Abstract
Hospital surfaces can harbour bacterial pathogens, which may disseminate and cause nosocomial infections, contributing towards mortality in low- and middle-income countries (LMICs). During the BARNARDS study, hospital surfaces from neonatal wards were sampled to assess the degree of environmental surface and patient care equipment colonisation by Gram-negative bacteria (GNB) carrying antibiotic resistance genes (ARGs). Here, we perform PCR screening for extended-spectrum β-lactamases (blaCTX-M-15) and carbapenemases (blaNDM, blaOXA-48-like and blaKPC), MALDI-TOF MS identification of GNB carrying ARGs, and further analysis by whole genome sequencing of bacterial isolates. We determine presence of consistently dominant clones and their relatedness to strains causing neonatal sepsis. Higher prevalence of carbapenemases is observed in Pakistan, Bangladesh, and Ethiopia, compared to other countries, and are mostly found in surfaces near the sink drain. Klebsiella pneumoniae, Enterobacter hormaechei, Acinetobacter baumannii, Serratia marcescens and Leclercia adecarboxylata are dominant; ST15 K. pneumoniae is identified from the same ward on multiple occasions suggesting clonal persistence within the same environment, and is found to be identical to isolates causing neonatal sepsis in Pakistan over similar time periods. Our data suggests persistence of dominant clones across multiple time points, highlighting the need for assessment of Infection Prevention and Control guidelines.
Similar content being viewed by others
Introduction
Environmental surfaces and patient care equipment in hospital settings are among the most critical factors in bacterial horizontal transmission events from high-touch surfaces to patients1,2,3, with higher hospital-acquired infection (HAIs) rates in low-income and middle-income countries (LMICs) compared to high-income countries (HICs)4. Unless appropriate infection prevention and control (IPC) guidelines are applied and include effective cleaning, disinfection and hygiene practices4,5,6, environmental reservoirs of multidrug-resistant (MDR) bacteria can develop and direct contact with these may contribute to HAIs, such as surgical site infections and bloodstream infections7,8,9,10,11,12,13,14. High levels of antimicrobial resistance (AMR) can lead to difficulties in the treatment of MDR infections, associated with high mortality15,16,17,18. Neonates are particularly at risk because of their underdeveloped immune system, and neonatal infection rates in LMICs are 3–20 times higher than in HICs19. Furthermore, AMR bacteria with tolerance to disinfectants may survive on environmental surfaces for months, depending on factors such as humidity, temperature, air ventilation and surface characteristics4,9,20, which will further increase transmission likelihood.
Empirical treatment using cephalosporins or carbapenems in LMIC healthcare settings has influenced the progressive emergence of extended-spectrum β-lactamase- (ESBL-) and carbapenemase-producing bacteria. Antibiotic resistance genes (ARGs) such as blaCTX-M-15, and blaNDM, blaOXA-48-like or blaKPC are often plasmid-borne and/or other mobile genetic element- (MGE) associated, enhancing potential for transmission8.
The most common ESBL- and carbapenemase-producing Gram-negative bacteria (GNB) in hospitals are Enterobacterales1,3,8,11,12,21,22. Few publications have shown data regarding the presence of GNB carrying β-lactamase genes on hospital surfaces in LMICs. Moreover, most of these studies performed in LMICs are single sites focused on specific countries, such as Bangladesh, Pakistan, Ethiopia, or Ghana1,23,24,25.
Here, we determine the prevalence and diversity of ESBL- and carbapenemase-carrying bacterial species colonising neonatal wards from six countries within the BARNARDS study (Burden of Antibiotic Resistance in Neonates from Developing Societies). Genetic lineages are assessed to determine whether transmission events occurred and whether they are related to bacteria causing neonatal sepsis in the BARNARDS study.
Results
GNB colonisation among countries and hospital sites
In total, 6290 hospital surface swabs (HSS) were processed from ten hospitals across six countries; and 60.7% HSS had GNB growth (Table 1). GNB colonisation varied significantly between the countries and hospital sites, with the highest growth in Bangladesh (92%).
ARG among countries and hospital sites
Overall, 839/6290 (13.3%) of HSS were positive for blaCTX-M-15, 338/6290 (5.4%) for blaNDM, and 74/6290 (1.2%) for blaOXA-48-like. blaKPC was not detected during this study, therefore, from this point, the term ARGs denotes blaCTX-M-15, blaNDM and blaOXA-48-like. Concomitant carriage of more than one ARG in the same sample was found in ~1% of samples (Supplementary Table 1).
The prevalence of blaCTX-M-15, blaNDM and blaOXA-48-like ranged between countries (Fig. 1 and Supplementary Table 2). Noticeable differences between hospitals were observed; for instance, in Bangladesh, almost twice as many blaNDM were detected in BK (17.3%) compared to BC (10.5%), whereas blaOXA-48-like was four times more frequent in BC (1.2%). Hospital surfaces in RU had higher blaNDM (2.2%) rates compared to RK, and conversely, blaCTX-M-15 and blaOXA-48-like were more prevalent (24.4% and 1.1%, respectively) in RK. In Nigeria, the prevalence of all the ARGs was highest in one site (NK).
Prevalence of blaCTX-M-15 (a), blaNDM (b), and blaOXA-48-like (c) genes per country and per hospital site. The total number of samples collected per country and per hospital site were used as denominators to calculate the prevalence of each antibiotic resistance gene (ARG) per country and per hospital site, respectively. Abbreviations for BARNARDS hospitals are detailed in the “Methods” section, map pins showing latitude and longitude, and coloured according to the ARG prevalence in that site. Source data are provided in Supplementary Table 2.
GNB colonisation and ARG detection among hospital surfaces
To study the correlation between the presence of ARG and surface type, n = 4126/6290 samples contained appropriate metadata following data cleaning (Table 2). The 4126 HSS were collected from 309 different surfaces (Supplementary Data 1) and classified into six categories (listed in Table 2; see methods section “Data cleaning and statistical analysis” for details).
Table 3a summarises the differences in GNB growth across surface categories observing the largest growth near sink drains (68.4%). HSS collected from medical equipment were the least contaminated with Gram-negative bacterial colonisation (45.9%). Furthermore, HSS positive for blaCTX-M-15 and blaOXA-48-like varied significantly (P < 0.001 and P = 0.004, respectively) between the six surface categories (Table 3b). Although the presence of blaNDM varied, this was not statistically significant (P = 0.071), and the ARG presence was higher in surfaces classified as near the sink drain (Table 3). For detailed ARG prevalence on the different 309 surface types collected and on surface categories among all hospital sites, see Supplementary Data 2 and 3, respectively.
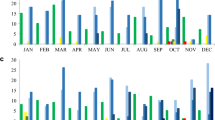
To study the correlation between the presence of ARG and a particular time of the year, n = 4662/6290 samples contained appropriate metadata following data cleaning. Temporal analysis revealed that the percentage of blaCTX-M-15, blaNDM and blaOXA-48-like varied significantly between the seven-time bands across the sampling period from November 2015 until January 2018 (P < 0.001 for blaCTX-M-15 and blaNDM, and P = 0.041 for blaOXA-48-like) (Table 4). GNB HSS colonisation appeared to be highest during the March–June and July–October periods.
GNB species carrying bla NDM and bla OXA-48-like
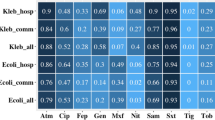
In total, 175 bacterial isolates from 151/6,290 HSS with a positive multiplex-PCR for a carbapenemase ARG were purified. MALDI-TOF MS identified 27 different bacterial species, with Klebsiella spp. and Enterobacter spp. carrying blaNDM and/or blaOXA-48-like equally dominant (n = 53/175, 30.3%). Within the Klebsiella spp., K. pneumoniae was most prevalent, accounting for 86.8% (n = 46/53). Similarly, within the Enterobacter spp. isolates recovered, E. hormaechei was the most frequent (n = 38/53, 71.7%), followed by E. cloacae (n = 8/53, 15.1%). Other GNBs identified include Acinetobacter baumannii (n = 13/175, 7.4%), Pseudomonas spp. (n = 14/175, 8%), Serratia marcescens (n = 12/175, 6.9%) and Leclercia adecarboxylata (n = 11/175, 6.3%) (Fig. 2). Within the dataset, only three E. coli isolates carrying carbapenemase ARG were recovered (n = 3/175, 1.7%, Supplementary Data 4 and Supplementary Table 3).
Bacterial species carrying blaNDM (a) and blaOXA-48-like (b) among countries and sites, according to PCR screening and MALDI-TOF MS identification (a total of 175 bacterial isolates from 27 bacterial species; and 3 “unidentified” isolates). Source data are provided in Supplementary Data 4.
blaNDM-producing K. pneumoniae and E. hormaechei isolates were mostly found in Pakistan and altogether represented over 60% of the 93 carbapenemase-producing isolates found in PP (Supplementary Data 4). Supplementary Table 3 shows detailed information on surfaces where these isolates were recovered from.
Bacterial diversity and whole genome sequencing (WGS) analysis of ARG variants
Whole genome sequencing (WGS) data was available for 128 carbapenemase-positive GNB isolates out of the 175 total isolates recovered from HSS (due to loss of growth or loss of carbapenemase gene upon regrowth), and of these, 18 bacterial species were identified (Supplementary Data 5).
Of the GNB isolates with WGS data, 122 carried at least one blaNDM variant (n = 114 blaNDM-1, n = 3 blaNDM-5, and n = 7 blaNDM-7 genes) and 17 isolates carried blaOXA-48-like genes (n = 14 blaOXA-181, n = 2 blaOXA-204, n = 1 blaOXA-48). The majority of carbapenemase ARGs were found to be plasmid-mediated (Fig. 3), however, for 16 isolates, the ARGs were chromosomally located (Supplementary Data 6).
There were n = 95 isolates with an identifiable Inc plasmid type (due to assembly fragmentation, it was not always possible to assemble and analyse whole plasmids). The carbapenemase variant NDM and OXA-48-like group are divided into variants, and the Inc type detected per ARG-variant is shown. Source data are provided in Supplementary Data 5 and 6 and as Source Data file.
Twelve isolates concomitantly carried variants of blaNDM and blaOXA-48-like. This was most notably detected within sequence type (ST) 15 K. pneumoniae (n = 10/16 from Pakistan), with n = 9/10 carrying both blaNDM-1 and blaOXA-181 on similar ~140 kbp IncA/C and ~50 kpb ColKP3-IncX3 hybrid plasmids respectively (Fig. 3, Supplementary Figs. 1–3); n = 1/10 carried both variants, but the plasmid type remained unknown due to assembly fragmentation.
Two ST11 K. pneumoniae isolates from Pakistan carried two blaNDM genes: one harboured two copies of blaNDM-7 on a 30 kbp IncX3 and 145 kbp IncA/C plasmids, and the other one carried blaNDM-1 (on a ~39 kpb IncX3 plasmid) and blaNDM-7 on a ~145 kpb IncA/C plasmid (Fig. 3).
The other two Klebsiella species that were identified were K. quasipneumoniae and K. michiganensis. Plasmids represented in Supplementary Figs. 1, 2 from were from multiple bacterial species, including K. pneumoniae, K. michiganensis and Enterobacter roggenkampii.
Of the six isolates with blaNDM-7, three were K. pneumoniae and three were E. hormaechei.
From the 47 Enterobacter isolates, E. hormaechei was dominant (n = 38). Supplementary Data 6 shows the distinct ST recovered, with 21 STs for Enterobacter alone, with the most frequent being ST120, ST231, ST316 and ST418, all E. hormaechei. Three E. hormaechei isolates with different STs carrying a blaNDM-5 on IncX3 plasmids (ranging from 46 to 55 kb; Fig. 3) were recovered from a single hospital (NK) in the same month in 2016 (Supplementary Data 5 shows the recovery date of the isolates with WGS data available). The two 46 kb plasmids (pNK-E166-IncX3 and pNK-E171A-IncX) shared 99.7% sequence homology. Supplementary Figs. 3, 4 show also that the 55 kb pNK-E179A-IncX3 was more genetically distant (30–31% aligned homologous sequences). Eight Enterobacter spp. isolates, mostly ST316 E. hormaechei (n = 5) co-carried blaNDM-1 and mcr-9 on a large 400kbp IncHI2 plasmid. S. marcescens was most often recovered in both BC and BK, all n = 9 isolates carried blaNDM-1, either on IncF or IncL/M plasmid types (Fig. 3).
One ST78 A. baumannii with two copies of blaNDM-1 and four ST52 A. baumannii isolates were recovered from RU and the hospital in South Africa (ZAT). Of the two E. coli isolates (ST448 and ST405) identified by WGS, both carried blaNDM-1 and one (ST405) co-carried blaNDM-1 and blaOXA-181.
WGS confirmed the presence of multiple aminoglycosides (aac, ant, aph), ESBLs (blaOXA-1, blaSHV-11, blaSHV-12, blaSHV-106, blaSHV-182 and blaSHV-187), fosfomycin (fosA) and tetracycline (tetA, tetB, tetD) ARGs. All isolates were additionally screened for genes conferring resistance to disinfectants, and MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexJK-OpmH were identified in three Pseudomonas spp. isolates, two from Bangladesh and one from Pakistan.
Evidence of local transmission and links to neonatal sepsis
From WGS and epidemiology analysis, nine potential cluster/transmission events were analysed by SNP analysis (Fig. 4). HSS isolates from two clusters shared the same ST of isolates from the same hospital site causing neonatal sepsis during BARNARDS (ST15 K. pneumoniae and ST52 A. baumannii isolates). WGS data from these sepsis isolates were included in the single nucleotide polymorphism analysis (total of n = 22 ST 15 K. pneumoniae isolates causing sepsis in Pakistan)26.
Sequence types (STs) and antimicrobial resistance genes (ARGs) are shown, and the highlighted background around isolates indicated these were within 10 pairwise SNPs. The different pink tone for ST20 K. pneumoniae indicates distinct sub-clusters identified among these isolates in Pakistan (over 1000 SNPs). Abbreviations for BARNARDS hospitals are detailed in the “Methods” section. The figure was created using Adobe Illustrator v26.5. Source data are provided in Supplementary Data 5 and 6.
For ST15 K. pneumoniae, there were 16 HSS isolates analysed alongside 22 ST15 K. pneumoniae isolates from neonatal blood cultures within PP during the same time-period (November 2015–November 2017). ST15 K. pneumoniae were recovered from HSS from emergency neonatal care (n = 5), patient’s zone (n = 4), ward furniture/surfaces (n = 1), medical equipment (n = 1), and NA (n = 5). SNP analysis revealed that, except for one isolate, ST15 K. pneumoniae from Pakistan from both HSS and neonatal sepsis blood cultures (n = 22) were within 10 pairwise SNPs. Interestingly, in eight HSS isolates, blaOXA-181 and the ColKP3–IncX3 hybrid replicon were not detected, indicating that some isolates might not have acquired the ColKP3-IncX3 plasmid or may have lost the plasmid during culture for WGS. Furthermore, the ST405 E. coli isolates from the same hospital wards in Pakistan carried genetically similar plasmids, both the IncA/C blaNDM-1 and the ColKP3-IncX3 blaOXA-181 plasmids suggesting plasmid transmission may have occurred between bacterial species colonising hospital surfaces. It is possible that the ST405 E. coli isolate acquired both blaNDM-1 and blaOXA-181 from ST15 K. pneumoniae, as our data indicates multiple hospital surfaces were contaminated with this strain between 2016 and 2017 (Supplementary Figs. 5–8).
From nine A. baumannii isolates with WGS data, five were ST52, and SNP analysis of ST52 revealed the sepsis isolate (identified in the same hospital in Rwanda (RU)26) to be genetically distant (>100 SNPs) from the isolates collected from HSS, which were within 10 SNPs.
Isolates of ST20 and ST1317 blaNDM-1 K. pneumoniae were within 10 pairwise SNPs among isolates from each cluster, further evidencing the colonisation and potential transmission across the hospital wards in Pakistan. For ST20 isolates, there were two small sub-clusters identified, with genetic grouping occurring for isolates collected in late 2016 (September–November) and a distinct cluster of over 1000 SNPs distinct for ST20 K. pneumoniae recovered between January and March 2017 (Fig. 4).
E. hormaechei, the dominant Enterobacter species, was mostly identified in Asia. Clusters of ST231 and ST316 (blaNDM-1 and mcr-9) E. hormaechei isolates recovered across surfaces in PP were within three and two pairwise SNPs. A cluster of seven ST418 E. hormaechei HSS isolates from BK, recovered during a 6-week period between October and November 2017, were within four pairwise SNPs.
A cluster of five BK S. marcescens isolates (within 3 SNPs) was over 10,000 SNPs distant from the S. marcescens isolates from NN and BC. Nine L. adecarboxylata isolates isolated between March and November 2017 from different surfaces in PP were closely related (within a single SNP distance) (Fig. 4).
Discussion
Herein, we report a high prevalence of bacteria carrying blaCTX-M-15, blaNDM and blaOXA-48-like genes colonising environmental surfaces and patient care equipment in 10 hospitals across six LMICs. To the best of our knowledge, this multinational study is the first to evidence transmission networks between hospital wards and neonates with sepsis in LMICs26. Importantly, a high-risk double carbapenemase ST15 K. pneumoniae clone8,27 was recovered from HSS and blood cultures from septic neonates in Pakistan over a 2-year period, suggesting this strain was colonising surfaces in the ward whilst simultaneously causing neonatal sepsis26. In our dataset, we identified a single E. coli isolate carrying a genetically similar blaNDM (IncAC plasmid type) plasmid to the ST15 K. pneumoniae strain isolated from the same hospital (PP). We evidence similarity between plasmids detected within Enterobacterales from multiple hospital surfaces sampled between May 2016 and November 2017, indicating possible horizontal transmission however, a larger genomics dataset would be needed to understand plasmid transmission dynamics upon hospital surface samples (e.g. plasmids harboured in E. hormaechei isolates from NK).
ST15 K. pneumoniae has been reported to be responsible for neonatal sepsis and high mortality in clinical settings8, is often linked with HAIs and hospital outbreaks28,29,30 and is frequently detected in the environment (wastewater and soil) in Africa and Asia31,32 Furthermore, multiple clusters of the same strain of different GNB species were detected from hospital surfaces in Bangladesh and Rwanda, suggesting that hospital surface colonisation by pathogenic bacteria is a significant concern in LMICs hospitals. WHO and Médecins Sans Frontières still lack AMR data from many LMIC settings, thus, global reports on IPC, burden of sepsis or HAIs contain information mostly generated from HICs. This data highlights the widespread bacterial colonisation and the transmission of bacteria carrying multiple ARG upon hospital surfaces, which could be useful to guide realistic approaches and support action plans for countries where IPC practices are limited13,18,19,22,33,34.
Previous studies report that frequently used medical equipment/touch surfaces in healthcare settings are crucial for the cross-transmission of AMR bacteria and HAIs, such as sepsis3,7,17,21,22,25,35. This is particularly true in institutions where resources to implement IPC programs are scarce4,13. Bacteria carrying ARGs appeared to be found most frequently on surfaces near the sink drain, which was concordant with the work by Firesbhat et al. in Ethiopia16. We also found contamination of medical equipment and/or ward furniture/surfaces, as detected in other African-based studies reporting on HAIs24,36,37,38.
Interestingly, ESBL- and carbapenemase-producing Enterobacterales were more frequently detected in HSS collected between March and October. Despite the potential contamination pattern during this period, due to inconsistent sampling throughout the hospital sites among time periods, detailed seasonal analysis per country could not be performed herein. Apart from unequal sample size, a variety of factors, including temperature, cleaning practices, or healthcare workers shifts, may be influencing bacterial reservoirs at particular times of the year.
Our findings are comparable with previous studies in Pakistan25, Nigeria36 and Ethiopia24,39, which report >60% bacterial growth on medical equipment and environmental surfaces. We observed large differences in GNB growth between countries or hospitals within the same country, emphasising that bacterial colonisation should be monitored in each hospital. Apart from the sampling limitations of this study, there are likely various factors contributing to the range of colonisation observed, including antibiotic accessibility40.
In this study, K. pneumoniae, E. hormaechei, A. baumannii, S. marcescens and L. adecarboxylata were the most prevalent bacterial species carrying blaCTX-M-15, blaNDM and blaOXA-48-like genes across all countries, which have been commonly described1,3,8,11,17,24,25,35,39. WGS revealed diversity within species showing multiple and co-occurring dominant STs of E. hormaechei and K. pneumoniae. MDR A. baumannii carrying OXA-carbapenemase genes were found among hospital surfaces in Bangladesh in 202123. In this study, however, E. hormaechei, Pseudomonas spp. and Shewanella putrefaciens were identified as the bacterial species carrying blaOXA-48-like genes in BK. Whilst we did detect A. baumannii (7.4%), this was mostly recovered from HSS collected in RU (ST52 carrying blaNDM-1), but none from Bangladesh. The present work confirms that blaOXA-48-like genes are widespread in African and South-Asian countries. Contrary to results from a hospital surface screen of multiple wards in a hospital in Ghana1, we showed a lower prevalence of blaNDM (n = 338/6290 samples, 5.4%). Nevertheless, the prevalence of this gene, particularly in Pakistan (n = 162/1033, 15.7%), was in line with a study from several hospitals in the same country41. blaKPC is not common in Africa or South Asia7,26, which this study also confirmed; however, blaKPC was detected from neonatal cots in Tanzania in 202117 and blaKPC K. pneumoniae was found on pillows and the floor in three hospitals in Bangladesh42.
Limitations of this study include a lack of consistency in swabbing areas, as their descriptions were, on occasions, imprecise and the samples varied across each hospital. Thus, we were cautious with data interpretation and did not present correlation data between GNB growth and/or ARG prevalence per surface per hospital or between surface type/category and bacterial species due to sampling limitations. A second limitation was not grouping swab locations to reduce the testing size, which resulted in a total of 309 different surfaces, causing difficulties when comparing across countries and hospitals. Thirdly, there were differences in the number of samples collected across hospital sites and time, as we did not confirm the exact number/type of samples per month. Moreover, there was a lack of written information accompanying some samples—not all the samples were well-described. Information related to cleaning and disinfection practices in each hospital site was not collected, which might explain some of the results obtained; PCR screening for other genes would have also given us a broader view of potential resistance to other antibiotics available in LMICs but not commonly used due to economic resources or accessibility.
To summarise, we have shown that ESBL- and carbapenemase-producing Enterobacterales were most prevalent in samples collected near the sink drain. Transmission events occurred across patient care equipment and environmental surfaces of the hospital wards, and worryingly, there was evidence that the same strains have caused neonatal sepsis. Moreover, and of particular concern, is the high prevalence of antibiotic resistance determinants and the diversity of ARG-containing bacteria among surfaces within the hospital facilities, presenting an increasing threat to the patients. Future work to determine the relative risk of inborn neonates developing sepsis with specific bacterial strains that are colonising HSS is warranted to fully understand the impact of bacterial transmission events across the wards in neonatal sepsis. Our results further emphasise the extent of hospital surface contamination with bacteria carrying multiple ARG in LMICs, calling for an urgent assessment of improved IPC practice compliance and tailored guidelines for each hospital site.
Methods
Settings, ethics, participants, and study design
The BARNARDS network included Bangladesh, Chittagong Maa-O-Shishu Hospital, Chattogram (BC), and Kumudini Women’s Medical College, Mirzapur (BK); Ethiopia, St. Paul’s Hospital Millennium Medical College, Addis Ababa (ES); India, Division of Bacteriology, ICMR-National Institute of Cholera and Enteric Diseases Beliaghata and Institute of Post-Graduate and Medical Education and Research, Kolkata (IN); Nigeria, National Hospital Abuja (NN), Wuse District Hospital, Abuja (NW), and Murtala Mohammad Specialist Hospital, Kano (NK); Pakistan, Pakistan Institute of Medical Sciences (PP) and Bhara Kahu Rural Health Centre, Bhara Kahu (PC); Rwanda, University Central Hospital of Kigali, Kigali (RU) and Kabgayi Hospital, Kabgayi (RK); South Africa, Tygerberg Hospital, Cape Town (ZAT). HSS were not collected in India or one of the hospitals in Pakistan (PC). The hospital site abbreviation names are used throughout this article; however, the country name is used when the results are applicable to all hospitals within that country.
Standard operating procedures (SOPs) were designed and adhered to throughout the network (https://www.ineosoxford.ox.ac.uk/research/areas-of-focus/amr-burden/barnards), and ethical approval was obtained from local ethics committees prior to the start of the study.
From November 2015 until January 2018, HSS were collected from different surfaces (Supplementary Data 1) within different wards (i.e. maternity and neonatal intensive care units). Frequently, information regarding wards was incomplete, thus, only the surface but not their location within the hospitals was considered for analysis. Samples from environmental surfaces and patient care equipment in hospital settings were collected with charcoal swabs and stored at 4 °C until transported to the UK under UN3373 conditions.
HSS processing
HSS were streaked on three chromogenic agar media plates (Liofilchem®, Italy) supplemented with vancomycin (10 mg/L) to promote the growth of GNB, vancomycin and cefotaxime (VC, 10 and 1 mg/L, respectively) to select for ESBL producers, and vancomycin and ertapenem (VE, 10 and 2 mg/L, respectively) to select for carbapenemase producers7. VC plates were tested for the presence of blaCTX-M-15 by PCR and VE plates were tested for blaNDM, blaOXA-48-like and blaKPC by multiplex-PCR. All bacterial cultures were preserved in TS/72 cryogenic beads (Technical Service Consultants, UK) and stored at −80 °C. When VE bacterial growth yielded multiplex-PCR positive results, phenotypically distinct bacterial colonies were isolated and screened for the carbapenemase genes in the study by multiplex-PCR. Those isolates with a positive result were identified by MALDI-TOF MS (Bruker Daltonik GmbH, Coventry, UK) and preserved as detailed above. As per the BARNARDS protocol, due to the high prevalence of blaCTX-M-15, we did not scrutinise samples for blaCTX-M-15 positive isolates. The study workflow and dataset are summarised in Fig. 5. ARG prevalence resulting from PCR screening was represented in Coloured maps, which were created using MapChart (https://www.mapchart.net).
Diagram detailing the total number of hospital surface swabs (HSS) collected, showing Gram-negative bacteria (GNB) growth, and screened for the presence of blaCTX-M-15, blaNDM, blaKPC and blaOXA-48-like antimicrobial resistance genes (ARGs), the number of GNB isolates recovered carrying carbapenemase genes, the number of isolates characterised by whole genome sequencing (WGS) and, where WGS data was sufficient, bioinformatic analysis was performed. Isolates for WGS were chosen after culture on VE (vancomycin, ertapenem) agar. Recoverable isolates after −80 °C preservation were selected for gDNA extraction and WGS. Data cleaning is also detailed; where data regarding hospital surfaces and collection dates was available, statistical analysis and data analysis were performed.
Whole genome sequencing (WGS)
WGS was carried out on GNB isolates positive for at least one of the carbapenemase genes. Briefly, and as detailed by Sands et al.26, gDNA was extracted using the QIAmp DNA mini kit (Qiagen, Germany) on the QIAcube platform (Qiagen, Germany) and quantified using the Qubit fluorometer 4.0. Short-read sequencing libraries were prepared using the Nextera XT v2 kit and paired-end sequenced on an Illumina MiSeq using the V3 chemistry to generate fragment lengths up to 300 bp (600 cycles). Long reads were prepared using Oxford Nanopore Technology (ONT) and libraries were generated using the 96-Rapid Barcoding Kit (SQK-RBK110.96; ONT). Sequencing was performed using MinION flow cells (R9.4.1) for a running time of 72 h within MinKnow.
Bioinformatic analysis
To determine whether bacterial transmission events occurred within the hospital wards and/or neonatal sepsis, bioinformatic analysis was performed as detailed by Sands K et al.26. ONT FAST5 reads were base called using Guppy v5.0.11 and NVIDIA V100 GPUs. Following QC on short reads (Illumina) and long reads (ONT) as described by Boostrom et al.43, reads were assembled using Unicycler (v0.4.9). Genome assembly metrics were generated using QUAST (v.5.2.0)44 and bacterial species were identified using Pathogenwatch45. Multilocus sequence typing (MLST), characterisation of genes conferring resistance to antibiotics and/or disinfectants genes and plasmid genomic profiles were performed using ABRicate (v0.9.7)46 and associated databases: NCBI47, PlasmidFinder48 and CARD49. Previously undefined alleles and sequence type (ST) profiles were submitted to Enterobase, BIGSbd and PubMLST for assignment50. Genomes were annotated using Prokka (v1.14.5)51. Following initial WGS analysis, four or more isolates with the matching ST/species from a single hospital site were considered a potential cluster and were subject to SNP analysis using snippy (v4.6.0)52 with BWA and freebayes mapping the reads and calling variants --min cov 20. To maximise SNP calling, a high-quality internal reference was used53. A pairwise SNP matrix was performed converting a FASTA alignment to an SNP distance matrix using Snp-dists (v0.8.2)54. Adobe Illustrator v26.5 was used to generate the isolate-relatedness timeline figure. The Sankey diagram was created using Sankeymatic.com.
For plasmid analysis for evidence of transmission between bacterial isolates/species, the contig containing the carbapenemase variant and plasmid was analysed using the Mauve aligner55 within Geneious (v2023.2.1). The size of the plasmid and whether the assembler theoretically denoted the contig as circular and complete were recorded. Following Mauve alignment, the plasmid sequences were annotated using Prokka (v1.14.5) and further annotated using a reference plasmid from a BLAST search within Geneious. Visualisations of annotated aligned plasmids were performed within Geneious. Further similarity matrices were generated using mash dist (v2.2)56 to estimate the genetic similarity between plasmid sequences. Sequence alignments were performed within Geneious using the MAFFT aligner plugin.
Data cleaning and statistical analysis
To analyse the frequency of GNB growth in hospital surfaces, GNB growth on plates supplemented with vancomycin was examined, and the total number of processed samples was used as a denominator for the prevalence of assessed ARGs. HSS was collected from different surfaces (Supplementary Data 1) and included both environmental surfaces and patient care equipment in hospital settings, which were classified into six different categories (Supplementary Data 7) based on WHO and CDC IPC guidelines and previous publications4,13,18,57, following data cleaning. In this study, environmental surfaces included the hands of healthcare workers, which were categorised into the patient’s zone as they can act as a source of transmission4,58,59. The category of patient care equipment was defined as medical equipment or mobile medical equipment, categories that include non-critical, semi-critical and noncritical patient care equipment. The six final categories were: surfaces from the patient’s zone (immediate environment), surfaces near the sink drain (including sink basin, faucet, faucet handles, and surrounding countertop)60,61, emergency neonatal care, ward furniture/surfaces, mobile medical equipment, and medical equipment. Due to unequal sample size (Table 2) as well as sample variety (Supplementary Data 1), statistical analyses were performed using the total number of samples included in each surface category and not per country/hospital. Furthermore, following data cleaning, the HSS collection dates were classified into seven-time bands (Supplementary Table 4) according to Climate Change Knowledge Portal (World Bank Group)62. Statistical analysis to study the ARG prevalence was performed considering HSS collected per time band as a denominator, inclusive of all countries and hospitals. No seasonal analysis per country was performed.
ARG frequencies per hospital, country and surface category and corresponding figures were assessed. RStudio ggplot2 package was used for figure creation (RStudio version 4.3.0 (2023-04-21) -- “Already Tomorrow”), and IBM SPSS Statistics (Version 25.0.0.1) (190) was used to perform the statistical analyses. Statistical analyses were performed to determine whether certain surfaces were at greater risk of colonisation with β-lactamase-producing bacteria. The relationship between ARG and a surface category, as well as the frequency of GNB colonisation of hospital surfaces among countries and hospital sites, and over the timeline, were analysed to obtain counts and percentages. Chi-Squared tests were conducted to test the independence of the variables on contingency tables to establish if the overall differences in frequencies between GNB colonisation and ARG prevalence over countries and surfaces were statistically significant at the P < 0.05 level. When such differences were seen, the individual proportions for each of the countries and surfaces were examined and compared to determine where the main differences lay.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Databases used for in silico analysis in this work are PlasmidFinder, CARD, Enterobase, and PubMLST. Coloured maps in the paper were created using MapChart (https://www.mapchart.net). The dataset generated in this study is deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.23790360). Source data are provided with this paper; available as Supplementary Data files or provided as a Source Data file. Genomes are available in the NCBI database under BioProject number PRJNA971772 (and accession codes/accessible links are provided in Supplementary Data 8). The plasmid analysis data generated in this study for evidence of transmission is available in Supplementary figures. Source data are provided with this paper.
References
Acolatse, J. E. E. et al. Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrob. Resist. Infect. Control 11, 49 (2022).
Dancer, S. J. The role of environmental cleaning in the control of hospital-acquired infection. J. Hosp. Infect. 73, 378–385 (2009).
Zahornacký, O., Porubčin, Š., Rovňáková, A. & Jarčuška, P. Gram-Negative rods on inanimate surfaces of selected hospital facilities and their nosocomial significance. Int. J. Environ. Res. Public. Health 19, 6039 (2022).
CDC and ICAN. Best Practices for Environmental Cleaning in Resource-Limited Healthcare Settings. A Healthcare Cleaning and Disinfection Guide for Healthcare Settings with Limited Resources (CDC and ICAN, 2023).
Boyce, J. M. Alcohols as surface disinfectants in healthcare settings. Infect. Control Hosp. Epidemiol. 39, 323–328 (2018).
World Health Organization. Standard Precautions for the Prevention and Control of Infections: Aide-Memoire (World Health Organization, 2022).
Carvalho, M. J. et al. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nat. Microbiol. 7, 1337–1347 (2022).
Mukherjee, S., Mitra, S., Dutta, S. & Basu, S. Neonatal sepsis: the impact of carbapenem-resistant and hypervirulent Klebsiella pneumoniae. Front. Med. 8, 634349 (2021).
Musoke, D. et al. The role of Environmental Health in preventing antimicrobial resistance in low- and middle-income countries. Environ. Health Prev. Med. 26, 100 (2021).
Rudd, K. E. et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020).
Sands, K. et al. Early-onset neonatal sepsis in low- and middle-income countries: current challenges and future opportunities. Infect. Drug Resist. 15, 933–946 (2022).
Shao, Y. et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121 (2019).
World Health Organization. Minimum Requirements for Infection Prevention and Control Programmes (World Health Organization, Geneva, 2019).
World Health Organization. Pocket Book of Hospital Care for Children. Guidelines for the Management of Common Childhood Illnesses (World Health Organization, Geneva, 2013).
Farzana, R. et al. Outbreak of hypervirulent multidrug-resistant Klebsiella variicola causing high mortality in neonates in Bangladesh. Clin. Infect. Dis. 68, 1225–1227 (2019).
Firesbhat, A., Tigabu, A., Tegene, B. & Gelaw, B. Bacterial profile of high-touch surfaces, leftover drugs and antiseptics together with their antimicrobial susceptibility patterns at University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Microbiol. 21, 309 (2021).
Silago, V. et al. Predominance of Acinetobacter spp., Harboring the blaIMP gene, contaminating the hospital environment in a tertiary hospital in Mwanza, Tanzania: a cross-sectional laboratory-based study. Pathogens 11, 63 (2022).
World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities (World Health Organization, Geneva, 2017).
World Health Organization. Report on the Burden of Endemic Health Care-Associated Infection Worldwide (World Health Organization, Geneva, 2011).
Chaoui, L., Mhand, R., Mellouki, F. & Rhallabi, N. Contamination of the surfaces of a health care environment by Multidrug-Resistant (MDR) bacteria. Int. J. Microbiol. 2019, 1–7 (2019).
Won, S. Y. et al. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase-producing enterobacteriaceae. Clin. Infect. Dis. 53, 532–540 (2011).
World Health Organization. Global Report on the Epidemiology and Burden of Sepsis. https://www.who.int/publications-detail-redirect/9789240010789 (World Health Organization, 2020).
Farzana, R., Swedberg, G., Giske, C. G. & Hasan, B. Molecular and genetic characterization of emerging carbapenemase-producing Acinetobacter baumannii strains from patients and hospital environments in Bangladesh. Infect. Prev. Pract. 4, 100215 (2022).
Darge, A., Kahsay, A. G., Hailekiros, H., Niguse, S. & Abdulkader, M. Bacterial contamination and antimicrobial susceptibility patterns of intensive care units medical equipment and inanimate surfaces at Ayder Comprehensive Specialized Hospital, Mekelle, Northern Ethiopia. BMC Res. Notes 12, 621 (2019).
Zubair, M. et al. Role of hospital surfaces in transmission of infectious diseases. Pak. J. Med. Health Sci. 12, 857–859 (2018).
Sands, K. et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 6, 512–523 (2021).
Farzana, R. et al. Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 5, e00023–20 (2020).
Lam, M. M. C. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188 (2021).
Wyres, K. L., Lam, M. M. C. & Holt, K. E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359 (2020).
Lam, M. M. C. et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 10, 77 (2018).
Ekwanzala, M. D., Dewar, J. B., Kamika, I. & Momba, M. N. B. Tracking the environmental dissemination of carbapenem-resistant Klebsiella pneumoniae using whole genome sequencing. Sci. Total Environ. 691, 80–92 (2019).
Obasi, A. et al. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae from pharmaceutical wastewaters in South-Western Nigeria. Microb. Drug Resist. 23, 1013–1018 (2017).
World Health Organization. Guidelines on Core Components of Infection Prevention and Control Programmes at the National and Acute Health Care Facility Level (World Health Organization, Geneva, 2016).
World Health Organization. Global Report on Infection Prevention and Control (World Health Organization, 2022).
Costa, D. M. et al. Biofilm contamination of high‐touched surfaces in intensive care units: epidemiology and potential impacts. Lett. Appl. Microbiol. 68, 269–276 (2019).
Yusha’u, M., Bukar, A., Aliyu, B. S. & Abdulkareem, A. Bacterial contamination of some hospital equipments in Kano, Nigeria. Hamdard Med. 55, 39–42 (2012).
Munyeshyaka, E., Cyuzuzo, P., Yadufashije, C. & Karemera, J. Contribution of medical wards contamination to wound infection among patients attending Ruhengeri Referral Hospital. Int. J. Microbiol. 2021, 7 (2021).
Owusu, E., Asane, F. W., Bediako-Bowan, A. A. & Afutu, E. Bacterial contamination of surgical instruments used at the surgery Department of a Major Teaching Hospital in a Resource-Limited Country: an observational study. Diseases 10, 81 (2022).
Shiferaw, T., Beyene, G., Kassa, T. & Sewunet, T. Bacterial contamination, bacterial profile and antimicrobial susceptibility pattern of isolates from stethoscopes at Jimma University Specialized Hospital. Ann. Clin. Microbiol. Antimicrob. 12, 39 (2013).
Thomson, K. M. et al. Effects of antibiotic resistance, drug target attainment, bacterial pathogenicity and virulence, and antibiotic access and affordability on outcomes in neonatal sepsis: an International Microbiology and Drug Evaluation Prospective substudy (BARNARDS). Lancet Infect. Dis. 21, 1677–1688 (2021).
Aleem, M. et al. Prevalence of bacteria and antimicrobial resistance genes in hospital water and surfaces. Cureus 13, 10 (2021).
Mahmud, Z. H. et al. Healthcare facilities as potential reservoirs of antimicrobial resistant Klebsiella pneumoniae: an emerging concern to public health in Bangladesh. Pharmaceuticals 15, 1116 (2022).
Boostrom, I., Portal, E. A. R., Spiller, O. B., Walsh, T. R. & Sands, K. Comparing long-read assemblers to explore the potential of a sustainable low-cost, low-infrastructure approach to sequence antimicrobial resistant bacteria with Oxford nanopore sequencing. Front. Microbiol. 13, 796465 (2022).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Pathogenwatch. A global platform for genomic Surveillance https://pathogen.watch/ (2023).
Seemann T. ABRicate. Mass Screening of Contigs for Antimicrobial and Virulence Genes https://github.com/tseemann/abricate (2016).
Feldgarden, M. et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype–phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483–19 (2019).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, 690–699 (2023).
Maiden, M. C. J. et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Snippy, S. T. Rapid Haploid Variant Calling and Core Genome Alignment https://github.com/tseemann/snippy (2015).
Bush, S. J. et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism–calling pipelines. GigaScience 9, 1–21 (2020).
GitHub—tseemann/snp-dists. Pairwise SNP distance matrix from a FASTA sequence alignment. https://github.com/tseemann/snp-dists (2021).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Ondov, B. D. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016).
Chemaly, R. F. et al. The role of the healthcare environment in the spread of multidrug-resistant organisms: update on current best practices for containment. Ther. Adv. Infect. Dis. 2, 79–90 (2014).
Russotto, V. et al. What healthcare workers should know about environmental bacterial contamination in the Intensive Care Unit. BioMed. Res. Int. 2017, 6905450 (2017).
World Health Organization & WHO Patient Safety. Hand Hygiene Technical Reference Manual: to be Used by Health-care Workers, Trainers and Observers of Hand Hygiene Practices (World Health Organization & WHO Patient Safety, 2009).
Centers for Disease Control and Prevention (CDC). Public Health Strategies to Prevent the Spread of Novel and Targeted Multidrug-resistant Organisms (MDROs) (Centers for Disease Control and Prevention (CDC), 2023).
Centers for Disease Control and Prevention (CDC). Preventing Healthcare-Associated Infections. Reduce Risk from Water (Centers for Disease Control and Prevention (CDC), 2023).
World Bank Climate Change Knowledge Portal. https://climateknowledgeportal.worldbank.org/ (2021).
Acknowledgements
We thank all enroled participants and their families. This work was supported by a combination of two research awards (nos. OPP1119772 and OP1191522) from the Bill & Melinda Gates Foundation. We thank Liofilchem® for their continued support in the distribution of their microbiology products to enable standardisation of standard operating procedures across the clinical sites. We thank J. Parkhill for their advice and guidance regarding the phylogenetic analyses. We thank Wales Gene Park and ARCCA for their continued bioinformatics support and infrastructure availability. Bioinformatics analysis was largely undertaken using the supercomputing facilities at CU operated by ARCCA on behalf of the Cardiff Supercomputing Facility and the HPC Wales and Supercomputing Wales projects. The latter is partly funded by the European Regional Development Fund via the Welsh Government. We thank the team of curators for the databases hosted on PubMLST https://pubmlst.org/databases. We thank the curators of the Institut Pasteur MLST and whole-genome MLST databases for curating the Klebsiella spp. data and making them publicly available at http://bigsdb.pasteur.fr. We thank M. Islam for providing access to the clinical sites and epidemiology data in Bangladesh. We would like to acknowledge R. Kamran, the microbiologist from PIMS, Pakistan, who sadly passed away in 2018. We thank the team within the Specialist Antimicrobial Chemotherapy Unit, University Hospital Wales, Public Health Wales, for their support for MALDI-TOF MS of bacterial isolates. We thank the BARNARDS group (Supplementary Table 5).
Author information
Authors and Affiliations
Consortia
Contributions
M.J.C. and K.S. designed and guided the study and analysis, M.N-R. and K.S. wrote the manuscript, and M.N.-R., K.S., and K.M.T. revised the manuscript. K.S., E.A.R.P. and I.B. performed the WGS experiments. M.J.C., K.S., K.M.T., E.A.R.P., J.M., C.D., C.A., P.H., H. Saif, A.D.S.F., M.N.-R., T.H., A.M., A.R., B.P., and L.R. performed the microbiology experiments. K.S., M.J.C. and R.A. performed the bioinformatics analysis. R.M., G.C., C.D. K.M.T., and T.R.W. designed and delivered the epidemiological aspects of the study. W.J.W. performed statistical analyses. R.Z., H. Shirazi., A.M., S.N.U., M.H.J., S.A., K.C.I., F.M., S.U., L.A., C.P.E., A.H.Y., A.A., A.S.M., J.B.M., A.R., L.G., S.M., A.N.H.B., A.W., L.R., D.B., S.S., M.A. and G.M. assisted in collecting hospital surface swab samples and transporting to the United Kingdom. G.C., K.C.I., R.Z., J.B.M. and S.M. facilitated the epidemiology data collection at the clinical sites. T.R.W., M.J.C., R.M., G.C., K.C.I., R.Z., J.B.M. and S.M. designed the BARNARDS study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nieto-Rosado, M., Sands, K., Portal, E.A.R. et al. Colonisation of hospital surfaces from low- and middle-income countries by extended spectrum β-lactamase- and carbapenemase-producing bacteria. Nat Commun 15, 2758 (2024). https://doi.org/10.1038/s41467-024-46684-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-46684-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.