Abstract
Modern, highly evolved nucleoside-processing enzymes are known to exhibit perfect regioselectivity over the glycosylation of purine nucleobases at N9. We herein report an exception to this paradigm. Wild-type nucleoside phosphorylases also furnish N7-xanthosine, a “non-native” ribosylation regioisomer of xanthosine. This unusual nucleoside possesses several atypical physicochemical properties such as redshifted absorption spectra, a high equilibrium constant of phosphorolysis and low acidity. Ultimately, the biosynthesis of this previously unknown natural product illustrates how even highly evolved, essential enzymes from primary metabolism are imperfect catalysts.
Similar content being viewed by others
Introduction
The modern natural nucleosides are N1-ribosides of pyrimidine nucleobases and N9-ribosides of purine nucleobases. Although, for instance, N7-ribosyl purines were likely readily available under prebiotic conditions1,2, nature elected to use N9-ribosides. As such, early metabolism was evolved to selectively furnish, degrade and re-use N9-ribosyl purines. As a result, modern (highly evolved) nucleoside-processing enzymes generally exhibit perfect control over the stereoselectivity of the attack at the anomeric center as well as the regioselectivity of the glycosylation. Although unusual glycosylation isomers of purine nucleobases are known in plants (where they serve as hormones)3,4, the other conceivable ribosylation isomers of the natural nucleobases do not occur as regular products of metabolism.
In modern nucleoside metabolism, nucleoside phosphorylases (NPs) are responsible for recycling nucleosides by cleaving them into the corresponding nucleobase and ribose 1-phosphate (Rib1P), which is funneled back into primary carbon metabolism. These enzymes are ubiquitous across all kingdoms of life and known for their high selectivity and robustness5,6,7. As their transformations are tightly thermodynamically controlled8, they also catalyze the corresponding reverse reactions (glycosylations) and thereby maintain balanced physiological concentrations of nucleosides and nucleobases. Any failure to do so generally results in disease (e.g. immune disorders)9. Thus, modern NPs are under considerable selection pressure to execute their physiological tasks fast and selectively. As group-specific enzymes, purine NPs are faced with the additional challenge of recognizing multiple different purine nucleosides (and nucleobases) as substrates and catalyzing transformations from and towards the natively occurring N9-ribosyl purines. Although purine NPs operate with astonishing selectivity, their ability to promiscuously degrade “non-native” glycosylation regioisomers of purines is documented by a handful of examples10,11,12,13. However, imperfect regioselectivity in glycosylation reactions with naturally occurring purines has not been reported yet for purine NPs.
Here, we report an exception to this paradigm and show that NPs can furnish N7-xanthosine (N7X, Fig. 1a), a “non-native” glycosylation regioisomer of xanthosine. Since N7-ribosides such as N7X are not known as natural products and remain uncharacterized, we herein report on the discovery of this unusual nucleoside as well as its atypical spectroscopic and physicochemical properties.
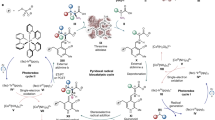
a Formation of N7X and/or N9X catalyzed by nucleoside phosphorylases. b Serendipitous discovery of N7X in an enzymatic cascade through (c) its unprecedented redshift compared to the free nucleobase. d–g Orthogonal data confirming N7X as the major product of the glycosylation of X by Geobacillus thermoglucosidasius purine NP (GtPNP) at pH 9. f Energy-minimized structure of N7X and key 2D NMR correlations. h UV absorption spectra of X and N7X at pH 10, where both species exist as monoanions. i pKa values as determined by UV spectroscopy. j, k Continuous monitoring of the X → N7X glycosylation at pH 10 (with 0.4 g L−1 GtPNP) to establish its equilibrium constant. By convention, these equilibrium constants are given for the phosphorolytic direction. l Michaelis-Menten kinetics for N7X formation by GtPNP. The relationship between kobs and [X] is non-linear (as supported by the Akaike information criterion, AIC), allowing us to estimate a Michaelis-Menten constant. m Formation of N9X and N7X by various wild-type nucleoside phosphorylases (with 500 µM X and 1 g L−1 enzyme). The bars in this panel each display the isomer distribution obtained in a single experiment (n = 1). Please see the Supplementary Information for experimental details and the externally hosted supplementary material for all raw data38.
Results
During our ongoing efforts to facilitate nucleoside synthesis with NPs14,15,16, we serendipitously discovered N7X as an unexpected byproduct of a reaction cascade involving a bacterial purine NP and a guanine deaminase (see Supplementary Fig. 1). With the former enzyme cleaving guanosine into guanine and Rib1P and the latter quantitatively hydrolyzing guanine into xanthine (X), the expected products of this sequence should be Rib1P and X (Fig. 1b). As N9-xanthosine (N9X, the “native” isomer) is typically not converted by bacterial nucleoside phosphorylases to any significant extent (this process requires a specialized enzyme: xanthosine phosphorylase), the reverse reaction, glycosylation of X, similarly should not proceed. Thus, Rib1P and X should be terminal products of this reaction sequence. However, UV absorption spectra obtained during this transformation proved inconsistent with this assumption (Fig. 1c). Similar results were obtained when X and Rib1P were directly incubated with the purine NP. We consistently observed absorption at higher wavelengths than anticipated for X and HPLC analysis of the reaction mixture showed a single additional peak corresponding to an unknown product (Fig. 1e). HRMS clearly supported the formation of a riboside of X but the retention times and absorption spectra of the reaction product evidently did not match N9X (Fig. 1c–e). We subsequently consulted DFT calculations to predict dominant tautomers and their UV spectra, which suggested N7X and O6-xanthosine (O6X) as sufficiently redshifted candidates, excluding O2-xanthosine (O2X, Fig. 1f, please see the Supplementary Information and Supplementary Fig. 3 for details). A detailed NMR-spectroscopic analysis of a partially purified sample of the reaction product in D2O revealed N7X as the likely product. A weak HMBC correlation between the anomeric position on the ribosyl unit and C5 of the X unit, as well as a strong correlation to C8, make N7 the most likely ribosylation site. NOESY correlations between the ribosyl hydrogens (1′, 2′ and 3′) and H-8 support this assignment (Fig. 1g). Subsequent freeze-drying of the sample, trituration with DMSO-d6 and comparison to Bridson’s 1998 NMR data (the only reported synthesis of N7X)17 unambiguously supported the formation of N7X (Supplementary Table 3). Notably, N7X can be distinguished well from N9X by 1H NMR through its significantly greater shift for H-8 and H-1′ as well as the much smaller H-1′,H-2′-coupling constant (Supplementary Table 4), likely caused by slight ring torsion introduced by intramolecular H-bonding (Fig. 1g).
N7X has several unusual physicochemical properties. First, N7X is the only known natural nucleoside whose UV absorption spectra are redshifted compared to those of the free nucleobase (Fig. 1h)18. N7X’s sweeping absorption spectra (particularly those of the anion, Supplementary Fig. 4) are more reminiscent of free pyrimidine nucleobases than purine nucleosides. Secondly, N7X has a pKa of around 8.5 (Fig. 1i and Supplementary Fig. 4), as determined by UV spectroscopy, which is notably higher than free X (ca. 7.3) and N9X (ca. 5.3)19. Both of these properties may originate from the unique electronic situation in N7X’s nucleobase. Although DFT calculations suggest that the negative charge in the anions of both N7X and N9X predominantly resides at O2 (Supplementary Fig. 3), only N7X can form an intramolecular H-bond between O6 and the 2′-OH (Fig. 1f). This grants N7X’s nucleobase more aromatic character and leads to a pronounced redshift in its absorption spectra, in line with the trends exhibited by pyrimidine nucleosides and -bases18. Similar, although much less pronounced, trends have been described for analogues of 8-azaguanosine20. Thirdly, N7X displays an equilibrium constant of phosphorolysis of ca. 0.5 at pH 10, which we determined by continuously monitoring the glycosylation of X by UV spectroscopy using principles of spectral unmixing (Fig. 1j, k)8,18,21,22. This unusually high value could be qualitatively confirmed by reactions monitored discontinuously by HPLC (Supplementary Table 4). As purine nucleosides typically have equilibrium constants of phosphorolysis of 0.01 − 0.1 (irrespective of the sugar moiety)8,23, this makes N7X’s phosphorolysis the most exothermic one known among purines. The next closest equilibrium constants of phosphorolysis of purines are those of N9X (0.2, determined by HPLC, Shugar reported 0.05)19 and inosine (0.1, determined by discontinuous spectral unmixing)8.
To our surprise, non-selective formation of N7X is a common reactivity across the entire family of NPs (Fig. 1m). We made our initial discovery using the purine NP from Geobacillus thermoglucosidasius (GtPNP), which almost exclusively furnishes N7X under alkaline conditions. Compared to its native reactions (e.g. the phosphorolysis/glycosylation of adenosine or guanosine), which proceed with rate constants of >1 s−1, the formation of N7X is a slow process. Under alkaline conditions, this transformation happens well over an order of magnitude slower and with a KM value exceeding the solubility limit of free X, as determined from kinetic experiments assessed by UV spectroscopy (Fig. 1l). Interestingly, at neutral pH values, this enzyme also yields N9X as a product of the same reaction and slowly degrades N7X (likely via hydrolysis, Fig. 1m). This process is pH-dependent (it does not happen at pH 10 to a relevant degree, Fig. 1j) and enzyme-catalyzed (purified N7X is stable, Supplementary Fig. 6). Other purine NPs, such as the ones from Thermus thermophilus (TtPNP), Escherichia coli and human, also make both N7X and N9X in different ratios, as do the xanthosine phosphorylase from E. coli (EcXP) and the pyrimidine NP from G. thermoglucosidasius (GtPyNP, albeit much slower, Fig. 1m and Supplementary Table 4). Although the elucidation of the exact mechanisms underpinning the observed selectivity preferences are beyond the scope of this work, we hypothesize that differences in H-bonding behavior may be responsible. Purine NPs preferentially yielding N7X possess a nucleobase-coordinating aspartate, while those preferentially making N9X have an asparagine at the analogous position (Supplementary Fig. 5).
In conclusion, we serendipitously found that NPs glycosylate X to yield both N7X and N9X in an enzyme- and pH-dependent fashion. N7X is a stable nucleoside, although its formation is thermodynamically disfavored and some purine NPs also catalyze its hydrolysis. We assume that the formation of N7X among NPs has previously gone unnoticed because i) N7X and N9X are indistinguishable by mass spectrometry, ii) N7X only forms slowly under somewhat unusual conditions (alkaline media and comparably high concentrations of Rib1P and X), and iii) NPs are usually screened in the phosphorolysis direction (obscuring potential glycosylation isomers only occurring in the reverse reaction). However, the synthesis of N7X by wild-type NPs likely carries no physiological relevance since both free X and Rib1P only occur in low micromolar concentrations in vivo (far below the KM values of this slow transformation)24, which precludes this transformation from happening to any relevant degree. This lack of physiological relevance is likely also responsible for the retention of this promiscuous activity in modern NPs. Collectively, this case study provides a notable exception to the paradigm that “native” purine nucleosides are N9-ribosides and further illustrates how even highly evolved enzymes are imperfect catalysts. Although catalytic promiscuity is an inherent trait of proteins25,26,27, it is comparably rare to observe a condition-dependent preference for the formation of a “non-native” reaction product in enzymes under evolutionary pressure for high selectivity. As NPs otherwise operate with perfect selectivity in their native target transformations, we hope that this report sparks future research into other “non-native” nucleoside isomers as well as the implications of this promiscuity for early metabolism and the convergent evolution of NPs6.
Methods
Accessibility statement
The data presented in this manuscript are depicted using scientific color maps. Since ca. 4% of the human population are color vision-deficient, we made a conscious effort to avoid unscientific uses of color, such as color ambiguities and other biases that could lead to misrepresentation, limited accessibility, or loss of information upon reduction of the color space28,29. Thus, all figures herein were created using the scientific color map plasma, which retains color-coded information for all readers.
Protein production and purification
All enzymes were heterologously produced in Escherichia coli as His6-tagged proteins through isopropyl β-D-1-thiogalactopyranoside (IPTG)-induced overexpression, as previously described21. For protein production, we employed two different strategies:
i) For production in Enpresso B medium (Enpresso GmbH, Berlin, Germany), which simulates a fed-batch culture in shake flasks, cells were washed from an agar plate of the expression strain (E. coli BL21g harboring the respective plasmid) with 1 mL 0.9% NaCl using a spatula, according to the manufacturer’s standard protocol. Then, OD600 was measured and 50 mL Enpresso B medium supplemented with antibiotic (100 mg L−1 ampicillin) was inoculated to an initial OD600 of 0.15. The resulting culture was grown at 37 °C and 250 rpm for 17 h before expression was induced by addition of IPTG to a final concentration of 0.1 mM (50 µL of a 0.1 M stock). The culture was incubated at 30 °C and 250 rpm for 24 h before the cells were harvested by centrifugation (8000 g, 10 min, 4 °C). The resulting cell pellet was either stored at −20 °C until further use or immediately subjected to lysis and purification.
ii) Alternatively, for GtPNP and GtPyNP, we employed a production workflow in traditional TB medium. To this end, a 20 mL preculture of the expression strain (E. coli JM109 harboring the respective plasmid) was grown in LB medium supplemented with antibiotic (50 mg L−1 ampicillin) overnight at 37 °C. This preculture was used to inoculate 250 mL TB medium (12 g L−1 tryptone, 24 g L−1 yeast extract, 5 g L−1 glycerol, 2.31 g L−1 KH2PO4, 12.54 g L−1 K2HPO4) containing antibiotic to an initial OD600 of around 0.15. This culture was incubated at 37 °C and 200 rpm until an OD600 of > 0.6 was reached, which typically happened after 2 − 3 h. At this point, protein production was induced by adding IPTG to a final concentration of 0.1 mM (25 µL of a 1 M stock in water). Expression was carried out overnight (ca. 20 h) at 37 °C and cells were harvested by centrifugation (4000 g, 20 min, 4 °C). The resulting cell pellet was either stored at −20 °C until further use or immediately subjected to lysis and purification.
For lysis and purification, pelleted cells were resuspended in binding buffer (20 mM KH2PO4, 500 mM NaCl, 20 mM imidazole) to a concentration of around 0.5 gpellet mL−1 and PierceTM protease inhibitor mini tables (Thermo Fisher Scientific) were added (one per 10 mL of suspension volume). Cells were disrupted by sonication (6 min, 10 s pulse, 10 s breaks, 60% amplitude or 10 min, 30 s pulse, 30 s breaks, 30% amplitude – both protocols performed similarly well). For some enzymes, the resulting lysate was heated for 20 − 30 min to precipitate E. coli proteins (60 °C for GtPNP and GtPyNP, 80 °C for TtPNP). Cell debris and precipitated protein was then removed by centrifugation (11000 rpm, 45 min, 4 °C) and filtration (0.45 µm pore size). The cell free extract was applied to a Ni Sepharose HistrapTM column (GE Healthcare) preequilibrated with binding buffer. Non-specifically bound proteins were removed by washing with approximately 10 column volumes (CV) of binding buffer and the target protein was eluted with elution buffer (20 mM KH2PO4, 500 mM NaCl, 500 mM imidazole) over ca. 3 CV. Fractions containing pure target protein (as assessed by SDS PAGE) were combined and, if necessary, concentrated by centrifugation (Vivaspin, Sartorius, Göttingen, Germany, molecular weight cut-off at 10 kDa). Afterwards, the protein was desalted into 20 mM MOPS buffer (pH 7, adjusted at 20 °C) or 20 mM taurine buffer (pH 9, adjusted at 20 °C) using a PD-10 desalting column (GE Healthcare). This desalting procedure was repeated once (for a total of two passes through a PD-10 column per protein preparation). Alternatively, the target protein obtained from the affinity column was directly dialyzed against 2 mM phosphate buffer (pH ≈ 9). The resulting protein preparations were stored at 4 °C. Alternatively, glycerol was added to a final concentration of 50% (v/v) to store the resulting protein stocks at −20 °C. Typical stock concentrations of pure protein ranged from 1 to 10 g L−1 (calculated with 1 AU at 280 nm being equal to a protein concentration of 1 g L−1).
Discovery of N7X
We discovered N7X via its unusual UV spectroscopic properties. In a reaction cascade featuring a purine nucleoside phosphorylase (purine NP, converting guanosine to guanine) and a deaminase (converting guanine to xanthine), we recorded UV spectra which did not match the expected products. For instance, we performed reactions with 200 µM guanosine, 400 µM phosphate, 100 µg mL−1 (ca. 3.8 µM) GtPNP and 1 µg mL−1 deaminase in 50 mM glycine buffer pH 9 in a total volume of 500 µL at 40 °C for 2 h. Then, 100 µL of reaction mixture were quenched in 100 µL 200 mM NaOH directly in wells of a UV-transparent 96-well plate. Subsequently, we recorded UV absorption spectra from 250 to 350 nm in steps of 1 nm with a platereader. Reference spectra for X and N9X were obtained from solutions of either compound in 50 mM glycine buffer and analogous dilution in aq. NaOH. This gave spectra such as those shown in Fig. 1c.
To check for the potential formation of glycosides of X, an analogous reaction mixture to the one described above was quenched 1:1 with MeOH, centrifuged to remove any precipitate (13,000 rpm, 10 min), and subjected to HRMS analysis (negative mode, as all relevant components in this mixture are anions at pH 9). This yielded clear evidence for the formation of a glycoside (Fig. 1d) with ions corresponding to xanthosine (N9X) or any of its isomers.
HPLC analyses
To further exclude the formation of N9X by GtPNP at pH 9, we analyzed the reaction products by HPLC. To this end, we performed a reaction containing 500 µM X, 500 µM Rib1P, and 1 g L−1 GtPNP in 50 mM glycine-KOH buffer pH 9. This reaction was started by the addition of the enzyme and incubated at 50 °C for 24 h. Afterwards, a 50 µL sample of the reaction mixture was quenched by addition to 50 µL ice cold methanol. The resulting sample was centrifuged (13,000 rpm, 10 min) and analyzed by HPLC, using an Agilent 2200 system with DAD detector at 260 nm. The eluents were a) 50 mM ammonium-acetate buffer pH 5 (from a 1 M stock adjusted to the correct pH with 100% acetic acid) and b) MeCN. The column was a 250 × 4.6 5 µm Kinetex Evo C18 Core Shell (Phenomenex), used at a flow rate of 1 mL min−1 and heated to 25 °C. The elution method consisted of a linear gradient from 3 to 40% MeCN over 10 min, followed by 4 min of re-equilibration at 3% MeCN. This experiment provided further evidence that the reaction product of the GtPNP-catalyzed glycosylation of X at pH 9 was not N9X. Indeed, under these HPLC conditions, the typical retention times were 3.4 − 3.5 min (X), 4.3 − 4.4 min (N7X), and 4.5 − 4.6 min (N9X), as confirmed with authentic standards of X and N9X, and purified N7X.
Computational prediction of UV absorption spectra
To narrow down possible glycosylation isomers of X, we first predicted UV spectra for all likely candidates by density functional theory (DFT). To this end, DFT calculations for all structures (Supplementary Fig. 3) were performed using the Amsterdam Modeling Suite (AMS) 2020.20330. For initial geometry optimizations the PBE31 XC functional with a TZP slater-type orbital basis set32 was applied and the COSMO33 model was used for solvation in water. The subsequent time-dependent DFT (TDDFT) single-point calculations were achieved using the B3LYP34,35,36 hybrid functional with a QZ4P basis set32. Solvation in water was again performed by COSMO and excitation energies were calculated according to the Davidson algorithm37. The generation of UV/Vis spectra was achieved in Python. For each spectrum, the contribution of the different calculated structures to the spectrum was determined based on the corresponding electronic energy applying the Boltzmann equation. The final spectra were subsequently convoluted applying peak broadening with Gaussians. The most redshifted peak of each spectrum was then normalized to its maximum and plotted as shown in Fig. 1f.
Purification of N7X and NMR analyses
N7X was produced by GtPNP-catalyzed glycosylation of X and purified by semi-preparative HPLC. To this end, 18.3 mg X, 119.2 mg Rib1P, and GtPNP (0.1 g L−1) were combined in 12 mL 50 mM HEPES buffer pH 7 (although pH 7 does decrease the stability of N7X in the presence of GtPNP this enzyme is much more active in this transformation than at pH 9, see above). This equals final concentrations of 10.5 mM X and 21 mM Rib1P in the reaction. The reaction mixture was incubated at 60 °C for 5 h and then frozen to stop the reaction. Next, the mixture was thawed and the denatured enzyme and precipitated X were removed by centrifugation at 4 °C. N7X was then purified by semi-preparative HPLC using a 250 × 10 mm MultoKrom100-10 C18 column (Chromatographie Service GmbH, Langerwehe, Germany). The purification method used water (ice-cold) and MeOH (room temperature) as eluents, a flow rate of 4.7 mL min−1 and an elution program starting with isocratic elution with 3% MeOH for 5 min, followed by a linear gradient to 10% MeOH over 2.5 min (7.5 min run time) and a drop back down to 3% MeOH over 0.5 min (8 min run time), which was held for 6 min (14 min run time) to re-equilibrate the column. Per run, 2 mL of sample were injected, for a total of 6 runs. N7X typically eluted between 3 and 5 min of run time, followed by X between 5.5 and 7.5 min. N9X was not detected in this experiment. This yielded a total of 12 mg partially purified N7X, which contained Rib1P and HEPES as major impurities (detected by NMR) due to its early elution from the HPLC column. Of this sample, 8 mg were dissolved in D2O (containing 0.05 wt% TSP as internal standard) for NMR analysis. Following the tentative identification of the nucleoside product as N7X (see the Supplementary Information for structure elucidation), this NMR sample was diluted with water, freeze-dried and triturated with DMSO-d6, which dissolved N7X, some of the HEPES, while leaving most of the Rib1P untouched. NMR analysis of the resulting sample unambiguously identified the product as N7X due to matching data with Bridson’s report (Supplementary Table 3)17. All NMR data collected in this study are freely available from the externally hosted supplementary information38.
Reference UV spectra of X and N7X at pH 10
At pH 10, X and N7X almost completely exist as their anions (see Fig. 1i). We therefore obtained reference spectra of both compounds at pH 10 to enable continuous reaction monitoring (see below). To this end, we prepared a 100 µM solution of X in 50 mM glycine/proline buffer pH 10 and measured its UV absorption spectrum in a platereader using UV-transparent 96-well plates. The Lambert-Beer law directly provided the extinction coefficients of X at all wavelengths. Since accurate preparation of exact concentrations of N7X proved difficult, we used isosbestic normalization based on the known isometric point of glycosylation to correct for concentration errors. Thus, we prepared a ca. 100 µM solution of purified N7X and obtained its UV absorption spectrum in analogy to X. Next, we normalized the spectra of X and N7X to the isosbestic point of glycosylation (288 nm) and used the scaling factor between the extinction coefficient of X and the normalized spectrum of X to calculate the extinction coefficients of N7X based on the normalized spectrum of X. The result is shown Fig. 1h and all experimental and calculated data are available from the externally hosted supplementary information38.
Absorption spectra of X, N9X and N7X as functions of pH
To determine the \({{pK}}_{a}\) values of X, N9X and N7X, we obtained UV absorption spectra from pH 2 − 12 and applied isosbestic normalization for fitting. To this end, we prepared ≈100 µM solutions of each compound in a universal buffer mix containing citrate (\({{pK}}_{a}\) = 3.1, 4.8, and 6.4), HEPES (\({{pK}}_{a}\) = 3.0 and 7.5), asparagine (\({{pK}}_{a}\) = 2.1 and 8.8) and proline (\({{pK}}_{a}\) = 2.0 and 10.6) (all adjusted to the respective pH with HCl and NaOH, all with 5 mM final concentration of each buffer component) and obtained UV absorption spectra of these solutions from 250 to 350 nm in steps of 1 nm in a platereader using UV-transparent 96-well plates. This revealed isosbestic points of deprotonation of 274 nm (X), 259/268 nm (N9X), and 279 nm (N7X). All spectra were then normalized to the absorbance at the isosbestic point (Supplementary Fig. 4) and the distribution of the neutral and deprotonated species was calculated as a function of pH as described in the Supplementary Information, giving the fits shown in Fig. 1i.
Equilibrium constant of phosphorolysis of N7X
To determine the equilibrium constant of phosphorolysis of N7X, we performed glycosylation experiments monitored by UV. To this end, we continuously monitored the glycosylation of X catalyzed by GtPNP at pH 10, where X and N7X almost exclusively exist as monoanions and GtPNP selectively yields N7X (and not N9X). Specifically, we performed glycosylation reactions with X (100 or 150 µM), Rib1P (0, 25, 50, 100, 200 or 500 µM, equivalent to 0, 0.25, 0.5, 1, 2 and 5 or 0.16, 0.33, 0.66, 1.33 and 3.33 equivalents) and 400 µg mL−1 GtPNP (equivalent to ca. 15.4 µM) in 50 mM glycine/proline buffer (containing 50 mM of both buffer components) pH 10 in wells of a UV-transparent 96-well plate in a total volume of 200 µL at room temperature (ca. 21 °C). These reactions were started by addition of 150 µL reaction mixture (containing all reagents except Rib1P) to 50 µL appropriately diluted solution of Rib1P. These reactions were monitored at 288 nm (the isosbestic point of phosphorolysis of this transformation at pH 10) and 300 nm (near the maximum positive difference of the extinction coefficients, \(\triangle {\varepsilon }_{300}\) = 3.6 mM−1 cm−1) for 4 h. The resulting absorption data were converted to conversions and equilibrium states as detailed in the Supplementary Information, giving the reaction courses and the relationship shown in Fig. 1j, k21,39.
Michaelis-Menten kinetics with GtPNP
To evaluate the affinity of GtPNP toward X under alkaline conditions (which essentially exclusively give N7X), we obtained Michaelis-Menten kinetics of this transformation at pH 10. To this end, we performed glycosylation reactions with X (20 − 500 µM), 200 µM Rib1P and 100 µg mL−1 GtPNP (equivalent to ca. 3.8 µM) in 50 mM glycine/proline buffer (containing 50 mM of both buffer components) pH 10 in wells of a UV-transparent 96-well plate in a total volume of 200 µL at room temperature (ca. 21 °C). These reactions were started by addition of 100 µL reaction mixture (containing all reagents except X) to 100 µL appropriately diluted solution of X. All reactions were run in duplicate and repeated with duplicates run with 50 µg mL−1 GtPNP (1.9 µM), for a total of four replicates per concentration of X. Reaction progress was monitored at 300 nm (\(\triangle {\varepsilon }_{300}\) = 3.6 mM−1 cm−1) for 20 min. The absorption change over time was approximated by linear fitting and converted into observed rate constants and kinetic parameters as detailed in the Supplementary Information, giving the Michaelis-Menten plot shown in Fig. 1l.
Synthesis of N7X/N9X by various nucleoside phosphorylases
To examine the ability of various NPs to synthesize N7X, we screened a small but diverse selection of NPs. To this end, we performed glycosylation reactions with 500 µM X, 500 µM Rib1P, and 1 g L−1 NP in 50 mM glycine-KOH buffer at pH 9 or 50 mM HEPES buffer at pH 7. These reactions were started by the addition of the enzyme and incubated at 50 °C for 1 or 24 h. Afterwards, a 50 µL sample of the reaction mixture was quenched by addition to 50 µL ice cold methanol and analyzed by HPLC as described above. For this experiment, we used bacterial (purine and pyrimidine NP from Geobacillus thermoglucosidasius, purine NP from Escherichia coli, and purine NP1 and NP2 from Thermus thermophilus) and mammalian NPs (human purine NP). These reactions yielded the conversions listed in Fig. 1m and Supplementary Table 5.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw and calculated data underlying this study are freely available from the externally hosted supplementary information at zenodo.org (https://doi.org/10.5281/zenodo.8382795)38. Specifically, this includes UV, NMR, HRMS, and DFT results.
References
Xu, J. et al. Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature 582, 60–66 (2020).
Fialho, D. M., Roche, T. P. & Hud, N. V. Prebiotic Syntheses of Noncanonical Nucleosides and Nucleotides. Chem. Rev. 120, 4806–4830 (2020).
Strnad, M. The aromatic cytokinins. Physiol. Plant 101, 674–688 (1997).
Sakakibara, H. CYTOKININS: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 57, 431–449 (2006).
Erion, M. D. et al. Purine Nucleoside Phosphorylase. 1. Structure−Function Studies. Biochemistry 36, 11725–11734 (1997).
Pugmire, M. J. & Ealick, S. E. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem. J. 361, 1–25 (2002).
Kamel, S., Thiele, I., Neubauer, P. & Wagner, A. Thermophilic nucleoside phosphorylases: Their properties, characteristics and applications. Biochimica et Biophysica Acta (BBA) - Proteins Proteomics 1868, 140304–140304 (2020).
Kaspar, F., Giessmann, R. T., Neubauer, P., Wagner, A. & Gimpel, M. Thermodynamic Reaction Control of Nucleoside Phosphorolysis. Adv. Synth. Catal. 362, 867–876 (2020).
Bzowska, A., Kulikowska, E. & Shugar, D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Therap. 88, 349–425 (2000).
Stachelska-Wierzchowska, A., Wierzchowski, J., Górka, M., Bzowska, A. & Wielgus-Kutrowska, B. Tri-Cyclic Nucleobase Analogs and Their Ribosides as Substrates of Purine-Nucleoside Phosphorylases. II Guanine and Isoguanine Derivatives. Molecules 24, 1493–1493 (2019).
Stachelska-Wierzchowska, A., Wierzchowski, J., Bzowska, A. & Wielgus-Kutrowska, B. Tricyclic nitrogen base 1,N6-ethenoadenine and its ribosides as substrates for purine-nucleoside phosphorylases: Spectroscopic and kinetic studies. Nucleosides, Nucleotides Nucleic Acids 37, 89–101 (2018).
Bzowska, A. et al. Purine nucleoside phosphorylase: Inhibition by purine N(7)- and N(9)-acyclonucleosides; and substrate properties of 7-β-D-ribofuranosylguanine and 7-β-D-ribofuranosylhypoxanthine. Biochem. Pharmacol. 48, 937–947 (1994).
Bzowska, A., Kulikowska, E., Poopeiko, N. E. & Shugar, D. Kinetics of Phosphorolysis of 3-(beta-d-Ribofuranosyl)Adenine and 3-(beta-d-Ribofuranosyl)Hypoxanthine, Non-Conventional Substrates of Purine-Nucleoside Phosphorylase. Eur. J. Biochem. 239, 229–234 (1996).
Kaspar, F. et al. Biased Borate Esterification during Nucleoside Phosphorylase‐Catalyzed Reactions: Apparent Equilibrium Shifts and Kinetic Implications. Angew. Chem. Int. Ed. 62, e202218492 (2023).
Kaspar, F., Stone, M. R. L., Neubauer, P. & Kurreck, A. Route efficiency assessment and review of the synthesis of β-nucleosides via N-glycosylation of nucleobases. Green Chemistry 23, 37–50 (2021).
Kaspar, F. et al. General Principles for Yield Optimization of Nucleoside Phosphorylase-Catalyzed Transglycosylations. Chem. Bio. Chem. 21, 1428–1432 (2020).
Bridson, P. K., Lin, X., Melman, N., Ji, X. & Jacobson, K. A. Synthesis AND Adenosine Receptor Affinity of 7-β-D-Ribofuranosylxanthine. Nucleosides Nucleotides 17, 759–768 (1998).
Kaspar, F. et al. Spectral Unmixing-Based Reaction Monitoring of Transformations between Nucleosides and Nucleobases. Chem. Bio. Chem. 21, 2604–2604 (2020).
Stoychev, G., Kierdaszuk, B. & Shugar, D. Xanthosine and xanthine: Substrate properties with purine nucleoside phosphorylases, and relevance to other enzyme systems. Eur. J. Biochem. 269, 4048–4057 (2002).
Elliott, R. D. & Montgomery, J. A. Analogs of 8-azaguanosine. J. Med. Chem. 19, 1186–1191 (1976).
Eilert, L., Schallmey, A. & Kaspar, F. UV-Spectroscopic Detection of (Pyro-)Phosphate with the PUB Module. Anal. Chem. 94, 3432–3435 (2022).
Kaspar, F., Ganskow, C. S. G., Eilert, L., Klahn, P. & Schallmey, A. Alternative Assay Reagents for UV-Spectroscopic Detection of (Pyro-)Phosphate with the PUB Module. Anal. Chem. 94, 8132–8135 (2022).
Kaspar, F. et al. Diversification of 4ʹ-Methylated Nucleosides by Nucleoside Phosphorylases. ACS Catal. 11, 10830–10835 (2021).
Traut, T. W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994).
Copley, S. D. An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 40, 72–78 (2015).
Leveson-Gower, R. B., Mayer, C. & Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. 3, 687–705 (2019).
Khersonsky, O. & Tawfik, D. S. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu. Rev. Biochem. 79, 471–505 (2010).
Kaspar, F. & Crameri, F. Coloring Chemistry—How Mindful Color Choices Improve Chemical Communication. Angew. Chem. Int. Ed. 61, e202114910 (2022).
Crameri, F., Shephard, G. E. & Heron, P. J. The misuse of colour in science communication. Nat. Commun. 11, 5444 (2020).
Software for Chemistry and Materials, AMS: Amsterdam Modelling Suite. http://www.scm.com-http://www.scm.com (2020).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 77, 3865–3868 (1996).
Van Lenthe, E. & Baerends, E. J. Optimized Slater-type basis sets for the elements 1–118. J. Comput. Chem. 24, 1142–1156 (2003).
Klamt, A. & Schüürmann, G. COSMO: a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc., Perkin Transactions 2, 799–805 (1993).
Stephens, P. J., Devlin, F. J., Chabalowski, C. F. & Frisch, M. J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 98, 11623–11627 (1994).
Becke, A. D. Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Lee, C., Yang, W. & Parr, R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988).
Davidson, E. R. The iterative calculation of a few of the lowest eigenvalues and corresponding eigenvectors of large real-symmetric matrices. J. Comput. Phys. 17, 87–94 (1975).
Kaspar, F. Supplementary Material N7X. zenodo https://doi.org/10.5281/zenodo.8382795 (2023).
Kaspar, F. Quality Data from Messy Spectra: How Isometric Points Increase Information Content in Highly Overlapping Spectra. ChemBioChem 24, e202200744 (2023).
Acknowledgements
The authors thank Samantha Voges (TU Berlin, NMR Department) for assistance with spectroscopic experiments and Dr. Maria Schlangen and Marc Griffel (TU Berlin, Center for Mass Spectrometry) for HRMS analyses. F.K. gratefully acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project number 492196858.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.K. and F.K.; Data curation, S.W., F.B., and F.K.; Formal analysis, S.W., F.B., S.K., and F.K.; Funding acquisition, A.K. and F.K.; Investigation, S.W., F.B., L.N., C.B., A.D., S.K., and F.K.; Methodology, F.K.; Project administration, A.K., F.K.; Resources, S.K., C.R.J., P.N., A.K., and F.K.; Software, -; Supervision, P.N., C.R.J., A.K., F.K.; Validation, -; Visualization, F.K.; Writing—original draft, F.K.; Writing—review & editing, all authors. Specifically regarding the experiments, A.D. and F.K. made the initial discovery. S.W. cloned the deaminase and the previously not reported nucleoside phosphorylases. S.W., L.N., C.B., A.D., and F.K. produced and purified the enzymes. S.W., L.N., C.B., and F.K. performed and analyzed the HPLC experiments (including the kinetics for the pH-dependent formation of N7X) and purified N7X. F.K. performed and analyzed the UV-spectroscopic experiments and carried out the physicochemical analyses. S.K. and F.K. carried out, and evaluated the NMR spectroscopic experiments. F.B. performed and evaluated the DFT calculations.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alan Michael Downey and Irina Varizhuk for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westarp, S., Brandt, F., Neumair, L. et al. Nucleoside Phosphorylases make N7-xanthosine. Nat Commun 15, 3625 (2024). https://doi.org/10.1038/s41467-024-47287-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-47287-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.