Abstract
Dominance hierarchies often form between species, especially at common feeding locations. Yet, relative to work focused on the factors that maintain stable dominance hierarchies within species, large-scale analyses of interspecific dominance hierarchies have been comparatively rare. Given that interspecific behavioral interference mediates access to resources, these dominance hierarchies likely play an important and understudied role in community assembly and behavioral evolution. To test alternative hypotheses about the formation and maintenance of interspecific dominance hierarchies, we employ an large, participatory science generated dataset of displacements observed at feeders in North America in the non-breeding season. Consistent with the hypothesis that agonistic interference can be an adaptive response to exploitative competition, we find that species with similar niches are more likely to engage in costly aggression over resources. Among interacting species, we find broad support for the hypothesis that familiarity (measured as fine-scale habitat overlap) predicts adherence to the structure of the dominance hierarchy and reduces aggression between species. Our findings suggest that the previously documented agonistic hierarchy in North American birds emerges from species-level adaptations and learned behaviors that result in the avoidance of costly aggression.
Similar content being viewed by others
Introduction
Interspecific behavioral interactions, such as aggressive contests, are important determinants of ecological and evolutionary processes1. Processes such as species range expansion2, range limits3, character displacement4,5, and species recognition4 are influenced by aggressive interactions between species. Indeed, rather than being ephemeral cases of mistaken recognition, abundant evidence suggests that aggression between species can arise as an adaptive response to competition for resources6,7. Consequently, the evolution and presence of interspecific aggression between species has multifaceted effects on species in certain areas and in certain communities8,9.
Interspecific aggression and behavioral interference are critical determinants of resource acquisition for many species. For example, the effects of interspecific aggression on space use has received considerable research attention10, especially in terms of range limits and expansion2,3,11. In addition to space use, aggression also influences access to foraging resources12,13. Although much of this interspecific research addresses interactions on a pairwise scale, such interactions unfold within the context of an entire community of potentially interacting species14. Aggressive interactions between multiple species at foraging locations are common15, and several studies document the presence of interspecific dominance hierarchies16,17. Consequently, the mechanisms explaining the formation and maintenance of such hierarchies may indeed have important resultant effects on community assembly and other ecological processes, yet these mechanisms remain largely unknown.
Dominance and dominance hierarchies within species have been recognized for a considerable amount of time18,19,20. A key explanation for the evolutionary stability of such hierarchies is that they minimize the frequency or intensity of aggression among conspecifics, particularly when the positions in the hierarchy are predictable and familiarity is high among individuals in a group21. Indeed, periods of instability or turnover in intraspecific dominance hierarchies are often associated with fitness costs. For instance, aggression was elevated in groups of domestic dogs (Canis familiaris) in parts of the dominance hierarchy that were least well-established22. Yet, dominance interactions are not limited to members of the same species16,23,24,25,26. It stands to reason that interspecific dominance hierarchies could be maintained by the same selection pressures that help generate intraspecific hierarchies: individuals evolve to avoid costly aggression, particularly when the outcome of dominance interactions is predictable;23 and to do so individuals recognize salient cues27. The cues in this case would be cues associated with identifying other species, rather than individuals within the same species28.
If species form, recognize, and abide by interspecific dominance hierarchies, one would expect that species pairs that co-occur frequently in space and time should adhere to the structure of the dominance hierarchy better than species pairs that are less familiar with one another. Alternatively, if the outcome of interspecific interactions is too unpredictable, or if the neurological mechanisms that mediate the recognition of rank in a within-species hierarchy are not tuned to heterospecific cues, then dominance hierarchies may instead be maintained as a simple consequence of asymmetries in size or other competitive abilities, with mere despotism generating and maintaining the structure of the hierarchy. If this second hypothesis is true, then the same dominance hierarchy should emerge regardless of how widespread the co-occurrence of members of the hierarchy is. These hypotheses have been difficult to test on a large, interspecific scale due to the intensive observational work required to generate a large interaction database.
To test these hypotheses, we amass and analyze an comparative dataset of interspecific avian interactions from feeders across North America, which adhere to a stable, interspecific dominance hierarchy16. We pair each aggressive interaction with multiple ecological, natural history, and social variables based on the species involved in the interaction to examine the factors that shape links in this dominance hierarchy. To dissect the evolutionary drivers of aggressive avian interactions, we generate three distinct variables of aggression that isolate specific aspects of interspecific aggression. These three measures allow us to determine if species pairs abide by an interspecific dominance hierarchy. The first measure is ‘displacement observed’, a simple presence/absence indicator of aggression. The binary displacement observed variable allows us to assess which evolutionary factors generate interspecific aggression. The second variable, deviation from expected encounters (‘DEE’, see below), is a measure of the extent of aggression that adjusts for species abundance. This second variable allows us to determine which factors lead to increased aggression above or below what one would expect if species were interacting at random based on abundance. The third variable, a measure of the directionality of aggression (i.e., the consistency with which individuals of one species ‘win’ in an interaction), allows us to isolate the factors that lead to interspecific dominance within a species pair. If species abide by an interspecific hierarchy, then we would expect species familiarity, in the form of spatial overlap, to lead to a higher likelihood of observing aggression between a pair of species. However, species that abide by an interspecific hierarchy would interact less than expected after controlling for abundance. Similarly, with increasing species familiarity, interactions between species would be resolved towards the dominant member of the pair more than expected.
Here, we show that the degree of species familiarity, as estimated by syntopic overlap, potentially drives the recognition of an interspecific dominance hierarchy. Within the hierarchy, we find that species pairs that show increasing overlap in space and time are more likely to be observed interacting at any point; however, more familiar species pairs interact less than expected after controlling for abundance. Not only do familiar species pairs interact less than expected, aggression between individuals in familiar species pairs more often resolve their aggression towards the relatively dominant species in the pair. Taken together, these effects suggest species recognize their relative position in the interspecific dominance hierarchy.
Results
FeederWatch data
There were a total of 88,988 interactions across all species in the full dataset. There were a total of 12,765 unique, co-occurring species pairs in the dataset. From the species pair dataset, we removed interactions with predatory birds and species pairs only found at one feeder location. The resulting data subset had 8,911 unique species pairs; in this subset, displacements were observed between 1,664 unique species pairs, with the sources of aggression consisting of 99 species from 63 genera, and the targets of aggression consisting of 176 species from 97 genera. Interactions were reported from across the United States and southern parts of Canada.
Likelihood of aggression increases with niche overlap
Several variables related to niche overlap predicted the presence of any aggressive displacements between species pairs (Fig. 1; Supplementary Table 1). Species pairs that share a diet (posterior mean = 0.37, 95% CI = 0.14–0.61, p = 0.002), have similar foraging ecologies (posterior mean = 0.54, 95% CI = 0.32–0.78, p < 0.001), and similar body masses (body mass difference posterior mean = −0.82, 95% CI = −0.97–−0.67, p < 0.001) were more likely to be observed in aggressive interactions. We also found that if both species were sedentary (i.e., non-migratory), they were more likely to be observed in an aggressive interaction (posterior mean = −0.50, 95% CI = 0.16–0.88, p = 0.006). When one member of a pair of species was generally more aggressive (i.e, had a high intraspecific DEE), the likelihood of observing aggressive interactions was higher (posterior mean = 0.88, 95% CI = 0.57–1.20, p < 0.001). Similarly, as species overlap more in range (sympatry posterior mean = 0.47, 95% CI = 0.33–0.61, p < 0.001) and specific habitat (syntopy posterior mean = 1.84, 95% CI = 1.66–2.01, p < 0.001), they are more likely to be observed in an aggressive displacement.
Black represents statistically significant effects, whereas gray represents effects not significantly different from zero. Dotted line is a horizontal intercept at 0. Effects generated from model on n = 1664 unique species pairs. A Coefficients from the phylogenetic logistic mixed model predicting the presence of aggression. B Coefficients from the phylogenetic linear mixed model predicting the extent of aggression compared to expected. C Coefficients from the phylogenetic linear mixed model predicting the directionality of aggression. Points represent the mean of the fixed effect posterior values and lines represent the 95% credibility intervals. Variables with credibility intervals that do not overlap 0 are depicted in black.
Unfamiliarity and body size drive interspecific aggression
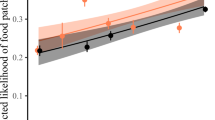
In support of the hypothesis that interspecific dominance hierarchies reduce the overall occurrence of costly interspecific aggression between species, we found that species that encounter each other infrequently and are therefore likely unfamiliar with one another engaged in aggressive displacements more frequently than expected by chance—syntopy was negatively associated with DEE (posterior mean = −0.85, 95% CI = −1.24–−0.46, p < 0.001; Supplementary Table 2). In these models we accounted for phylogeny. Specifically, patristic distance was negatively associated with DEE in interspecific interactions (posterior mean = −3.62, 95% CI = −6.33–−1.26, p = 0.008; Fig. 2); as species become more distantly related, they are less likely to interact based on abundance. Finally, with increasing difference in body size, there was a lower likelihood that species will interact based on abundance (posterior mean = −1.07, 95% CI = −1.46–−0.68, p < 0.001).
Plotted are the lowest 10% (a) and highest 10% (b) of deviation from expected encounters (DEE) values, representing species pairs which interact far less frequently than would be expected if interactions occurred at random and species pairs which interact far more frequently than random expectations, respectively. Dotted connections are representative examples. Example species pair in A are a dark-eyed junco (Junco hyemalis – illustrated by David Quinn) and mourning dove (Zenaida macroura – illustrated by Martin Elliott); example species pair in B are a blue jay (Cyanocitta cristata – illustrated by Brian Small) and Woodhouse’s scrub jay (Aphelocoma woodhouseii – illustrated by Brian Small). Illustrations used with permission, © Birds of the World | Cornell Lab of Ornithology.
Unfamiliarity, body size, and foraging ecology lead to unstructured interactions
The increased rate of interaction between unfamiliar species was driven, at least in part, by a breakdown in the interspecific hierarchy at feeders. Species that encounter each other more frequently exhibit more structured interactions—with increasing syntopy, species pairs exhibit higher directionality in aggressive encounters (posterior mean = 0.10, 95% CI = 0.08 – 0.12, p < 0.001; Fig. 3); that is, the identities of the winner and the loser were more consistent in these interactions. Yet, other factors impact the structure of interactions at feeders as well (Supplementary Table 3). For instance, species pairs with similar niches exhibit more symmetrical interactions (foraging niche overlap: posterior mean = 0.04, 95% CI = 0.02–0.08, p = 0.02; body mass difference: posterior mean = 0.10, 95% CI = 0.07–0.12, p < 0.001;). Similarly, species pairs involving a more aggressive species (i.e., those with higher intraspecific DEE values) exhibited higher directionality (posterior mean = 0.08, 95% CI = 0.05–0.11, p < 0.001). In these models we account for phylogeny, with more distantly related species exhibited lower directionality (posterior mean = −0.06, 95% CI = −0.10–−0.02, p < 0.001). Finally, our analyses lent some support to the observation that the role of mass in determining interaction structure breaks down over evolutionary time—there was an interaction between mass and patristic distance that influenced directionality in the full dataset (p < 0.05); though we only recovered a trend in the data subsets (p = 0.05 and p = 0.06).
Bird figures in each panel represent different species, and arrows represent directed aggression. Black and gray colors represent different hypothetical species. Thryothorus ludovicianus silhouette from phylopic.org [CC0 1.0]. A Increasing syntopy increases the likelihood of observing interspecific aggression. B Increasing syntopy increases directionality. C Increasing syntopy decreases DEE. Trendlines are presented from (generalized) linear models, with standard errors depicted in gray.
Discussion
Our analyses provide further evidence that aggressive interspecific interactions are more likely to arise between ecologically similar species. We also found support for the hypothesis that familiarity plays a key role in the formation and maintenance of interspecific dominance hierarchies. Species pairs that are unfamiliar with one another engage in more frequent and less structured aggressive interactions at feeders, likely increasing the time and energy spent in such agonistic encounters.
While previous findings have identified mechanisms that reduce aggression within species (e.g. intraspecific dominance hierarchies, dear enemy effects29), the results presented here suggest that individuals across many avian species are adjusting their behavior based on heterospecific recognition cues30. Recent evidence in reef fish suggests that heterospecifics recognize one another and are more likely to resolve contests with agonistic signaling when resources are relatively abundant31. Similarly, analyses of mixed-species groups of parrots at clay licks in the Amazon basin demonstrate that heterospecific recognition influences decisions to join mixed groups, thereby determining species compositions32, further underscoring the ability of individuals to respond to complex social tapestries across species boundaries. Our results show that there is a clear benefit to doing so, yet there is likely to be further, as yet unstudied variation in the degree to which species respond to heterospecific cues. Recognizing heterospecifics may be especially valuable for the smaller species in a species pair33, for instance, and individuals may quickly learn their relative dominance position compared to familiar species34.
In addition to familiarity between species, we also found that niche similarity influenced the likelihood of observing interspecific aggression. First, large differences in body mass were negatively correlated with observing interspecific aggression. Unsurprisingly, species that differed considerably in body size did not enter aggressive contests as readily. That species with large differences in size avoid conflict is in line with previous results26, though we did not find an effect of evolutionary distance influencing whether species engaged in contests. However, evolutionary distance was negatively associated with DEE; in other words, more closely related species engage in aggressive contests more often than expected based on abundance. In terms of ecological variables both shared foraging strategy and shared diet led to an increased likelihood of species engaging in aggressive contests. Consequently, species that overlap in resources, in addition to space, tend to have more contests than species with differing foraging ecologies—a result echoed in a previous study on interspecific territoriality during the breeding season7. Similarly, species pairs that are both sedentary tend to have a higher likelihood of engaging in aggressive contests over food. However, this result may be due to the structure of the data, as FeederWatch primarily operates during the northern hemisphere winter. Further research into the temporal dynamics of interspecific dominance hierarchies would likely shed additional light on the mechanisms driving their formation and maintenance.
One aspect of the study worth considering is how closely the feeders and associated avian behavior at feeders correspond to interactions over resources in more natural habitats. Specifically, do bird feeders induce increased and different behavioral dynamics than other areas? In some areas, fruiting trees produce concentrated resources that attract a large congregation of birds in both species and abundance35. Similarly, feeders may increase aggression above what would be experienced if feeders were not present. However, the aggression and dominance hierarchy present at feeders is a likely representation of these dynamics in other situations. While feeders may represent a relatively novel resource for animals, such concentrated resources are increasing over time. For instance, bird feeding and bird watching are increasing;36 additionally, landfills are often areas where larger species congregate37. These concentrated resources are increasing with urbanization, and will likely bring more species, including non-native species38, into close proximity over time. The facilitation of non-native species at these resources could very well modify the current dominance hierarchies in these areas.
Interspecific dominance at feeders may be modified by other variables in addition to the variables included here. For instance, both intraspecific and interspecific sociality could modify the competitive nature of interactions14. Intriguingly, individuals in species that form intraspecific flocks39, and fight more amongst conspecifics40, tend to be less socially dominant. However, the presence of conspecifics may increase the competitiveness of the group at the expense of competitors41.
The combination of findings presented here suggests that individuals may adapt to interspecific hierarchies. These results compliment findings from intraspecific dominance hierarchies42,43,44, suggesting that selection to reduce costly aggression will evolve among familiar individuals both within and among species. Deviations from the patterns presented here might provide fruitful opportunities to understand why certain familiar species engage in more aggression than expected. Specifically, studies on targeted species pairs may illuminate potential traits that produce more constant aggression among familiar individuals14. Aggression among species is common among many groups of animals, and given the capacity for heterospecific recognition across many of the same groups of animals, the presence of interspecific dominance hierarchies may be more widespread than traditionally believed.
Methods
FeederWatch data
To quantify dominance interactions between species, we employed a large dataset of interspecific displacements at feeders derived from data from Project FeederWatch. Project FeederWatch is a program run by the Cornell Laboratory of Ornithology where participants can submit observed interactions between birds at bird feeders45. Specifically, observers submitted information about displacements, identifying the species of the winner and loser of each successful displacement. We incorporate interaction data from 2015-2020 with submitted interactions from across North America.
Database
In calculating certain variables (e.g. DEE, see below), we found that some rare species pairs yielded extreme values. These extreme values sometimes had high leverage in the models, and to generate a conservative dataset and subsequent set of analyses, we subset the data in multiple ways for the analysis described below. Specifically, we removed species pairs that were found at only one single feeder location; several of these singleton pairs had high DEE values and we removed them to eliminate the effects of their high statistical leverage. We also removed any species pairs that include a predatory bird (e.g. a hawk or falcon). Since predatory birds can be observed around feeders they are in the overall dataset, but are removed from the focal subset of data. We ran all analyses on the full dataset, the dataset with species pairs observed at more than one feeder, and a focal dataset removing both predatory birds and species pairs observed at more than one feeder. We report the results of the most conservative analysis (i.e. the analysis with both predators and singleton pairs removed) here and the results of the other datasets in the supplementary information (Supplementary Tables 4–9). Importantly, the main results of all three sets of analyses were qualitatively identical.
Aggression variables
From this dataset, we calculated several variables to characterize the network of agonistic interactions between species. First, to identify factors that predict which species pairs exhibited interspecific aggression, and therefore contribute to the formation of interspecific dominance hierarchies, we created a simple binary variable to indicate whether two species were ever observed in a displacement, for which we restricted the data to species that ever co-occurred at any feeder and therefore could have conceivably been observed interacting.
Next, to test the key prediction of the alternative hypotheses for the maintenance of interspecific dominance hierarchies, we used two indices to characterize the adherence to the dominance hierarchy. The first—a standardized effect size (SES) value following refs. 28,46—is an index of the deviation of the frequency of observed displacements from the number expected if species interact at random. To improve clarity, we now refer to this SES as ‘Deviation from Expected Encounters’, or DEE. We used abundance data from FeederWatch counts to simulate interaction datasets where, for each feeder, we simulated the same number of displacements as were recorded in the observed dataset, sampling two interacting individuals in proportion to their abundances at the observed feeder. We then aggregated simulated fights across all feeders in the empirical dataset, creating a matrix of simulated displacements. We repeated this 10,000 times and calculated the mean and standard deviation of the number of simulated fights across these 10,000 simulated matrices. DEE values were then calculated by dividing the difference between the observed number of pairwise interactions and the mean expected number of such interactions by the standard deviation of the simulated number of interactions. Consequently, positive values of DEE represent more interactions than expected based on abundances, whereas negative values of DEE represent fewer interactions than expected based on abundances. Notably, while each species pair has an interspecific DEE value, each species has its own, intraspecific DEE value, as well (generated from expected extent of aggression towards conspecifics). Secondly, we calculated an index of directionality to capture the asymmetry in aggressive interactions in each species pair using the index developed by ref. 26
where Nwins by dominant is the number of displacements won by the species in the pair which won the highest number of displacements (i.e., the dominant species), and Nwins by subordinate is the number of displacements won by the species in the pair which won the fewest number of displacements (i.e., the subordinate species). Zero values, therefore, indicate that members of both species displace individuals of the other species at equal rates (i.e., that interspecific aggression is symmetrical), while high values correspond to asymmetric cases where the dominant species primarily displaces the subordinate species.
Trait data
To identify factors that predict the occurrence of interspecific aggression, we collected data on numerous morphological and ecological traits. Data on species body mass, diet, foraging behavior, and migration strategy were obtained from ref. 47. To test whether species with similar niches are more likely to engage in interspecific aggression at feeders, we generated several variables using the trait data for species pairs. Specifically, we determined whether species pairs shared the same general diet, the same foraging strategy, the same migratory status, and whether both species were passerines or not, coding these as binomial variables (i.e., yes = 1, no = 0).
Sympatry and syntopy indices from eBird
To test whether the magnitude of aggressive interactions in dominance hierarchies was influenced by species familiarity with one another, we calculated two estimates of spatial overlap for each species pair. Estimates of species pair range overlap (i.e., sympatry) and habitat overlap (i.e., syntopy) were produced from data collected from eBird.org48, following the approach developed in ref. 7 To this end, data from eBird were downloaded for each species in our analyses, restricting data to complete checklists with observations collected in the United States or Canada between the months of November to March (the months in which the FeederWatch project runs) for the five nonbreeding seasons between 2015 and 2020. These data were further filtered to only include lists collected by observers who traveled less than 0.25 miles to ensure that all birds in each checklist occurred in similar areas.
We used these filtered eBird observations to calculate an index of non-breeding range overlap (sympatry) for each species pair following ref. 7. Briefly, this index is similar to the commonly used Szymkiewicz-Simpson overlap index and was calculated using the formula:
where N1,2i refers to the number of unique locations where species 1 was observed in sympatry with (within 24.5 miles of) species 2 in year I, N1i refers to the number of unique locations where species 1 was observed in year i, and Y refers to the total number of years (5, in this case).
Similarly, we calculated an index of fine-scale non-breeding habitat overlap (‘syntopy’) for each species pair using the filtered eBird observations, calculated using the formula:
where n1,2i refers to the number of unique locations where species 1 was observed in syntopy with (within 0.25 miles of) species 2 in year i, and other terms are as above (as in ref. 7). Species pairs with high syntopy values, therefore, are more likely to be found in close proximity and, crucially, be familiar with each other.
Phylogeny
To account for the non-independence among species resulting from their shared evolutionary history, we conducted analyses that incorporate the avian phylogeny. We used a maximum clade credibility tree constructed from the posterior distribution of trees available on birdtree.org49, using the taxonomic backbone from Hackett et al.50. To this phylogeny, we added recent splits (winter wren [T. hiemalis] and Pacific wren [T. pacificus]; California scrub jay [Aphelocoma californica] and Woodhouse’s scrub jay [A. woodhouseii]) following published estimates for the timing of these splits51,52 (Supplementary Data 1). We then used the R package ape53 to calculate patristic distance for each species pair (the branch length separating them; i.e., twice the time separating each species in the pair from their most recent common ancestor) from this phylogeny.
Statistical analyses
All analyses were conducted using R v. 4.1.2 (). Given that each observation in our dataset is derived from a species pair, we used phylogenetic linear mixed models (PLMMs) adapted for species interaction data54,55 to identify which factors predict the occurrence, strength, and direction of interactions observed by FeederWatch participants. Each model was fit using the R package MCMCglmm56, including species identity and the phylogeny as random effects, specifying which node in the phylogeny represent the most recent common ancestor linking each species pair. We used an uninformative, inverse Wishart distribution as a prior distribution for the random effects, fixing the residual variance at 1 for binomial models. To fit the models, we ran MCMC chains for 5 ×105 generations, logging the output every 500 generations and ignoring the first 1,000 generations as burn-in. For each model we used uninformative inverse-gamma priors. We fit each model four times, verified convergence by visually inspecting trace plots and confirming that values Gelman-Rubin values were close to 1 (1-1.01) in the R-package coda57,58, and then merged the converged chains. From these PLMMs, we calculated the phylogenetic signal, λ, from the random effects as the phylogenetic intraclass correlation59. All statistical tests were two-sided tests.
To test the hypothesis that the effect of body size on the directionality of interactions varies as a function of evolutionary relatedness, we added an interaction term between the body mass difference and patristic distance for the PLMM fit to the directionality dataset. When analyzing the unexplained extent of aggressive interactions (DEE) between species and directionality, we subset the dataset to only those species pairs where aggressive interactions were observed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The species pair data generated in this study can be found in the figshare repository DOI: 10.6084/m9.figshare.24309697. The data are under no restriction and can be accessed freely. Raw individual interaction data are provided as a Source Data file. Source data are provided with this paper.
Code availability
Code is available in the figshare repository DOI: 10.6084/m9.figshare.24309697.
References
Grether, G. F., Peiman, K. S., Tobias, J. A. & Robinson, B. W. Causes and Consequences of Behavioral Interference between Species. Trends Ecol. Evol. 32, 760–772 (2017).
Duckworth, R. A. & Badyaev, A. V. Coupling of dispersal and aggression facilitates the rapid range expansion of a passerine bird. Proc. Natl Acad. Sci. 104, 15017–15017 (2007).
Freeman, B. G., Strimas-Mackey, M. & Miller, E. T. Interspecific competition limits bird species’ ranges in tropical mountains. Science 377, 416–420 (2022).
Grether, G. F., Losin, N., Anderson, C. N. & Okamoto, K. The role of interspecific interference competition in character displacement and the evolution of competitor recognition. Biol. Rev. 84, 617–635 (2009).
Kirschel, A. N. G., Blumstein, D. T. & Smith, T. B. Character displacement of song and morphology in African tinkerbirds. Proc. Natl Acad. Sci. 106, 8256–8256 (2009).
Cowen, M. C., Drury, J. P. & Grether, G. F. Multiple routes to interspecific territoriality in sister species of North American perching birds. Evolution 74, 2134–2148 (2020).
Drury, J. P., Cowen, M. C. & Grether, G. F. Competition and hybridization drive interspecific territoriality in birds. PNAS 117, 12923–12930 (2020).
Anderson, T. L. & Whiteman, H. H. Asymmetric effects of intra- and interspecific competition on a pond-breeding salamander. Ecology 96, 1681–1690 (2015).
Anderson, T. L. & Whiteman, H. H. Non-additive effects of intra- and interspecific competition between two larval salamanders. J. Anim. Ecol. 84, 765–772 (2015).
Peiman, K. S. & Robinson, B. W. Ecology and evolution of resource-related heterospecific aggression. Q. Rev. Biol. 85, 133–158 (2010).
Pasch, B., Bolker, B. M., Phelps, S. M., Gaillard, A. E. J.-M. & Bronstein, E. J. L. Interspecific Dominance Via Vocal Interactions Mediates Altitudinal Zonation in Neotropical Singing Mice. Am. Naturalist 182, E161–E173 (2013).
Schoener, T. W. Field Experiments on Interspecific Competition. Am. Naturalist 122, 240–285 (1983).
Balfour, N. J., Gandy, S. & Ratnieks, F. L. W. Exploitative competition alters bee foraging and flower choice. Behav. Ecol. Sociobiol. 69, 1731–1738 (2015).
Coppinger, B. A., Carlson, N. V., Freeberg, T. M. & Sieving, K. E. Mixed-species groups and the question of dominance in the social ecosystem. Philos. Trans. R. Soc. B Biol. Sci. 378, 20220097 (2023).
Rome, M. S. & Ellis, J. C. Foraging Ecology and Interactions between Herring Gulls and Great Black-backed Gulls in New England. COWA 27, 200–210 (2004).
Miller, E. T. et al. Fighting over food unites the birds of North America in a continental dominance hierarchy. Behav. Ecol. 28, 1454–1463 (2017).
Renaud, T. & Root-Bernstein, M. Flower visitor insects display an interspecific dominance hierarchy on flowers. Ecology n/a, e3958 (2023).
Schjelderup-Ebbe, T. Beiträge zur Sozialpsychologie des Haushuhns. [Observation on the social psychology of domestic fowls.]. Z. f.ür. Psychologie und Physiologie der Sinnesorgane. Abt. 1. Z. f.ür. Psychologie 88, 225–252 (1922).
Tinbergen, N. The Functions of Territory. Bird. Study 4, 14–27 (1957).
Maynard Smith, J. The theory of games and the evolution of animal conflicts. J. Theor. Biol. 47, 209–221 (1974).
Tibbetts, E. A., Pardo-Sanchez, J. & Weise, C. The establishment and maintenance of dominance hierarchies. Philos. Trans. R. Soc. B: Biol. Sci. 377, 20200450 (2022).
Silk, M. J., Cant, M. A., Cafazzo, S., Natoli, E. & McDonald, R. A. Elevated aggression is associated with uncertainty in a network of dog dominance interactions. Proc. R. Soc. B: Biol. Sci. 286, 20190536 (2019).
Morse, D. H. Niche Breadth as a Function of Social Dominance. Am. Naturalis 108, 818–830 (1974).
Wallace, M. P. & Temple, S. A. Competitive Interactions within and between Species in a Guild of Avian Scavengers. The Auk 104, 290–295 (1987).
Chock, R. Y., Shier, D. M. & Grether, G. F. Body size, not phylogenetic relationship or residency, drives interspecific dominance in a little pocket mouse community. Anim. Behav. 137, 197–204 (2018).
Martin, P. R. & Ghalambor, C. K. When David beats Goliath: the advantage of large size in interspecific aggressive contests declines over evolutionary time. Plos One 9, e108741–e108741 (2014).
Kenyon, H. L. & Martin, P. R. Aggressive signaling among competing species of birds. PeerJ 10, e13431 (2022).
Leighton, G. M., Lees, A. C. & Miller, E. T. The hairy–downy game revisited: an empirical test of the interspecific social dominance mimicry hypothesis. Anim. Behav. 137, 141–148 (2018).
Whiting, M. J. When to be neighbourly: Differential agonistic responses in the lizard Platysaurus broadleyi. Behav. Ecol. Sociobiol. 46, 210–214 (1999).
Matyjasiak, P. Birds associate species-specific acoustic and visual cues: recognition of heterospecific rivals by male blackcaps. Behav. Ecol. 16, 467–471 (2005).
Keith, S. A., Hobbs, J.-Pa, Boström-Einarsson, L., Hartley, I. R. & Sanders, N. J. Rapid resource depletion on coral reefs disrupts competitor recognition processes among butterflyfish species. Proc. R. Soc. B Biol. Sci. 290, 20222158 (2023).
Ferdinand, V., Pattenden, E., Brightsmith, D. J. & Hobson, E. A. Inferring the decision rules that drive co-foraging affiliations in wild mixed-species parrot groups. Philos. Trans. R. Soc. B Biol. Sci. 378, 20220101 (2023).
Martin, P. R. & Ghalambor, C. K. A Case for the “Competitive Exclusion–Tolerance Rule” as a General Cause of Species Turnover along Environmental Gradients. Am. Naturalist 000–000 (2023) https://doi.org/10.1086/724683.
Jedlikowski, J., Polak, M. & Ręk, P. Dear-enemy effect between two sympatric bird species. Anim. Behav. 184, 19–26 (2022).
Eaton, D. P. et al. Citizen scientists help unravel the nature of cattle impacts on native mammals and birds visiting fruiting trees in Brazil’s southern Pantanal. Biol. Conserv. 208, 29–39 (2017).
McKinley, D. C. et al. Investing in citizen science can improve natural resource management and environmental protection. Issues Ecol. 19, 27 (2015).
Lato, K. A., Madigan, D. J., Veit, R. R. & Thorne, L. H. Closely related gull species show contrasting foraging strategies in an urban environment. Sci. Rep. 11, 23619 (2021).
Camacho-Cervantes, M., Keller, R. P. & Vilà, M. Could non-native species boost their chances of invasion success by socializing with natives? Philos. Trans. R. Soc. B Biol. Sci. 378, 20220106 (2023).
Berberi, I., Miller, E. T. & Dakin, R. The effect of sociality on competitive interactions among birds. Proc. R. Soc. B: Biol. Sci. 290, 20221894 (2023).
Leighton, G. M., Lamour, D., Malcolm, K. & Miller, E. T. Both morphological and behavioral traits predict interspecific social dominance in birds. J. Ornithol. 164, 163–169 (2023).
Freeman, B. G. & Miller, E. T. Why do crows attack ravens? The roles of predation threat, resource competition, and social behavior. The Auk 135, 857–867 (2018).
Beaulieu, M., Mboumba, S., Willaume, E., Kappeler, P. M. & Charpentier, M. J. E. The oxidative cost of unstable social dominance. J. Exp. Biol. 217, 2629–2632 (2014).
Mendonça-Furtado, O. et al. Does hierarchy stability influence testosterone and cortisol levels of bearded capuchin monkeys (Sapajus libidinosus) adult males? A comparison between two wild groups. Behav. Process. 109 Pt A, 79–88 (2014).
Holekamp, K. E. & Strauss, E. D. Aggression and dominance: an interdisciplinary overview. Curr. Opin. Behav. Sci. 12, 44–51 (2016).
Bonter, D. N. & Greig, E.I. Over 30 Years of Standardized Bird Counts at Supplementary Feeding Stations in North America: A Citizen Science Data Report for Project FeederWatch. Front. Ecol. Evol. 9, 1–6 (2021).
Miller, E. T., Mac Aodha, O., Greig, E. I., Bonter, D. N. & Hochachka, W. M. Congeneric predators fill discrete niches created by the relative abundances of their prey species. J. Avian Biol. n/a, e02934 (2022).
Tobias, J. A. & Pigot, A. L. Integrating behaviour and ecology into global biodiversity conservation strategies. Philos. Trans. R. Soc. B: Biol. Sci. 374, 20190012–20190012 (2019).
Sullivan, B. L. et al. eBird: A citizen-based bird observation network in the biological sciences. Biol. Conserv. 142, 2282–2292 (2009).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Hackett, S. J. et al. A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1768 (2008).
Toews, D. P. L. & Irwin, D. E. Cryptic speciation in a Holarctic passerine revealed by genetic and bioacoustic analyses. Mol. Ecol. 17, 2691–2705 (2008).
McCormack, J. E., Heled, J., Delaney, K. S., Peterson, A. T. & Knowles, L. L. Calibrating divergence times on species trees versus gene trees: Implications for speciation history of Aphelocoma Jays. Evolution 65, 184–202 (2011).
Paradis, E., Claude, J. & Strimmer, K. APE: analysis of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Tobias, J. A., Planqué, R., Cram, D. L. & Seddon, N. Species interactions and the structure of complex communication networks. Proc. Natl Acad. Sci. 111, 1020–1025 (2014).
Drury, J. P., Grether, G. F., Garland, T. Jr. & Morlon, H. An Assessment of Phylogenetic Tools for Analyzing the Interplay Between Interspecific Interactions and Phenotypic Evolution. Syst. Biol. 67, 413–427 (2018).
Hadfield, J. D. MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R Package. J. Stat. Softw. 33, 1–22 (2010).
Gelman, A. & Rubin, D. B. Inference from iterative simulation using multiple sequences (with discussion). Stat. Sci. 7, 457–472 (1992).
Plummer, M., Best, N., Cowles, K. & Vines, K. CODA: Convergence diagnosis and output analysis for MCMC. R. N. 6, 7–10 (2006).
Freckleton, R. P., Harvey, P. H. & Pagel, M. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Naturalist 160, 712–726 (2002).
Acknowledgements
We thank the many participatory scientists that have submitted observations in the FeederWatch database. G.M.L. thanks SUNY Buffalo State for support during the project. This work was funded by NSFDEB-NERC-2040883 (J.P.D.). E.T.M. acknowledges support from Schmidt Futures via an Eric and Wendy Schmidt AI in Science Postdoctoral Fellowship to Cornell University. We thank Christophe Patterson, Dan Nesbit, and Erandi Bonillas-Monge, Wesley Hochachka, and David Bonter for helpful feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
J.P.D., E.T.M., and G.M.L. conceived of the project. E.T.M., J.P.D., and J.S. prepared the data. J.P.D., J.S., E.T.M., and G.M.L. analyzed the data. J.P.D., E.T.M., G.M.L., and J.S. interpreted the results. J.P.D., G.M.L., and E.T.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Scott Robinson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leighton, G.M., Drury, J.P., Small, J. et al. Unfamiliarity generates costly aggression in interspecific avian dominance hierarchies. Nat Commun 15, 335 (2024). https://doi.org/10.1038/s41467-023-44613-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-44613-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.