates measured for both strains under O2−respiring conditions. Biomass yields Yx/m (g DW⋅mol−1 N2O or O2 reduced) for the methanol-oxidizing cultures of strains T4 and IT6 reducing N2O as the sole electron acceptor were also lower than for cells reducing O2 as the sole electron acceptor. The biomass yield of Methylocella tundrae T4 cells grown anaerobically on N2O (4.64 ± 0.04 g DW⋅mol−1 N2O reduced) was approximately 45% of that of aerobically grown cells (10.41 ± 0.04 g DW⋅mol−1 O2 reduced). Similarly, Methylacidiphilum caldifontis IT6 had a biomass yield when grown anoxically on N2O (2.36 ± 0.04 g DW⋅mol−1 N2O reduced), which was only about 38% of that achieved by aerobically grown cells (6.27 ± 0.14 g DW⋅mol−1 O2 reduced). This improved molar yield on O2 is expected despite the higher reduction potential of N2O (see Eqs. [1] and [2]), since O2 respiration accepts twice as many electrons as N2O respiration (Eq. 1 and 2)53. In addition, the aerobic terminal oxidases of both strains are proton pumps and conserve energy (Supplementary Datasets 5, 6)54,55, whereas N2OR does neither56. To our knowledge, our results constitute the first report of N2O reduction coupled with anaerobic growth in any methanotroph.
It is well known that N2O reduction is generally inhibited at acidic pH (<6.0)57, resulting in N2O accumulation in acidic environments28,58. However, the current study revealed that two acidophilic methanotrophs, Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 can reduce N2O in moderately acidic (pH 5.5) and extremely acidic (pH 2.0) conditions, respectively. The existence of acid-tolerant N2O reducers (pH 4.0 to 6.0) has been proposed in soil microcosm and enrichment experiments59,60. So far, the only isolate implicated in N2O reduction at an acidic pH (5.7) is Rhodanobacter sp. C01 isolated from acidic soil in Norway61. Our study reveals that N2O reduction can occur even at an extremely acidic pH of 2.0. Furthermore, the conditions required for N2O reduction in the N2OR-containing Methylocystis strains remain unresolved. Perhaps some unknown growth or environmental factors are required to stimulate N2O respiration in these methanotrophs, which will require further investigation.
Nitrate and nitrite reduction in Methylocella species
No anoxic growth of Methylocella species with CH3OH and NO3 −
We next tested if the presence of denitrification enzymes in Methylocella tundrae T4 (nitrate reductase [NAR], nitric oxide reductase [NOR] and N2OR) and Methylocella silvestris BL2 (NAR, nitrite reductase [NIR], and NOR) (Supplementary Dataset 1) can equate to growth when NO3− or NO2− is used as the sole terminal electron acceptor. Indeed, the presence of NAR (and NIR) in these methanotrophs resulted in NO3− (and NO2−) reduction when methanol was provided as the sole electron donor. However, growth was barely detected under these conditions (Fig. 3A, B). Strain T4, which lacks a canonical NIR, reduced all the provided NO3− stoichiometrically to NO2− when provided with methanol as the sole electron donor (Fig. 3A). Under the same condition, strain BL2, a NAR and NIR-containing methanotroph, initially reduced the provided NO3− to NO2− and eventually, all the accumulated NO2− was stoichiometrically reduced to N2O towards the end of the incubation (Fig. 3B). These results demonstrate that these methanotrophs have a functional NAR and or NIR and can utilize NO3− and/or NO2− instead of O2 as a terminal electron acceptor. Nevertheless, these methanotrophs do not appear to rely on these activities for growth. Likewise, other aerobic methanotrophs have demonstrated denitrification activities under suboxic conditions. For example, the gammaproteobacterial methanotrophs Methylomonas denitrificans FJG1 and Methylomicrobium album BG8 were discovered to couple the oxidation of diverse electron donors to NO3− and NO2− reduction, respectively21,22. However, none of these strains was demonstrated to couple this activity to growth, prompting us to investigate the possible reasons behind the lack of growth (see below). It should be noted that the genomes of all known Methylacidiphilum strains lack genes encoding a respiratory NAR (Supplementary Dataset 1).
Methylocella tundrae T4 and Methylocella silvestris BL2 cells were grown in LSM medium supplemented with 30 mM methanol and 2–4 mM NO3−. NH4+ (2 mM) was supplied as the N-source. Anaerobic growth of Methylocella tundrae T4 (A) and Methylocella silvestris BL2 (B) cells on methanol as the sole electron donor with NO3− as the sole electron acceptor. Anaerobic growth of Methylocella tundrae T4 (C) and Methylocella silvestris BL2 (D) cells on pyruvate as the sole electron donor with NO3− as the sole electron acceptor. N2O produced from NO3− reduction by cells of Methylocella silvestris BL2 grown on methanol or pyruvate is shown as an inset plot within each figure. N2O production was not observed in strain T4, hence inset plots for N2O production were not displayed. Lower NO3− (ca. 2.0 mM) was used in the case of methanol (A) to avoid NO2− toxicity. Growth was determined by optical density measurements at 600 nm, followed by measurements of NO3- and NO2− concentrations. Data are presented as mean ± 1 SD of triplicate experiments, and the error bars are hidden when they are smaller than the width of the symbols. Source data are provided as Source Data file.
Toxicity of reactive nitrogen species for Methylocella species
Considering that methanol oxidation was coupled to N2O reduction and led to obvious growth in the N2OR-containing methanotrophs (Figs 2B, H), the lack of growth during NO3− reduction by these microorganisms is suspected to be caused by the accumulation of growth-arresting reactive nitrogen species (RNS) like NO2− and NO (refs. 62,63). Consistent with this hypothesis, the accumulation of NO2− in suboxic cultures of Vibrio cholerae and other bacterial species was found to limit population expansion but nitrate reduction still promoted cell viability64. NO2− typically accumulates due to a lack of functional NIR as observed for strain T4 (Fig. 3A) and, to some degree, even transiently accumulates in the presence of a functional NIR, as observed for strain BL2 (Fig. 3B). The impact of NO2− accumulated from NO3− reduction might be more severe in acidic environments since protonation of NO2− leads to the formation of free nitrous acid (FNA), a known inhibitor of microbial anabolic and catabolic processes65. In addition, chemodenitrification of NO2− (ref. 66) could result in an accumulation of NO in the cell environment, which is highly toxic to microbial life67. To further support the hypothesis of RNS toxicity, strain T4 was cultivated under N2O-reducing conditions with methanol as the sole electron donor and supplied with NO3− instead of NH4+ as the N source in the medium (Supplementary Fig. 5). Consistent with the idea that NO2− accumulation results in growth arrest, the culture growth plateaued at approximately the same time NO2− accumulated (≥ 0.3 mM NO2−) (Supplementary Fig. 5A), whereas in control cultures containing NH4+ instead of NO3− as the N-source, NO2− accumulation was not observed, and the cells were able to reach higher cell densities (Supplementary Fig. 5B). Furthermore, the effect of NO2− stress induced in strain T4 was verified by adding varying NO2− concentrations (0, 0.01, 0.03, 0.1, 0.3, and 1 mM) to aerobic (Supplementary Fig. 6A) and anaerobic N2O-respiring cultures (Supplementary Fig. 6B). Nitrite, particularly at concentrations higher than 0.3 mM at pH 5.5, induced stress in Methylocella tundrae T4, resulting in growth inhibition (Supplementary Fig. 6). These results are comparable to that of Methylophaga nitratireducenticrescens JAM1, a facultative methylotroph, which, when grown aerobically on methanol at pH 7.4, had a four-fold decrease in biomass in the presence of 0.36 mM NO2− and did not grow in the presence of 0.71 mM NO2− (ref. 68). Taken together, our data suggest that the failure of NO3−/NO2−-reducing methanotrophs to grow on methanol may result from RNS toxicity. On the other hand, when N2O is reduced to N2 by N2O-reducing methanotrophs, the creation of these RNS is avoided, which may explain the disparity in growth with N2O as the terminal electron acceptor compared to NO3− and NO2−.
Toxicity of C1 metabolites in nitrate-reducing Methylocella species
Aside from the inhibitory effects of RNS, toxic intermediates from methanol metabolism might synergistically contribute to the inability of methanotrophs to grow when respiring NO3−/NO2−. Although formaldehyde is a key intermediate in the C1 metabolic pathway in many methylotrophs, it is highly toxic69. Therefore, in situations where biomass production is limited due to RNS toxicity, it is likely that formaldehyde further retards the growth of denitrifying methanotrophs. To investigate this mechanism, we grew Methylocella strains under NO3−-reducing conditions using a C-C electron donor, pyruvate, which does not generate formaldehyde as a major metabolite (Figs. 3C, 3D). Eventually, nearly all the supplied NO3− was stoichiometrically converted to NO2− and N2O in strains T4 and BL2, respectively. In contrast to the lack of growth on methanol, pyruvate supported the growth of both Methylocella strains under NO3−-reducing conditions (Fig. 3C, D). Growth was more pronounced in strain BL2 than in strain T4 (Fig. 3C, D), possibly due to the presence of NIR and NOR in addition to NAR in strain BL2, which limited NO2− accumulation (Fig. 3D). Nonetheless, no further growth on pyruvate was observed in strain BL2 after day 5, despite reduction of the accumulated NO2− (~2.5 mM) to N2O (Fig. 3D). It is worth noting that the accumulated NO2− concentration (Fig. 3D) is higher than the 0.3 mM concentration that inhibited Methylocella tundrae T4 (Supplementary Fig. 6) and may also be responsible for the lack of growth in strain BL2.
Overall, these results demonstrate that in the tested Methylocella strains: (i) RNS have a major inhibitory effect on growth under denitrifying conditions; (ii) there are no growth benefits from methanol oxidation coupled to NO3− reduction, probably due to toxic C1 metabolic intermediates as well as RNS; and (iii) anaerobic growth is observed when NO3− reduction is coupled to the oxidation of pyruvate, a C-C electron donor; although the amount of growth is dependent on the completeness of the denitrification pathway and the accumulation of RNS. These propositions are supported by increased expression of genes involved in RNS and C1 metabolite detoxification under denitrifying conditions (see transcriptomic analysis below). Taken together, these results may explain why methanotrophs that couple methanol oxidation to NO3− or NO2− reduction show no clear signs of growth due to this process. Most methanotrophs can only utilize methane and its C1 derivatives as energy sources70 and thus should not be able to grow under denitrifying conditions21,22. On the other hand, versatile facultative methanotrophs of the genus Methylocella are potentially able to grow in strictly anoxic habitats when alternative multi-carbon substrates are available. In terrestrial environments, various nitrogen oxides, originating from nitrification and denitrification processes, coexist and are spatiotemporally dynamic71. Thus, depending on the versatility of NO2− and NO reduction potential of methanotrophs as well as their coexistence with other NO2− and NO-reducing microorganisms, N2O respiration can be supported or compromised (see Supplementary Figs 5, 6).
N2O reduction coupled with CH3OH or CH4 oxidation
N2O reduction kinetics
We investigated N2O respiration kinetics using resting cells of anaerobic N2O-respiring cultures (CH3OH + N2O) in a microrespirometry (MR) chamber. Harvested cells of strains Methylacidiphilum caldifontis IT6 and Methylocella tundrae T4 were dispensed into a closed 10-mL MR chamber outfitted with O2 and N2O-detecting microsensors, supplied with CH3OH (2 mM) and N2O as a sole electron donor and acceptor, respectively, and incubated anoxically. The N2O respiration kinetics followed Michaelis-Menten kinetics (Supplementary Fig. 7, Supplementary Note 2). The cells of strains T4 and IT6 grown at anoxic CH3OH + N2O conditions reduced N2O at a maximum rate of 1.122 ± 0.005 mmol N2O·h−1·g DW−1 (Supplementary Fig. 7A) and 0.414 ± 0.003 mmol N2O·h−1·g DW−1 (Supplementary Fig. 7B), respectively. The molar ratios of CH3OH to O2 and CH3OH to N2O consumed were approximately 1:1.0 ( ± 0.05; n = 3) and 1:2.04 ( ± 0.17; n = 3), respectively, which coincide with the theoretical values obtained from Eqs. 3 and 4.
Sensitivity of N2OR to O2
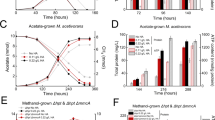
While O2 is well known to impair N2OR activity72, some bacterial strains have been reported to reduce N2O in the presence of O2 (refs. 73,74). We therefore tested the capacity of strains IT6 and T4 to reduce N2O in the presence of O2 by using resting cells of anoxic CH3OH + N2O cultures. After spiking O2 to strain IT6 cells respiring N2O in the anoxic MR chamber, N2O-respiration ceased: dropping from the maximum (0.4–0.5 mmol N2O·h−1·g DW−1) to zero (Fig. 4A, Table 1). N2O reduction activity only started when the dissolved O2 concentration was below ca. 3 µM, suggesting the N2O reduction activity of this strain is highly sensitive to O2. In contrast, when O2 ( ~ 14 and 30 µM) was added to N2O-respiring cells of strain T4, simultaneous reduction of N2O and O2 was observed (Fig. 4B). However, the N2O respiration rates dropped to 0.24 and 0.13 mmol N2O·h−1·g DW−1 after spiking ~14 and 30 µM O2, respectively, which were approximately 34 and 20% of the maximum rate before O2 introduction (0.64–0.68 mmol N2O·h−1·g DW−1). These results suggest that in contrast to strain IT6, N2O reduction in strain T4 is not highly impaired by O2. N2OR activity fully recovered in both strains after O2 was depleted. Because the N2OR of strain IT6 was found to be highly sensitive to O2, further characterization of methanotroph N2OR activity in response to O2 exposure was limited to strain T4.
N2O and O2 reduction by cells of Methylacidiphilum caldifontis IT6 (A) and Methylocella tundrae T4 (B) during methanol oxidation. Filled blue dots represent dissolved N2O, filled orange dots represent dissolved O2, and filled black dots represent N2O reduction rates. Experiments were performed in a microrespirometry (MR) chamber fitted with O2 and N2O microsensors. The red arrows mark the addition of 14–33 µM O2 into the MR chamber. The red- and green-marked numbers close to the red and green lines represent the N2O reduction rates before and during O2 reduction (gray-shaded area) in the MR chamber, respectively. Source data are provided as Source Data file.
Considering these results, we set out to see if cells of strain T4 could continue N2O respiration while using O2 for CH4 oxidation in the MR chamber. The cells used for this experiment were cultured in suboxic conditions with starting gas mixing ratios (v/v) of 1% O2, 5% N2O, and 20% CH4 (i.e., CH4 + O2 + N2O condition). Similar to the anoxic CH3OH + N2O-adapted cells described above, the suboxic CH4 + O2 + N2O-adapted cells co-respired O2 and N2O after injecting CH4 (~406 µM) into a 5-mL MR chamber containing O2 (~30 µM) and N2O (~480 µM) (Fig. 5A). Interestingly, the maximum N2O respiration rates during each O2 spike were 1.4 to 2 times higher (1.58–2.47 mmol N2O·h−1·g DW−1) in the suboxic CH4 + O2 + N2O-adapted cells (Fig. 5B, Table 1) than in the anoxic CH3OH + N2O-adapted cells (1.12 ± 0.01 mmol N2O·h−1·g DW−1) (Table 1, Supplementary Fig. 7B), suggesting that the cells can modulate the rates of N2O reduction in response to O2 availability.
A MR experiment showing the simultaneous reduction of N2O and O2 by Methylocella tundrae T4 cells during CH4 oxidation. B N2O and O2 reduction rates by cells of strain T4 during CH4 oxidation calculated from (A). The filled orange and blue dots in the upper (A) represent the concentrations of dissolved O2 and N2O, respectively. The filled orange and blue dots in the bottom (B) represent the rates of O2 and N2O reduction, respectively. Experiments were performed in a MR chamber fitted with O2 and N2O microsensors. The red arrow marks the addition of CH4 (~406 µM) into the MR chamber. The black arrow marks the addition of ~26 µM or ~60 µM O2 into the MR chamber. The gray-shaded area represents points where N2O and O2 are reduced simultaneously. C Growth experiment showing Methylocella tundrae T4 cells reducing N2O and O2 simultaneously during CH4 oxidation. The culture was grown in 2-liter sealed bottles (triplicates) containing 60 mL of LSM medium with 2 mM NH4+ as the N-source. The headspace of the bottles was composed of CH4 (5%, v/v), O2 (0.5%, v/v), N2O (1.4%, v/v), and CO2 (5%, v/v) and supplemented with additional O2 (~0.5%, v/v) before its depletion. The incubation period shown in (C) is after the initial 20-day incubation period. After the depletion of O2, additional O2 was spiked to observe the simultaneous reduction of O2 and N2O during CH4 oxidation. Data are presented as the mean ± 1 SD of a triplicate experiment, and the error bars are hidden when they are smaller than the width of the symbols. Source data are provided as Source Data file.
Accordingly, the maximum rates of N2O reduction (1.58–2.47 mmol N2O·h−1·g DW−1) and O2 reduction (0.98–2.37 mmol O2·h−1·g DW−1) by the suboxic CH4 + O2 + N2O-adapted cells were comparable (Fig. 5B, Table 1). As the O2 concentration and reduction rate decreased, the N2O reduction rate also decreased (Fig. 5A, B), revealing that activation of CH4 by O2 is required for stimulating N2O respiration by CH4 + O2 + N2O-adapted cells. Based on these results, we conclude that, under suboxic conditions, both aerobic CH4 oxidation and N2O reduction were operating in concert: O2 was needed for the monooxygenase, but the N2OR remained active and was able to accept electrons released downstream in the C1 oxidation pathway. This adds to the evidence that aerobic N2O respiration occurs in strain T4 and is linked to aerobic CH4 oxidation.
Finally, we estimated the O2 concentration range at which the suboxic CH4 + O2 + N2O-adapted cells of strain T4 show N2O-reducing activity. At a O2 concentration of 170 µM, O2 and N2O were reduced simultaneously (Supplementary Fig. 8A, B). The maximum N2O reduction rate (Table 1) was nearly constant (1.32 ± 0.25 mmol N2O·h−1·g DW−1) across the O2 concentration range of 5–170 µM (Supplementary Fig. 8B, C) and was about 1.4 times higher than the maximum O2 reduction rates (0.95 ± 0.09 mmol O2·h−1·g DW−1). This means that even when exposed to high levels of O2, the N2OR in the suboxic CH4 + O2 + N2O-adapted cells remained functional and could reduce N2O at high rates. Other bacterial strains’ N2OR activities have been reported at O2 concentrations between 100 and 260 µM (refs. 73,74), indicating that their N2OR activity is similarly O2-tolerant73 as that of strain T4. According to the findings of Wang and colleagues73, N2O reducers with an O2 tolerant N2OR maintain low internal O2 concentrations in their cells by rapidly consuming O2, allowing the N2OR to remain active. However, it remains unclear if Methylocella tundrae T4 employs a similar strategy to maintain an O2-tolerant N2OR.
Improved methanotrophic growth of Methylocella tundrae in the presence of N2O
Based on the MR experiments showing the simultaneous reduction of O2 and N2O by CH4-fed cells of strain T4, alongside the clear N2O-dependent anaerobic growth, we hypothesized that strain T4 growth can be enhanced when it oxidizes CH4 by simultaneously reducing O2 and N2O under suboxic conditions. Using fed-batch growth experiments, we verified that strain T4 grows by CH4 oxidation coupled with co-respiration of N2O and O2 (Fig. 5C, Table 2), strongly supporting the MR results above. Cells grown under the suboxic CH4 + O2 + N2O condition consumed roughly the same amount of O2 and N2O (Fig. 5C, Table 2), and these values were comparable to what CH4 + O2 + N2O-grown cells consumed in the MR experiments (see Fig. 5A). Consequently, our results demonstrate that in an O2-limited environment, the cells can benefit energetically by directing more O2 to the monooxygenase step of CH4 oxidation, and simultaneously running a hybrid (O2 + N2O) electron transport system as shown in Table 2 and Fig. 5C.
The data showed unequivocally that the total electron equivalents released during CH4 oxidation to CO2 could account for the total electron acceptor (O2 + N2O) reduced. Based on a CH4 to O2 ratio of 1:1.57 (ref. 75), the total amount of O2 reduced (13.96 mmol·L-1) by the suboxic CH4 + O2 + N2O cultures could theoretically only account for 8.89 mmol·L-1 oxidized CH4. However, a larger total of 12.19 mmol·L-1 CH4 was oxidized by this culture (Table 2), and the excess 3.29 mmol·L-1 must have required an additional electron acceptor (i.e., N2O). Consistently, about 10.15 mmol·L-1 N2O was reduced by the suboxic CH4 + O2 + N2O cells, equivalent to 5.08 mmol·L-1 O2, since half as many electrons are consumed per mol during N2O reduction to N2 compared to O2 reduction to H2O. By running the N2O respiration system, the cells lower their O2-demand for respiration by the aerobic terminal oxidase and maximize O2 use by the methane monooxygenase76. Due to having more CH4 oxidized per O2 reduced (~37%) when N2O is present, higher cell densities (OD600) per O2 reduced (~34%) were reached in the suboxic CH4 + N2O + O2 cultures than in the O2-replete CH4 + O2 cultures (Table 2), further demonstrating the beneficial contribution of N2O reduction to growth on CH4 at suboxic conditions.
Transcriptomics
The overall regulation of key genes involved in denitrification and methane oxidation is depicted in Fig. 6 as well as in the supplementary material (Supplementary Figs. 9, 10, 11, Supplementary Datasets 5, 6, 7). Differences in expression were considered significant if the Log2FC was higher than [0.85] or lower than [-1.0] with an adjusted p ≤ 0.05.
The genes used to reconstruct the metabolic pathway are listed in Table S5. The gene products are shaded according to the relative fold change (Log2FC) in gene expression between cells grown under anoxic (CH3OH + N2O) and O2-replete (CH3OH + O2) conditions. Genes up-regulated in CH3OH + N2O-grown cells are shown in teal green, while those up-regulated in CH3OH + O2-grown cells are shown in purple. Note that proteins are not drawn to scale. Methanol oxidation: Methanol is oxidized to formaldehyde in the periplasmic space by the PQQ-dependent methanol dehydrogenase (Xox- and Mxa-type), T4_03519–21, T4_00353–5, and T4_01862–76. The NAD(P)+-dependent alcohol dehydrogenase (T4_03199) may also be involved in methanol oxidation to formaldehyde in the cytoplasmic space during anaerobic growth on methanol. Formaldehyde oxidation to formate then proceeds via the tetrahydromethanopterin (H4MPT) pathway, and C1 incorporation into the serine cycle is mediated by the tetrahydrofolate (H4F) carbon assimilation pathway. The Calvin-Benson-Bassham pathway is also a possible route for CO2 fixation. Nitrous oxide reduction: N2O is reduced to N2 through the activity of nitrous oxide reductase in the periplasmic space. Electron transfer to NosZ occurs via cytochrome c from the cytochrome bc1 (Qcr) complex136,137. Electron transfer to the NosZ may also involve direct interaction with methanol dehydrogenase C-type cytochrome (XoxG, MxaG). The NosR protein may be involved in the transfer of electrons to NosZ (refs. 136,137).
N2OR (O2 replete vs. anoxic conditions)
The transcript levels of the N2OR-encoding genes (T4_03941–7), nosRZDFYLX, were 2- to 4.7-fold higher in strain T4 cells respiring N2O in the anoxic CH3OH + N2O conditions compared to strain T4 cells respiring O2 in the O2-replete CH3OH + O2 conditions (Fig. 6 Supplementary Datasets 5, 6). Cells of strain IT6 respiring N2O in the anoxic CH3OH + N2O conditions showed transcriptional upregulation (1.9–6.7-fold) of four nos genes (nosC1BZC2) under anoxic conditions (Supplementary Fig. 10, Supplementary Dataset 7). Other nos operon genes (nosYFDL; IT6_00904–11) were expressed constitutively under both the anoxic CH3OH + N2O and O2-replete CH3OH + O2 conditions. The NosC1 and NosC2 proteins of Wolinella succinogenes were predicted to facilitate electron transfer from menaquinol to the periplasmic NosZ during the reduction of N2O to N2 (ref. 47) and are likely to play a similar role in strain IT6. Although the exact function of NosB has yet to be elucidated, Hein and colleagues77 used a non-polar nosB deletion mutant of Wolinella succinogenes to show that it is necessary for N2O respiration. Overall, increased expression of N2OR-encoding genes in Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 cells during anaerobic growth indicates that the N2OR is functional in these methanotrophs and supports their ability to respire and grow using N2O as a terminal electron acceptor.
N2OR (O2 replete vs. suboxic conditions)
Transcript levels of N2OR-encoding genes were 2–10.7-fold higher in strain T4 cells grown under suboxic CH4 + O2 + N2O conditions than in cells grown under O2-replete CH4 + O2 conditions (Supplementary Fig. 9, Supplementary Datasets 5, 6). This finding is consistent with the N2O respiration activity and growth of strain T4 under suboxic CH4 + O2 + N2O conditions (Fig. 5), in which the cells can efficiently oxidize more CH4 (see Table 2), most likely because the use of N2O for cellular respiration allows them to devote more O2 to CH4 oxygenation.
Methanol dehydrogenase (O2 replete vs. anoxic conditions)
In methanotrophs, methanol oxidation occurs in the periplasmic space by PQQ (pyrroloquinoline quinone)-dependent methanol dehydrogenase (MDH). Seven PQQ-dependent alcohol dehydrogenases (ADHs)78 are encoded in the genome of strain T4 (Supplementary Dataset 5). Five are type I ADHs (quinoproteins), which include one calcium-dependent MDH (MxaF-type MDH), and four lanthanide-dependent MDHs (XoxF-type MDH), divided into clades 1 (XoxF1), 3 (XoxF3), and 5 (XoxF5; 2 copies) (Supplementary Fig. 12). The other two are type II ADHs (quinohemoproteins). In addition to the PQQ-dependent ADH, Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 genomes contain genes encoding cytosolic Zn2+-dependent ADH, which are part of a large family of enzymes that oxidize alcohols to aldehydes or ketones and reduce NAD(P)+ or a similar cofactor79 (Supplementary Datasets 5, 6).
Among the four XoxF-type MDHs encoded in the genome of strain T4, genes in a xoxFGJ operon (T4_03519–21), which include a gene encoding a XoxF5 enzyme, were found to be constitutively transcribed at high levels in cells grown under both O2-replete CH3OH + O2 and anoxic CH3OH + N2O conditions (Fig. 6, Supplementary Datasets 5, 6). Thus, the xoxF5 gene likely encodes the predominant MDH used by strain T4 in both O2-respiring and N2O-respiring cells. The other singleton xoxF5 gene (T4_03691) and a xoxF3 gene found in a separate xoxFGJ cluster (T4_00353–5) were also significantly upregulated in cells grown under anoxic CH3OH + N2O conditions in comparison to cells grown under O2-replete CH3OH + O2 conditions (Fig. 6, Supplementary Datasets 5, 6). Furthermore, we observed a significant upregulation (2- to 22-fold) of the genes encoding MxaFI-type MDH (T4_01872–6) in the anoxic CH3OH + N2O-grown cells (Fig. 6, Supplementary Datasets 5, 6). Thus, our results indicate the use of various MDHs by strain T4 during anaerobic growth. In strain IT6 a xoxF gene encoding a XoxF2-type MDH is present as part of the xoxGJF operon (IT6_00336–8) (Supplementary Dataset 7) and the expression of the xoxF2 gene was 2-fold upregulated in the N2O-respiring cells (Supplementary Fig. 10, Supplementary Dataset 7).
A cytosolic Zn2+-dependent ADH bound to NAD(P)+ is known to perform the oxidation of methanol in Gram-positive methylotrophs80. A Zn2+-dependent ADH (T4_03199) of strain T4 was significantly upregulated (13.8-fold) in the anoxic CH3OH + N2O-grown cells compared to the O2-replete CH3OH + O2-grown cells (Fig. 6, Supplementary Datasets 5, 6). Strain IT6 genome also contained three copies of genes encoding enzymes annotated as Zn2+-dependent ADH (Supplementary Dataset 7). The expression of two of these genes (IT6_01501 and IT6_01931) were 3.9-fold and 2.5-fold upregulated in N2O-respiring cells compared to cells respiring O2 (Supplementary Fig. 10, Supplementary Dataset 7). Even though PQQ-dependent MDHs have a high affinity for and activity with methanol as a substrate, their use in strictly anoxic conditions will be limited because PQQ biosynthesis requires molecular oxygen81. Thus, PQQ-dependent MDHs are suggested to be functional at completely anoxic conditions only when PQQ is carried over from an aerobic growth stage or provided externally82. On the other hand, Zn2+-dependent MDHs have the advantage of utilizing a ubiquitous cofactor, NAD(P)+, and can be functional during anaerobic growth83. This finding raises the possibility that strains T4 and IT6 can employ alternative ADHs such as the Zn2+-dependent ADH to facilitate methanol oxidation in strict anoxia. Some genes required for the subsequent steps of C1 metabolism, i.e., formaldehyde and formate dehydrogenases, were also upregulated in strain T4 (but not IT6) growing anaerobically. These are depicted in Fig. 6 and supplementary materials (Supplementary Fig. 9, Supplementary Dataset 5).
Methanol dehydrogenase (O2 replete vs. suboxic conditions)
Furthermore, we also examined expression levels of genes encoding MDHs in strain T4 cells grown under suboxic CH4 + O2 + N2O conditions (Supplementary Fig. 9, Supplementary Datasets 5, 6). Genes in the cluster T4_01862–76, which encodes the calcium-dependent MDH (MxaF-type MDH), had the highest transcript expression among all MDH-encoding genes in CH4-oxidizing cells grown under suboxic CH4 + O2 + N2O conditions. When compared to O2-replete CH4 + O2 conditions, the expression of genes within this cluster was 1.8- to 371.5-fold upregulated (Supplementary Fig. 9, Supplementary Datasets 5, 6). This is unexpected since genes encoding the Mxa-type MDH are typically downregulated in the presence of lanthanides84; which we also included (2 µM each of cerium and lanthanum) in the growth medium. Their apparent upregulation (even when lanthanides are present) suggests that this enzyme might play an important role in CH4 metabolism in the presence of N2O and suboxic conditions. As observed above, genes in the xoxFGJ operon (T4_03519–21) were also highly expressed at the suboxic conditions (Supplementary Fig. 9, Supplementary Datasets 5, 6), suggesting that this key MDH is used by strain T4 in all three conditions. Genes in the cluster T4_01892–4 including the gene encoding the XoxF1 MDH were also significantly upregulated (18- to -35-fold) in the suboxic CH4 + O2 + N2O-grown cells compared to the O2-replete CH4 + O2-grown cells. The operon T4_02097–8, which encodes a cytochrome c550 (T4_02097) and a type II ADH (T4_02098), exhibited 6-fold and 30.6-fold upregulation, respectively, in cells grown under suboxic CH4 + O2 + N2O conditions as opposed to cells grown under O2-replete CH4 + O2 conditions. In addition, two Zn2+-dependent ADHs (T4_03097 and T4_03199) were significantly upregulated (3.5-fold and 46-fold, respectively) in strain T4 cells grown under suboxic CH4 + O2 + N2O conditions compared to cells grown under O2-replete CH4 + O2 conditions. Overall, it appears that cells oxidizing methanol under anoxia (CH3OH + N2O-grown cells) or those oxidizing methane under suboxia (CH4 + O2 + N2O-grown cells) use a distinct set of MDHs from those they use during O2 respiration.
Methane monooxygenase
The genomes of Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 contain genes that encode sMMO and pMMO, respectively. In the suboxic CH4 + O2 + N2O conditions, all the genes (mmoXYBZDCRG) in the gene cluster T4_01946–54 displayed a high degree of transcriptional upregulation (18.7–96-fold) compared to O2-replete CH4 conditions (Supplementary Fig. 9, Supplementary Datasets 5, 6). In a previous study85, Methylosinus trichosporium OB3b sMMO activity and protein expression were found to be significantly elevated under hypoxic conditions (24 µM) compared to higher O2 conditions (188 µM). Furthermore, Methylosinus trichosporium OB3b sMMO’s catalytic activity in the degradation of dichloroethane was enhanced at low O2 levels and impaired at elevated O2 levels86. Thus, in methanotrophs, upregulation of methane monooxygenase genes under O2 limiting conditions might be a strategy to produce more methane monooxygenase. This will lead to increased methane oxidation and thus provide stronger competition for the limited O2 with the terminal oxidase. Aside from the methane monooxygenase genes, group II and III truncated hemoglobin encoding genes were upregulated in Methylocella tundrae T4 (T4_02445, T4_02637, and T4_00400; 4- to 12-fold) and Methylacidiphilum caldifontis IT6 (IT6_00149; 3-fold) cells in response to suboxia or anoxia (Supplementary Datasets 5, 6). These truncated hemoglobins are thought to transport O2 to the methane monooxygenase22. Compared to methane, methanol resulted in lower transcript levels of sMMO genes in Methylocella tundrae T4 (Supplementary Fig. 11, Supplementary Dataset 5), with much lower levels in the O2 replete CH3OH + O2 conditions compared to the anoxic CH3OH + N2O conditions (Fig. 6, Supplementary Datasets 5, 6). Transcriptional repression of sMMO genes by growth substrates other than methane has been observed in Methylocella silvestris BL2 (refs. 87,88). The expression of genes encoding denitrification enzymes, their transcriptional regulators, and terminal oxidase is described in Supplementary Note 3.
Ecological relevance
Our findings revealed that certain methanotrophic strains, particularly those from the genera Methylocella and Methylocystis, which are commonly found in acidic and neutral terrestrial environments based on ecological meta-data from the BacDive database89,90, have the ability to reduce N2O. Wetlands, such as acidic peatlands and paddy fields, are significant contributors to the release of CH4 and N2O (refs. 27,91,92). Although active N2O consumption has been observed in acidic wetlands93, little is known about the microbial mechanisms that drive these processes. In a recent study27 wherein active N2O consumption was observed in peatlands (pH 6.4–3.7) located in Central and South America, Methylocystis species accounted for over 20% of the N2O-reducing microbial community based on nosZ gene amplicon sequence variants. This implies that N2OR-containing methanotrophs might make significant contributions to N2O reduction in these environments. The current prevailing perception of N2OR containing methanotrophs as a phylogenetically narrow group with limited ecological impact might be heavily biased by the scarcity of cultured methanotrophs with such metabolic capabilities. Thus, additional in situ and ecogenomic-based investigations are needed to more precisely quantify the contribution of known methanotrophs to N2O reduction as well as to uncover other novel N2O-reducing methanotrophs, such as those belonging to the Gemmatimonadota phylum50.
Short-term or seasonal water table fluctuations caused by either natural or anthropogenic desiccation influence the transition zone from oxic to anoxic conditions in wetlands94,95,96. In the deeper, water-filled anoxic layer of wetlands97, and even in oxygenated wetland soils98, methanogens produce CH4. N2O can be produced from denitrification processes, especially by incomplete denitrifiers which are frequently abundant in environments30,31,32. Nitrifiers also produce a significant amount of N2O as a byproduct of ammonia oxidation in the suboxic layers99. Furthermore, NO2− produced from nitrogen cycling processes can be abiotically reduced to N2O through chemodenitrification due to the stability of Fe2+ in acidic peat soils. At the oxic-anoxic interface, where CH4 and O2 gradients overlap, N2O-respiring methanotrophs will have simultaneous access to both CH4 and N2O. Although the CH4-O2 counter gradient is dynamic and O2-respiring organisms can rapidly deplete the limited O2, these N2O-respiring methanotrophs can use a growth strategy that involves respiring both N2O and O2 and coupling it to CH4 oxidation. This unique lifestyle, combined with the potential ability to respire N2O solely with non-methane substrates such as C1, C-C compounds51,100 as well as H2 (refs. 52,100), can confer a selective growth advantage, facilitate their niche expansion to suboxic and anoxic zones, and make them resilient in such environments.
In conclusion, we revealed that sMMO- and pMMO-containing acidophilic methanotrophs of the genera Methylocella and Methylacidiphilum can grow anoxically by respiring N2O using clade I and II NosZ, respectively. N2O reduction was detected at an extremely acidic pH of 2.0, which is by far the lowest pH reported for this process27,92. Further, N2O reduction can improve the growth yields of these bacteria under O2-limiting conditions and provide a competitive advantage. This study significantly expands our perception of the potential ecological niches of aerobic methanotrophs. In addition to mitigating CH4 and CO2 emissions, aerobic methanotrophs potentially play a role in reducing the emission of the climate-active and ozone-depleting gas N2O, particularly in low pH environments.
Methods
Bacterial strains and growth conditions
The methanotrophic bacterial strains used for the experiments include Methylacidiphilum caldifontis IT6, Methylacidiphilum infernorum IT5, Methylocella tundrae T4 ( = KCTC 52858 T), Methylocella silvestris BL2 ( = KCTC 52857 T), Methylocystis sp. SC2, and three in-house Methylocystis echinoides-like isolates (strains IM2, IM3, and IM4). The Methylacidiphilum strains are also in-house strains isolated previously from a mud-water mixture taken from Pisciarelli hot spring in Pozzuoli, Italy44. The Methylocella strains were obtained from the Korean Collection for Type Cultures (KCTC). Growth of the bacterial strains was performed using a low salt mineral (LSM) medium. The medium contained 0.4 mM MgSO4·7H2O, 0.2 mM K2SO4, and 0.1 mM CaCl2·2H2O and was supplemented with filter-sterilized solutions of 2 mM (NH4)2SO4, 0.1 mM KH2PO4, 1 µM CeCl3, 1 µM LaCl3, 1 mL (1×) vitamin, and 1 mL (1×) trace element solutions101 per liter. The pH of the medium was adjusted to pH 2.0 with concentrated sulfuric acid (filter-sterilized) for the Methylacidiphilum strains and to pH 5.5 with 20 mM 2-morpholinoethanesulfonic acid (filter-sterilized) for the Methylocella strains. The cultures were incubated at 52 °C for Methylacidiphilum strains (IT5 and IT6) and 28 °C for Methylocella strains (T4 and BL2) with shaking at 160 rpm. Unless stated otherwise, ammonium sulfate, (NH4)2SO4, was used as the nitrogen source.
Enrichment and isolation of Methylocystis strains
The N2OR-containing Methylocystis strains were isolated from an acidic forest soil in Chungcheongbuk-do, South Korea (36°55'31” N 127°54'86” E). The soil sample preparation and initial enrichment of the methanotrophs102, as well as the isolation of methanotrophic strains through repeated serial dilution of the enrichment cultures103, have all been described previously. Briefly, the most diluted culture exhibiting methane oxidation was serially diluted and filtered through 0.2-μm Track-Etch membrane polycarbonate filters (Whatman). The filters were placed on LSM medium (pH 5.5) in Petri dishes and incubated at 30 °C in airtight containers containing CH4 (10%, v/v) and CO2 (5%, v/v). Colonies that appeared on the filters after 3 weeks of incubation were transferred to fresh LSM medium in 160-mL serum vials with the same gas composition. Three individual methanotrophic isolates were identified by sequencing the 16 S rRNA gene with the 27 F/1492 R primer set104. The purity of the isolates was confirmed by seeding aliquots of the CH4-grown cultures into the LSM medium with 0.05% (w/v) yeast extract, tryptic soy broth, and Luria-Bertani broth without CH4 and incubating at 30 °C. Three methanotrophic isolates, IM2, IM3, and IM4, shared 99.46% 16 S ribosomal RNA (rRNA) gene-sequence identity with the alphaproteobacterial methanotroph Methylocystis echinoides LMG27198. The three strains share average nucleotide identity values ranging from 81.85–81.93 with Methylocystis echinoides LMG27198, implying that they represent a new species in the genus Methylocystis.
DNA isolation, genomic and phylogenetic analyses
High-molecular-weight genomic DNA was extracted using a modified CTAB method105, from 200 mL amounts of Methylocella tundrae T4 grown in methanol, and the Methylocystis isolates (strain s IM2, IM3, and IM4) grown in CH4. The genomes of Methylocella tundrae T4 and Methylocystis sp. IM3 were sequenced at LabGenomics (Seongnam, Republic of Korea) and Macrogen (Seoul, Republic of Korea) using the PacBio RS II (long-read sequencing) and Illumina HiSeq (2 × 150 bp) platforms, respectively. The genomes of Methylocystis sp. IM2 and Methylocystis sp. IM4 were sequenced using a MinION R10.4.1 flow cell (FLO-MIN114, Oxford Nanopore Technologies). The PacBio reads were assembled with the Trycycler pipeline (v0.5.4)106. Filtered reads were subsampled and assembled using Miniasm/Minpolish (v0.3-r179)107, Flye (v2.9.2)108, and Raven (v1.8.3)109 assemblers. The consensus contigs were polished with Illumina short reads using Polypolish (v0.5.0)110 and POLCA (v4.0.5)111. The circularity was confirmed during the Trycycler pipeline assembly and again by mapping the Illumina reads backward. De novo genome assembly of the MinION long reads was accomplished using Flye (v2.9.2)108. Annotation of methanotrophs’ genomes was performed with the Prokka annotation pipeline (v1.14.6)112 and NCBI Prokaryotic Genome Annotation Pipeline (PGAP; v4.2)113. Functional assignment of the predicted genes was improved using a set of public databases (InterPro114, GO115,116, PFAM117, CDD118, TIGRFAM119, and EggNOG120). Prediction of signal peptides and transmembrane helices was performed using the web-based services SignalP (v6.0)121 and TMHMM (v2.0)122 with default settings.
The distribution of denitrification genes in methanotroph isolates or metagenome-assembled genomes (MAG) (meeting the following CheckM (v1.2.2) criteria: completeness > 60% and contamination <10%) was examined using genomic data from the NCBI assembly database. Reference protein sequences of denitrification enzymes (NapA, NapB, NarG, NarH, NarI, NirS, NirK, NorB, NorC, and NosZ) were obtained from the NCyc123 and BV-BRC124 databases. The annotated protein sequences of methanotrophs were re-annotated against the obtained reference sequences from the NCyc123 and BV-BRC124 databases. The identities of the obtained denitrification protein sequences in methanotrophs were verified using manual alignment and tree-building procedures with reference sequences. Sequences incorrectly annotated as denitrification genes were removed, and only candidate genes that clustered with reference sequences were counted as true hits.
For phylogenetic analyses of the NosZ proteins and methanol dehydrogenases of strains T4 and IT6, representative amino acid sequences of the genes of related taxa were obtained from NCBI. The derived amino acid sequences of the NosZ and methanol dehydrogenases (XoxF and MxaF) were aligned using MAFFT (v7.511)125. Maximum-likelihood trees were inferred with IQ-TREE (v1.6.12). The constructed trees and operon arrangements were visualized using iTOL (v.6.7.2)126 and used for annotation. Genomic islands were predicted using the IslandViewer web server127.
Anoxic growth coupled with N2O reduction
To demonstrate the ability of N2OR-containing methanotrophs to grow using N2O as the electron acceptor, we established anoxic batch cultures of Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6 in 160-mL bottles containing 20 mL of LSM media and inoculated with 1–5% (v/v) actively growing-cells from the log phase (starting OD600 values ≤ 0.05). To remove oxygen, nitrogen gas (N2, purity >99.999%) was introduced into the bottles via a long needle (18 G). Following that, the bottles were flushed with N2 gas for 20 min before being sealed with gas-tight butyl rubber stoppers and aluminum crimp seals to prevent O2 leakage. We used contactless trace-range oxygen sensor spots (TROXSP5) to monitor O2 contamination (<0.10%, v/v) in the culture bottles incubated after N2-flushing (see Analytical methods, for details). These spots have a detection limit of 20 nM O2. Chemical-reducing agents, Na2S (0.5, 1, and 2 mM), cysteine (0.5 mM), DTT (0.5 mM), and titanium citrate (0.5 and 1 mM) in the media resulted in severe cell toxicity, hindering their use in this study as previously reported for N2OR reducer Anaeromyxobacter dehalogenans128. When the cultures were incubated without the chemical-reducing agents, the cells completely depleted the trace O2 concentration present in the culture bottles in less than 24 h as measured by the oxygen sensor spots.
The N2OR-lacking methanotrophs Methylocella silvestris BL2 and Methylacidiphilum infernorum IT5, which are closely related to Methylocella tundrae T4 and Methylacidiphilum caldifontis IT6, respectively, were used as negative controls. Methanol (30 mM), N2O (5%, v/v), and CO2 (5%, v/v) were used as the energy source, electron acceptor, and carbon source, respectively. In addition, pyruvate (10 mM) and hydroxyacetone (acetol) (10 mM) were tested as the sole C-C electron donors in strains T4 and IT6, respectively. Furthermore, strain IT6 cells were investigated to grow chemolithoautotrophically in sealed 1-liter bottles (duplicate) containing 20 mL of LSM medium at pH 2.0 on H2 (10% v/v) with and without N2O (5% v/v). As part of the control experiments, we incubated cells from the four strains in LSM media under anoxic conditions (without N2O) to assess the contribution of the initial trace O2 present in the culture bottles to biomass increase. The increase in biomass as OD600 by the trace O2 in the control cultures was negligible when compared to cultures growing with N2O as the sole electron acceptor (see Fig. 2C, F, I, L). Positive control experiments with methanol (30 mM) and O2 (5%, v/v) as the electron donor and electron acceptor, respectively, were conducted for each strain. The concentrations of H2, O2, N2O, NO3−, and NO2− were monitored at intervals during incubations (described in Analytical methods). Cell growth was also evaluated using optical density measurements (λ = 600 nm), direct microscopic cell counts, and real-time quantification of 16 S rRNA gene abundance (described in Analytical methods). All growth experiments were performed in triplicates unless otherwise stated.
Next, we checked the anoxic growth of Methylocella strains on NO3− (2 to 4 mM KNO3) as the terminal electron acceptor instead of N2O. Methanol (30 mM) was used as the sole electron donor and 2 mM NH4+ was used as the N-source. To compare the effect of electron donors on NO3− and NO2− reduction, Methylocella strains were also anoxically grown in LSM medium containing a C-C substrate, pyruvate (10 mM). Cells of strain T4 were grown under O2-replete (O2; 21%, v/v) or anoxic conditions (O2; 0%, v/v, N2O; 5%, v/v) for the NO2− toxicity test (triplicates) with varying NO2− (KNO2) concentrations (0, 0.01, 0.03, 0.1, 0.3, and 1 mM).
Analytical methods
A YL 6100 gas chromatograph (YL Instrument Co., Anyang, South Korea) with a flame ionization detector (FID) and a thermal conductivity detector (TCD) was used to analyze the mixing ratios of CH4, N2O, and H2 in the headspace of the sealed bottles used to cultivate the Methylocella and Methylacidiphilum strains. Using a Hamilton glass syringe, 100 µL of the sealed bottle headspaces were injected into a gas chromatograph equipped with MolSieve 5 A column (3Ft, 1/8, 2 mm, 60/80 SST, Agilent Technologies, Inc., CA, USA; for separating H2, O2, and N2O) and Haysep N column (7Ft, 1/8, 2 mm, 60/80 SST, Agilent Technologies, Inc., CA, USA; for separating CO2 and CH4) to determine the gases present. Helium was used as the carrier gas, with a flow rate of 15 mL·min−1. By utilizing pure gases of known concentrations, a calibration curve of the gases used as substrates was generated. The bottles were fitted with contactless trace range oxygen sensor spots (TROXSP5, PyroScience, Germany) calibrated at 0% and ambient air (21% oxygen), and a FireSting-Pro multi-analyte meter (FSPRO-4, PyroScience, Germany) was used to measure the O2 concentration in the sealed bottles. Acidic Griess reagent and VCl2/Griess reagent were used for photometric quantification of NO2− and NO3− concentrations129, respectively, using a SpectraMax M2 microplate reader (Molecular Devices, USA). Cell growth was assessed by measuring changes in OD600 using a spectrophotometer (Optizen 2120UV, Mecasys Co., Daejeon, Korea). Real-time quantification of the 16 S rRNA gene was performed using the 518 F/786 R primer set130. The total cell number was determined by counting cells stained with DAPI (4,6-diamidino-2-phenylindole) using an epifluorescence microscope (AxioScope.A1; Carl Zeiss, Oberkochen, Germany).
Kinetic analysis using microrespirometry (MR)
For kinetic analysis using microrespirometry (MR), Methylocella tundrae T4 cells were grown under three different O2 conditions: O2-replete (CH3OH + O2), suboxic (CH4 + O2 + N2O), and anoxic (CH3OH + N2O). Methylacidiphilum caldifontis IT6 cells were grown under O2-replete (CH3OH + O2) and anoxic (CH3OH + N2O) conditions. The O2-replete growth conditions included ambient air (21% O2, v/v) and CH3OH (30 mM) as the sole electron donor. The suboxic cell cultures were grown under a condition that included CH4 (5%, v/v) as the sole electron donor and O2 (0.5%, v/v) with N2O (1%, v/v) as terminal electron acceptors. O2 (0.5%, v/v) was resupplied intermittently before its depletion. Anaerobically grown cells were cultured in bottles containing 30 mM CH3OH as the sole electron donor and 5% (v/v) N2O as the terminal electron acceptor. The cultures were monitored daily and harvested as soon as active consumption of electron donors and acceptors was detected. After being collected by centrifugation (5000 × g, 30 min, 25 °C), the cells were washed twice with substrate- and N-source-free MES-buffered LSM (20 mM MES; pH 5.5) or H2SO4-buffered LSM (4 mM H2SO4; pH 2.0) and then resuspended in 20 mL of the same media without electron donors and acceptors. In the cultures grown under anoxic and suboxic conditions, the cell suspensions were transferred to sealed 20-mL bottles and flushed with nitrogen gas (N2, purity >99.999%) before use. The cell suspensions were dispensed into a double-port MR chamber (no headspace) with a capacity of 5 or 10 mL outfitted with O2 and N2O-detecting microsensors, two MR injection lids, and two glass-coated stir bars. Kinetics and stoichiometry of N2O and O2 reduction coupled to CH3OH oxidation were estimated using anoxic CH3OH + N2O- and oxic CH3OH + O2-grown cells, respectively. Anoxic CH3OH + N2O-grown cells were used to test CH3OH-dependent O2 and N2O uptake by strains IT6 (starting OD600 = 0.96) and T4 (starting OD600 = 0.79). The effect of O2 to N2OR activities of strains T4 and IT6 was determined by spiking varying O2 to the N2O respiring cells. In a 5-mL MR chamber, suboxic CH4 + O2 + N2O-grown cells of strain T4 (starting OD600 = 1.0) were used to test the CH4-dependent simultaneous respiration of O2 and N2O.
All MR experiments were performed in a recirculating water bath at 27 °C and 50 °C for strains T4 and IT6, respectively. A 10-µL or 50-µL syringe (Hamilton, Reno, USA) fitted with a 26 G needle was used to inject the substrate (CH4, CH3OH, N2O, or O2) into the chamber via an injection port. Concentrations of O2 and N2O were measured using an OX-MR oxygen microsensor (OX-MR-202142, Unisense, Aarhus, Denmark) and a N2O-MR sensor (N2O-MR-303088, Unisense), respectively. The detection limits of the OX-MR and N2O-MR microsensors are 0.3 µM O2 and 0.1 µM N2O, respectively. The OX-MR and N2O-MR microsensors were directly plugged into a microsensor multimeter before being polarized for more than a day and calibrated according to the manufacturer’s instructions. All data from the microsensor multimeter was logged onto a laptop using SensorTrace Suite software (v.3.3.0, Unisense). Anoxically prepared aliquots of N2O, CH4, and CH3OH were injected into the MR chamber via the injection port with a 10-µL syringe (Hamilton, Reno, USA). Anoxic substrate-free LSM media (at pH 2.0 and 5.5) were prepared by sparging the solutions with N2 gas for 1 h before use. Anoxic saturated-aqueous CH4 and N2O solutions were made in capped 160-mL bottles containing 100 mL of LSM medium and pressurized with CH4 or N2O (1, 2, or 3 atm; 100%, v/v). Saturated-aqueous O2 solutions were prepared in capped 160-mL bottles containing 100 mL of LSM medium and pressurized with O2 (1, 2, and 3 atm; 100%, v/v).
Growth based on CH4 oxidation coupled with co-respiration of O2 and N2O
Suboxic cultivations were carried out to investigate the growth of Methylocella tundrae T4 by oxidizing methane with simultaneous respiration of O2 and N2O. The experiments were conducted in N2-flushed 2-liter sealed bottles containing 60 mL of LSM medium with 2 mM NH4+ as the N-source. The headspace of the bottles was composed of CH4 (5%, v/v), O2 (0.5%, v/v), N2O (1%, v/v), and CO2 (5%, v/v) and supplemented with additional O2 (~0.5%, v/v) before its depletion. The headspace gas (CH4, N2O, and O2) mixing ratios were monitored at intervals during incubations as described above in Analytical methods. To investigate the growth benefits of cells of strain T4 respiring N2O in tandem with O2 during CH4 oxidation, an O2-replete culture was included for comparison (triplicates). The apparent increase in cell densities of both growth conditions was compared using OD600 measurements.
Transcriptome analysis
Cells of strains T4 and IT6 were cultured in 60 mL of LSM medium at pH 5.5 and pH 2.0 in sealed 2-liter bottles (4 or 5 replicates) for transcriptome analyses. Strain T4 cells were cultured under three different O2 levels, with the first setting being O2-replete (CH4 + O2 and CH3OH + O2), the second being suboxic (CH4 + O2 + N2O), and the third being anoxic (CH3OH + N2O). Strain IT6 was cultivated in O2-replete CH3OH + O2 and anoxic CH3OH + N2O conditions. Cells were grown anaerobically in bottles containing 30 mM CH3OH as the sole electron donor and 5% N2O as the terminal electron acceptor. The O2-replete growth conditions were made up of ambient air (21% O2, v/v) with CH4 (5%, v/v) or CH3OH (30 mM) serving as the sole electron donor. The suboxic growth conditions were made up of a mixture of CH4 (5% v/v) as the sole electron donor and O2 (0.5% v/v) and N2O (1% v/v) as terminal electron acceptors. Before the depletion of O2, additional O2 was resupplied intermittently at a mixing ratio of 0.5% (v/v). Contactless trace-range oxygen sensor spots (TROXSP5) were installed into the culture bottles to monitor O2 concentration.
The cells were harvested during the mid-exponential phase by centrifugation at 5000 × g for 10 min at 25 °C. Total RNA was extracted from the cells in four replicates using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA quality was checked with the Agilent 2100 Expert Bioanalyzer (Agilent), and cDNA libraries were prepared from the RNA samples using the Nugen Universal Prokaryotic RNA-Seq Library Preparation Kit. The cDNA libraries were sequenced using NovaSeq6000 (Illumina) at LabGenomics (Seongnam, Korea). Read quality was evaluated with FastQC (v0.11.8)131. Trimmomatic (v0.36)132 was used to trim reads with the options: SLIDINGWINDOW:4:15 LEADING:3 TRAILING:3 MINLEN:38 HEADCROP:13. Reads mapped to strains T4 and IT6 rRNA sequences were removed with SortMeRNA (v4.3.6)133. The remaining reads were aligned to the genomes of strains T4 and IT6 using Bowtie2 (v2.4.4)134, and the reads mapped to each gene were counted using HTSeq (v0.12.3)135. Expression values are presented as transcripts per kilobase million (TPM). The statistical analysis of differentially expressed genes was performed using the DESeq2 package in R (v4.3.2). A two-sided Wald test was used to calculate the p values, and multiple-comparison adjustments were made using the Benjamini-Hochberg method by default in DESeq2 (v1.40.2).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All numerical data used to make the figures is provided in source data. The complete genome sequence of strain T4 was deposited in the National Center for Biotechnology Information (NCBI) GenBank (accession nos. CP139089 (Chromosome), CP139088 (Plasmid 1), and CP139087 (Plasmid 2)). The genomic sequences and genome annotations of Methylocystis species (strains IM2, IM3, and IM4) and ‘Ca. Methylotropicum kingii’ are available on Figshare (https://doi.org/10.6084/m9.figshare.25521913.v2). All previously sequenced genomes analyzed in this study are available in the NCBI Database with the GenBank accession numbers listed in Supplementary Dataset 1. The whole transcriptome data was deposited in the NCBI BioProject database under the accession number PRJNA1050235. The following are the publicly available databases/datasets used in the study: NCBI NR [https://www.ncbi.nlm.nih.gov/refseq/], BV-BRC, NCyc [https://github.com/qichao1984/NCyc], Pfam [https://pfam.xfam.org/], InterPro [https://www.ebi.ac.uk/interpro/], GO [https://geneontology.org/], CDD, TIGRFAM, and EggNOG [https://tigrfams.jcvi.org/cgi-bin/index.cgi]. Source data are provided with this paper.
References
IPCC. Summary for Policymakers. In: Climate Change 2021 – The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (ed Intergovernmental Panel on Climate C). (Cambridge University Press, 2023).
Forster, P. et al. The Earth’s Energy Budget, Climate Feedbacks and Climate Sensitivity. In Climate Change 2021: The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (eds Masson-Delmotte, V., P. Zhai, A. Pirani, S.L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M.I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J.B.R. Matthews, T.K. Maycock, T. Waterfield, O. Yelekçi, R. Yu, and B. Zhou) (Cambridge University Press United Kingdom and New York, NY, USA, 2023).
Prinn, R. G. et al. Evidence for variability of atmospheric hydroxyl radicals over the past quarter century. Geophys. Res. Lett. 32, L07809 (2005).
Myhre, G. et al. Anthropogenic and natural radiative forcing. in Climate change 2013: The physical science basis. Contribution of working group I to the fifth assessment report of the Intergovernmental Panel on Climate Change (eds Stocker T. F., et al.) (Cambridge University Press, 2013).
Szopa, S. et al. Short-Lived Climate Forcers. In: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (eds Masson-Delmotte V., et al.) (Cambridge University Press, 2021).
Prather, M. J. et al. Measuring and modeling the lifetime of nitrous oxide including its variability. J. Geophys. Res. Atmos. 120, 5693–5705 (2015).
Ravishankara, A. R., Daniel, J. S. & Portmann, R. W. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009).
Montzka, S. A., Dlugokencky, E. J. & Butler, J. H. Non-CO2 greenhouse gases and climate change. Nature 476, 43–50 (2011).
Canadell, J. G. et al. Global Carbon and Other Biogeochemical Cycles and Feedbacks. In Climate Change 2021:The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (eds Masson-Delmotte, V., P. Zhai, A. Pirani, S.L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M.I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J.B.R. Matthews, T.K. Maycock, T. Waterfield, O. Yelekçi, R. Yu, and B. Zhou) (Cambridge University Press, 2021).
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R. & Zechmeister-Boltenstern, S. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130122 (2013).
Ringeval, B. et al. Climate-CH4 feedback from wetlands and its interaction with the climate-CO2 feedback. Biogeosciences 8, 2137–2157 (2011).
Beaulieu, J. J., DelSontro, T. & Downing, J. A. Eutrophication will increase methane emissions from lakes and impoundments during the 21st century. Nat. Commun. 10, 1375 (2019).
Murrell, J. C. & Jetten, M. S. The microbial methane cycle. Environ. Microbiol. Rep. 1, 279–284 (2009).
Conrad, R. The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292 (2009).
Bürgmann, H. Methane oxidation (aerobic). in Encyclopedia of Geobiology, (eds Reitner, J., Thiel, V.) (Springer Netherlands, 2011).
Leu, A. O. et al. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 14, 1030–1041 (2020).
Haroon, M. F. et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567 (2013).
Scheller, S., Yu, H., Chadwick, G. L., McGlynn, S. E. & Orphan, V. J. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707 (2016).
Ettwig, K. F. et al. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl Acad. Sci. USA 113, 12792–12796 (2016).
Ettwig, K. F. et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548 (2010).
Kits, K. D., Campbell, D. J., Rosana, A. R. & Stein, L. Y. Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front. Microbiol. 6, 1072–1072 (2015).
Kits, K. D., Klotz, M. G. & Stein, L. Y. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 17, 3219–3232 (2015).
Dam, B., Kube, M., Dam, S., Reinhardt, R. & Liesack, W. Complete sequence analysis of two methanotroph-specific repABC-containing plasmids from Methylocystis sp. strain SC2. Appl. Environ. Microbiol. 78, 4373–4379 (2012).
Kox, M. A. R. et al. Complete genome sequence of the aerobic facultative methanotroph Methylocella tundrae strain T4. Microbiol. Resour. Announc. 8, e00286–00219 (2019).
Tian, H. et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature 586, 248–256 (2020).
Thomson, A. J., Giannopoulos, G., Pretty, J., Baggs, E. M. & Richardson, D. J. Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1157–1168 (2012).
Buessecker, S. et al. Coupled abiotic-biotic cycling of nitrous oxide in tropical peatlands. Nat. Ecol. Evol. 6, 1881–1890 (2022).
Su, Q., Domingo-Félez, C., Jensen, M. M. & Smets, B. F. Abiotic nitrous oxide (N2O) production Is strongly pH dependent, but contributes little to overall N2O emissions in biological nitrogen removal systems. Environ. Sci. Technol. 53, 3508–3516 (2019).
Zumft, W. G. & Kroneck, P. M. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52, 107–227 (2007).
Graf, D. R., Jones, C. M. & Hallin, S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLOS ONE 9, e114118 (2014).
Sanford, R. A. et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl Acad. Sci. USA 109, 19709–19714 (2012).
Hallin, S., Philippot, L., Löffler, F. E., Sanford, R. A. & Jones, C. M. Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 26, 43–55 (2018).
Payne, W. J., Grant, M. A., Shapleigh, J. & Hoffman, P. Nitrogen oxide reduction in Wolinella succinogenes and Campylobacter species. J. Bacteriol. 152, 915–918 (1982).
Yoon, S., Nissen, S., Park, D., Sanford, R. A. & Löffler, F. E. Nitrous oxide reduction kinetics distinguish bacteria harboring clade I NosZ from those harboring clade II NosZ. Appl. Environ. Microbiol. 82, 3793–3800 (2016).
Park, D., Kim, H. & Yoon, S. Nitrous oxide reduction by an obligate aerobic bacterium, Gemmatimonas aurantiaca strain T-27. Appl. Environ. Microbiol. 83, e00502–00517 (2017).
Dam, B., Dam, S., Blom, J. & Liesack, W. Genome analysis coupled with physiological studies reveals a diverse nitrogen metabolism in Methylocystis sp. strain SC2. PLOS ONE 8, e74767 (2013).
Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 6, 1346 (2015).
Reim, A., Lüke, C., Krause, S., Pratscher, J. & Frenzel, P. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic–anoxic interface in a flooded paddy soil. ISME J. 6, 2128–2139 (2012).
Farhan Ul Haque, M., Crombie, A. T. & Murrell, J. C. Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome 7, 134 (2019).
Kantor, R. S., Miller, S. E. & Nelson, K. L. The water microbiome through a pilot scale advanced treatment facility for direct potable reuse. Front. Microbiol. 10, 993 (2019).
McGuirl, M. A., Bollinger, J. A., Cosper, N., Scott, R. A. & Dooley, D. M. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. 6, 189–195 (2001).
Kang, C. S., Dunfield, P. F. & Semrau, J. D. The origin of aerobic methanotrophy within the Proteobacteria. FEMS Microbiol. Lett. 366, fnz096 (2019).
Berks, B. C., Palmer, T. & Sargent, F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8, 174–181 (2005).
Awala, S. I. et al. Verrucomicrobial methanotrophs grow on diverse C3 compounds and use a homolog of particulate methane monooxygenase to oxidize acetone. ISME J. 15, 3636–3647 (2021).
Zhang, L., Trncik, C., Andrade, S. L. A. & Einsle, O. The flavinyl transferase ApbE of Pseudomonas stutzeri matures the NosR protein required for nitrous oxide reduction. Biochim. Biophys. Acta Bioenerg. 1858, 95–102 (2017).
Honisch, U. & Zumft, W. G. Operon structure and regulation of the nos gene region of Pseudomonas stutzeri, encoding an ABC-Type ATPase for maturation of nitrous oxide reductase. J. Bacteriol. 185, 1895–1902 (2003).
Simon, J., Einsle, O., Kroneck, P. M. & Zumft, W. G. The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569, 7–12 (2004).
Suzuki, M., Cui, Z. J., Ishii, M. & Igarashi, Y. Nitrate respiratory metabolism in an obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus TK-6. Arch. Microbiol. 175, 75–78 (2001).
Sharp, C. E., den Camp, H.J.M.O., Tamas, I., Dunfield P. F. Unusual members of the PVC superphylum: the methanotrophic Verrucomicrobia genus “Methylacidiphilum”. in Planctomycetes: cell structure, origins and biology (ed Fuerst, J. A.) (Humana Press, 2013).
Bay, S. K. et al. Trace gas oxidizers are widespread and active members of soil microbial communities. Nat. Microbiol. 6, 246–256 (2021).
Dedysh, S. N., Knief, C. & Dunfield, P. F. Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670 (2005).
Awala, S. I. et al. Methylacidiphilum caldifontis gen. nov., sp. nov., a thermoacidophilic methane-oxidizing bacterium from an acidic geothermal environment, and descriptions of the family Methylacidiphilaceae fam. nov. and order Methylacidiphilales ord. nov. Int. J. Syst. Evol. Microbiol. 73, 006085 (2023).
Thauer, R. K., Jungermann, K. & Decker, K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180 (1977).
Svensson-Ek, M. et al. The X-ray crystal structures of wild-type and EQ (I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 321, 329–339 (2002).
Iwata, S., Ostermeier, C., Ludwig, B. & Michel, H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 (1995).
Chen, J. & Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta Bioenerg. 1827, 136–144 (2013).
Bergaust, L., Mao, Y., Bakken, L. R. & Frostegård, A. Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396 (2010).
Van den Heuvel, R. N., Bakker, S. E., Jetten, M. S. & Hefting, M. M. Decreased N2O reduction by low soil pH causes high N2O emissions in a riparian ecosystem. Geobiology 9, 294–300 (2011).
Palmer, K., Drake, H. L. & Horn, M. A. Association of novel and highly diverse acid-tolerant denitrifiers with N2O fluxes of an acidic fen. Appl. Environ. Microbiol. 76, 1125–1134 (2010).
Brenzinger, K., Dörsch, P. & Braker, G. pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front. Microbiol. 6, 961 (2015).
Lycus, P. et al. Phenotypic and genotypic richness of denitrifiers revealed by a novel isolation strategy. ISME J. 11, 2219–2232 (2017).
Almeida, J. S., Júlio, S. M., Reis, M. A. & Carrondo, M. J. Nitrite inhibition of denitrification by Pseudomonas fluorescens. Biotechnol. Bioeng. 46, 194–201 (1995).
Fang, F. C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832 (2004).
Bueno, E., Sit, B., Waldor, M. K. & Cava, F. Anaerobic nitrate reduction divergently governs population expansion of the enteropathogen Vibrio cholerae. Nat. Microbiol. 3, 1346–1353 (2018).
Vadivelu, V. M., Keller, J. & Yuan, Z. Free ammonia and free nitrous acid inhibition on the anabolic and catabolic processes of Nitrosomonas and Nitrobacter. Water Sci Technol. 56, 89–97 (2007).
Zhu-Barker, X., Cavazos, A. R., Ostrom, N. E., Horwath, W. R. & Glass, J. B. The importance of abiotic reactions for nitrous oxide production. Biogeochemistry 126, 251–267 (2015).
Cole, J. Anaerobic bacterial response to nitric oxide stress: Widespread misconceptions and physiologically relevant responses. Mol. Microbiol. 116, 29–40 (2021).
Auclair, J., Lépine, F., Parent, S. & Villemur, R. Dissimilatory reduction of nitrate in seawater by a Methylophaga strain containing two highly divergent narG sequences. ISME J. 4, 1302–1313 (2010).
Kamps, J. J., Hopkinson, R. J., Schofield, C. J. & Claridge, T. D. How formaldehyde reacts with amino acids. Commun. Chem. 2, 126 (2019).
Dedysh, S. N. & Dunfield, P. F. Facultative and obligate methanotrophs:how to identify and differentiate them.Methods Enzymol. 495, 31–44 (2011).
Purchase, M. L., Bending, G. D. & Mushinski, R. M. Spatiotemporal variations of soil reactive nitrogen oxide fluxes across the anthropogenic landscape. Environ. Sci. Technol. 57, 16348–16360 (2023).
Pauleta, S. R., Dell’Acqua, S. & Moura, I. Nitrous oxide reductase. Coord. Chem. Rev. 257, 332–349 (2013).
Wang, Z., Vishwanathan, N., Kowaliczko, S. & Ishii, S. Clarifying microbial nitrous oxide reduction under aerobic conditions: tolerant, intolerant, and sensitive. Microbiol. Spectr. 11, e0470922 (2023).
Suenaga, T., Riya, S., Hosomi, M. & Terada, A. Biokinetic characterization and activities of N2O-reducing bacteria in response to various oxygen levels. Front. Microbiol. 9, 697 (2018).
Hilgeri, H. & Humer, M. Biotic landfill cover treatments for mitigating methane emissions. Environ. Monit. Assess. 84, 71–84 (2003).
Ross, M. O. & Rosenzweig, A. C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 22, 307–319 (2017).
Hein, S., Witt, S. & Simon, J. Clade II nitrous oxide respiration of Wolinella succinogenes depends on the NosG, -C1, -C2, -H electron transport module, NosB and a Rieske/cytochrome bc complex. Environ. Microbiol. 19, 4913–4925 (2017).
Keltjens, J. T., Pol, A., Reimann, J. & Op den Camp, H. J. M. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98, 6163–6183 (2014).
Sirota, F. L., Maurer-Stroh, S., Li, Z., Eisenhaber, F. & Eisenhaber, B. Functional classification of super-large families of enzymes based on substrate binding pocket residues for biocatalysis and enzyme engineering applications. Front. Bioeng. Biotechnol. 9, 701120 (2021).
Le, T.-K., Lee, Y.-J., Han, G. H. & Yeom, S.-J. Methanol dehydrogenases as a key biocatalysts for synthetic methylotrophy. Front. Bioeng. Biotechnol. 9, 787791 (2021).
Bonnot, F., Iavarone, A. T. & Klinman, J. P. Multistep, eight-electron oxidation catalyzed by the cofactorless oxidase, PqqC: identification of chemical intermediates and their dependence on molecular oxygen. Biochemistry 52, 4667–4675 (2013).
Matsushita, K. et al. Escherichia coli is unable to produce pyrroloquinoline quinone (PQQ). Microbiology 143, 3149–3156 (1997).
Zhang, W. et al. Guidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy. RSC Adv. 7, 4083–4091 (2017).
Chu, F. & Lidstrom, M. E. XoxF Acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J .Bacteriol. 198, 1317–1325 (2016).
Kim, H. J. & Graham, D. W. Effect of oxygen level on simultaneous nitrogenase and sMMO expression and activity in Methylosinus trichosporium OB3b and its sMMOC mutant, PP319: aerotolerant N2 fixation in PP319. FEMS Microbiol. Lett. 201, 133–138 (2001).
Kim, H. J. & Graham, D. W. Effects of oxygen and nitrogen conditions on the transformation kinetics of 1,2-dichloroethenes by Methylosinus trichosporium OB3b and its sMMOC mutant. Biodegradation 14, 407–414 (2003).
Smirnova, A. V. & Dunfield, P. F. Differential transcriptional activation of genes encoding soluble methane monooxygenase in a facultative versus an obligate methanotroph. Microorganisms 6, 20 (2018).
Theisen, A. R. et al. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 58, 682–692 (2005).
Reimer, L. C., Sarda Carbasse, J., Koblitz, J., Podstawka, A. & Overmann, J. Methylocella tundrae Dedysh et al. 2004. DSMZ. https://doi.org/10.13145/bacdive1659.20230509.8.1 (2023).
Reimer, L. C., Sarda Carbasse, J., Koblitz J., Podstawka, A., Overmann, J. Methylocystis echinoides (ex Gal’chenko et al. 1977) DSMZ. https://doi.org/10.13145/bacdive169085.20230509.8.1 (2023).
Qian, H. et al. Greenhouse gas emissions and mitigation in rice agriculture. Nat. Rev. Earth Environ. 4, 716–732 (2023).
Kolb, S. & Horn, M. A. Microbial CH4 and N2O consumption in acidic wetlands. Front. Microbiol. 3, 78 (2012).
Ishii, S., Ohno, H., Tsuboi, M., Otsuka, S. & Senoo, K. Identification and isolation of active N2O reducers in rice paddy soil. ISME J. 5, 1936–1945 (2011).
Taminskas, J. et al. Climate change and water table fluctuation: Implications for raised bog surface variability. Geomorphology 304, 40–49 (2018).
Ratcliffe, J. L., Campbell, D. I., Clarkson, B. R., Wall, A. M. & Schipper, L. A. Water table fluctuations control CO2 exchange in wet and dry bogs through different mechanisms. Sci. Total Environ. 655, 1037–1046 (2019).
Evans, C. D. et al. Overriding water table control on managed peatland greenhouse gas emissions. Nature 593, 548–552 (2021).
Kotsyurbenko O. R., Glagolev M. V., Merkel A. Y., Sabrekov A. F., Terentieva I. E. Methanogenesis in Soils, Wetlands, and Peat. in Biogenesis of Hydrocarbons (eds Stams A. J. M., Sousa D. Z.) (Springer International Publishing, 2019).
Angle, J. C. et al. Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions. Nat. Commun. 8, 1567 (2017).
Zhu, X., Burger, M., Doane, T. A. & Horwath, W. R. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl Acad. Sci. USA 110, 6328–6333 (2013).
Hakobyan, A., Zhu, J., Glatter, T., Paczia, N. & Liesack, W. Hydrogen utilization by Methylocystis sp. strain SC2 expands the known metabolic versatility of type IIa methanotrophs. Metab. Eng. 61, 181–196 (2020).
Widdel, F., Bak, F. Gram-negative mesophilic sulfate-reducing bacteria. In The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications (eds Balows, A., Trüper, H. G., Dworkin, M., Harder, W., Schleifer, K.-H.) (Springer International Publishing, 1992).
Bellosillo, L. A. Effects of environmental factors on methanotroph communities from a forest soil, lake sediment and a landfill soil. (Chungbuk National University, 2020).
Awala, S. I. et al. Methylococcus geothermalis sp. nov., a methanotroph isolated from a geothermal field in the Republic of Korea. Int. J. Syst. Evol. Microbiol. 70, 5520–5530 (2020).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Hurt, R. A. et al. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67, 4495–4503 (2001).
Wick, R. R. et al. Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol. 22, 266 (2021).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Vaser, R. & Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 1, 332–336 (2021).
Wick, R. R. & Holt, K. E. Polypolish: short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 18, e1009802 (2022).
Zimin, A. V. & Salzberg, S. L. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput. Biol. 16, e1007981 (2020).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016).
Hunter, S. et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 (2009).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Consortium, T. G. O. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–d334 (2021).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Lu, S. et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–d268 (2020).
Haft, D. H., Selengut, J. D. & White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 31, 371–373 (2003).
Huerta-Cepas, J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019).
Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Tu, Q., Lin, L., Cheng, L., Deng, Y. & He, Z. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics 35, 1040–1048 (2018).
Olson, R. D. et al. Introducing the bacterial and viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689 (2022).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Bertelli, C. et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–w35 (2017).
Onley, J. R., Ahsan, S., Sanford, R. A. & Löffler, F. E. Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking the nitrite reductase genes nirS and nirK. Appl. Environ. Microbiol. 84, e01985–01917 (2018).
Miranda, K. M., Espey, M. G. & Wink, D. A. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5, 62–71 (2001).
Muyzer, G., De Waal, E. C. & Uitterlinden, A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993).
Andrews, S. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2014).
Hein, S. & Simon, J. Bacterial nitrous oxide respiration: electron transport chains and copper transfer reactions. Adv .Microb. Physiol. 75, 137–175 (2019).
Torres, M. J. et al. Nitrous oxide metabolism in nitrate-reducing bacteria: physiology and regulatory mechanisms. Adv. Microb. Physiol. 68, 353–432 (2016).
Acknowledgements
This work was supported by the NRF (National Research Foundation of Korea) grant funded by the Korean government (Ministry of Science and ICT) (2021R1A2C3004015), the Basic Science Research Program through NRF funded by the Ministry of Education (2020R1A6A1A06046235), and the National Institute of Agricultural Science, Ministry of Rural Development Administration, Republic of Korea (research project PJ01700703). J.-H.G. was supported by the NRF grant funded by the Korean government (Ministry of Science and ICT) (RS-2023-00213601). M.-Y.J. was supported by the NRF grant funded by the Korean government (Ministry of Science and ICT) (2021R1C1C1008303 and 2022R1A4A503144711). P.F.D. was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (2019-06265). M.W. was supported by the Austrian Science Fund FWF Cluster of Excellence “Microbiomes drive planetary health” COE7.
Author information
Authors and Affiliations
Contributions
S.I.A., J.-H.G., and S.-K.R. designed research. S.I.A., J.-H.G., M.-Y.J., and Y.K. performed research. S.I.A., J.-H.G., Y.K., M.-Y.J., P.F.D., and S.-K.R. analyzed data. S.I.A., J.-H.G., M.-Y.J., P.F.D., M.W., and S.-K.R. wrote the manuscript with contributions and comments from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Simon Guerrero-Cruz, Lisa Stein and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Awala, S.I., Gwak, JH., Kim, Y. et al. Nitrous oxide respiration in acidophilic methanotrophs. Nat Commun 15, 4226 (2024). https://doi.org/10.1038/s41467-024-48161-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48161-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.