Abstract
Organ regeneration necessitates precise coordination of accelerators and brakes to restore organ function. However, the mechanisms underlying this intricate molecular crosstalk remain elusive. In this study, the level of proenkephalin-A (PENK-A), expressed by renal proximal tubular epithelial cells, decreases significantly with the loss of renal proximal tubules and increased at the termination phase of zebrafish kidney regeneration. Notably, this change contrasts with the role of hydrogen peroxide (H2O2), which acts as an accelerator in kidney regeneration. Through experiments with penka mutants and pharmaceutical treatments, we demonstrate that PENK-A inhibits H2O2 production in a dose-dependent manner, suggesting its involvement in regulating the rate and termination of regeneration. Furthermore, H2O2 influences the expression of tcf21, a vital factor in the formation of renal progenitor cell aggregates, by remodeling H3K4me3 in renal cells. Overall, our findings highlight the regulatory role of PENK-A as a brake in kidney regeneration.
Similar content being viewed by others
Introduction
Multiple organs can recover their original functions through regeneration after injury. For example, the human kidneys can be partially repaired after renal tubular injury, as surviving tubular epithelial cells can repair various injuries via proliferation and migration1. The human liver can recover its original size and function following partial hepatectomy (PHx)2. During organ regeneration, many kinds of accelerators promote regeneration3. However, excessive activation of regeneration accelerators can lead to disease4, highlighting the importance of precise regulation. Thus, the body must finely control the levels of these accelerators to ensure successful organ regeneration.
Considerable efforts have been dedicated to investigating the mechanisms triggering organ regeneration. Calcium (Ca2+) signaling5,6, reactive oxygen species (ROS)7,8,9, inflammation10,11,12,13, and nerve-related factors14,15 have been demonstrated to act as accelerators in the regeneration of various organs. However, the understanding of regeneration “brakes” remains limited16. For instance, integrin-linked kinase (ILK) has been established as a termination signal in liver regeneration. Mice with liver-specific ILK ablation exhibit 58% larger liver following PHx than controls17. Glypican 318, C/EBPα19, and HNF4α20 are also related to the suppression of liver regeneration. In the case of heart regeneration in adult mice, the Hippo pathway serves as a brake; blocking Hippo signaling components enables adult cardiomyocyte renewal after myocardial infarction21. Nonetheless, the molecular brake involved in kidney regeneration remains elusive.
Zebrafish kidney development consists of two stages, namely the pronephros and mesonephros stages. During the embryonic stage, the zebrafish pronephros consists of two nephrons, whereas the adult zebrafish mesonephros comprises approximately 500 nephrons22. Unlike mammals, zebrafish possess a robust capacity for kidney regeneration. Intraperitoneal injection of excessive gentamicin (Gent) in adult zebrafish leads to apoptosis of renal tubular epithelial cells, resulting in acute kidney injury (AKI)22,23,24. However, zebrafish can rapidly regenerate a significant number of new nephrons to repair kidney damage22. lhx1a positive (lhx1a+) renal progenitor cells (RPCs) in the renal medulla aggregate at the distal segments of the renal tubules to produce RPC aggregates (RPCAs) after AKI. Subsequently, these cell aggregates can differentiate into mature nephrons22,25. Therefore, the zebrafish is a promising animal model for investigating kidney regeneration. While some mechanisms of kidney regeneration have been elucidated in this model25,26,27,28, further clarification is needed regarding the molecular mechanism that acts as a brake during kidney regeneration.
Previous investigations from our laboratory have demonstrated that the zebrafish kidney exhibits a pronounced ability to generate substantial quantities of hydrogen peroxide (H2O2) in response to injury29. In the initial stage of kidney regeneration, suppressing H2O2 production effectively diminishes the formation of lhx1a+ RPCAs and impedes the onset of kidney regeneration29. Thus, H2O2 functions as an initial signaling molecule in kidney regeneration. However, the effect of H2O2 on the kidney is dual-sided. High concentrations of H2O2 can cause renal injury and fibrosis30,31. Consequently, the levels of H2O2 in the kidney necessitate meticulous regulation. However, the precise mechanism underlying this process remains incompletely understood.
Human proenkephalin (PENK) is an endogenous opioid polypeptide hormone. Upon proteolytic cleavage, PENK generates enkephalin peptides, predominantly Met-enkephalin (Met-ENK) and, to a lesser extent, Leu-enkephalin (Leu-ENK)32,33. These enkephalin peptides can activate opioid receptors, influencing physiological functions such as pain perception and stress responses34. PENK has emerged as a potential biomarker for assessing kidney function, showing a robust negative correlation with estimated glomerular filtration rate (eGFR), thus serving as a kidney biomarker of glomerular function32,33. In addition, increased plasma PENK concentrations have also been associated with long-term outcomes in AKI and cardiac diseases35. In addition, PENK is reportedly expressed in the renal tubules of rats36, suggesting that intrarenal PENK may participate in normal kidney function and kidney diseases. However, research on these topics is still limited.
In the present study, we observed a significant correlation between the expression of penka (an ortholog of human PENK) in zebrafish proximal tubule epithelial cells (PTECs) and the state of kidney regeneration. Specifically, penka levels decreased during the loss of proximal tubules (PTs) and increased upon their recovery at the end of the kidney regeneration process. Interestingly, these changes in penka levels were in contrast to the levels of H2O2. Through the use of penka mutants and antagonists, we demonstrated that inhibiting the PENK-A signaling pathway led to increased H2O2 production and accelerated kidney regeneration. Conversely, activation of PENK-A reduced H2O2 production and resulted in earlier termination of kidney regeneration. Moreover, our findings revealed that transcription factor 21 (tcf21), a critical factor in RPCA formation, was regulated by H2O2. H2O2 promoted the expression of tcf21 by remodeling trimethylation at the 4th lysine residue of the histone H3 protein (H3K4me3) in renal cells. Collectively, our results highlight the role of PENK-A as a negative feedback regulator in kidney regeneration.
Results
Specific expression of penka in PTECs
AKI is defined as sudden damage to a significant number of nephrons (the functional units of the kidneys)37. However, it is not clear whether nephron cells affect their own regeneration. To answer this question, we reanalyzed the previously published single-cell RNA sequencing (scRNA-seq) data of adult zebrafish kidneys38. Our analysis revealed that penka was specifically expressed in PTECs (Fig. 1a). The zebrafish penka gene encodes an enkephalin precursor that undergoes processing to produce four Met-ENKs, as well as one Met-enkephalin-Ile and one Met-enkephalin-Asp39. To confirm the expression pattern of penka, we initially utilized immunofluorescence staining for a marker for renal tubules, Pax2a40, combined with penka fluorescence in situ hybridization (FISH). We observed that penka is likely expressed in the PTs (Fig. 1b). PTs in zebrafish consist of proximal convoluted tubules (PCTs) and proximal straight tubules (PSTs)41. By using FISH with markers for PCT and PST42, namely, slc20a1a and trmp7, respectively, we observed that the Met-ENK immunofluorescence signal co-localized with the signals of slc20a1a and trmp7 (Fig. 1c), confirming the expression of penka in the PTs of adult zebrafish kidneys. Furthermore, we investigated PENK expression in normal human kidney samples and samples from patients with AKI. Through FISH analysis, we observed high PENK expression in the human PTs (Fig. 1d), as indicated by colocalization with lotus tetragonolobus lectin (LTL, a marker for PTs)43 staining. Importantly, in AKI patients, who experienced loss of PTs, PENK expression was significantly decreased (Fig. 1d). These findings suggest a potential role for PENK in AKI. Therefore, further exploration of PENK-A’s involvement in kidney injury using the zebrafish model holds promise for providing valuable insights for clinical treatment.
a scRNA-seq analysis revealed that penka was specifically expressed in zebrafish PTECs. t-SNE plots showing zebrafish kidney cell clusters and the expression of penka. VECs, vascular endothelial cells; DTECs, distal tubular epithelial cells; Mφ, macrophages; HSCs, hematopoietic stem cells; MSCs, mucin-secreting cells; and RICs, renal interstitial cells. b Confocal images showing double labeling of FISH-penka and anti-Pax2a in un-injured (Un-Inj) adult zebrafish kidney sections (n = 3). c Confocal images showing triple labeling of FISH-slc20a1a, FISH-trpm7, and anti-Met-ENK in Un-Inj adult zebrafish kidney sections. The Met-ENK signal co-localized with the signals of slc20a1a and trmp7, which are markers of PCT and PST, respectively (n = 3). d Confocal images of combined FISH-PENK and LTL staining in kidney sections of patients with AKI and patients with no detectable lesions (Un-Inj). Human PENK was expressed in PTs and downregulated after AKI (n = 3). e, f RT-PCR (e) and qRT-PCR (f) analyses of penka in zebrafish kidneys during Gent-induced AKI (n = 3). penka expression was decreased by 1 dpi and reached its lowest level at 7 dpi and returned to its un-injured level at 15 dpi. The data in (f) were analyzed by two-sided t-test and are presented as mean values ± SD. Scale bars in (b), (c), and (d), 50 μm. Source data are provided as a Source data file.
Next, we examined the expression of penka during kidney regeneration in an adult zebrafish AKI model. To establish the model, we intraperitoneally injected Gent (2.7 μg/μL, 20 μL per fish) into adult zebrafish22,25. Gent can accumulate in the lysosomes of PTECs, and high doses can induce PTEC apoptosis, leading to AKI44. The kidney injuries mentioned subsequently in this study were all induced using this established adult zebrafish AKI model. Subsequently, we used reverse transcription polymerase chain reaction (RT-PCR) (Fig. 1e) and quantitative Real-time PCR (qRT-PCR) (Fig. 1f) to examine the changes in penka expression during the kidney regeneration process in this model. Our results showed that penka expression in kidneys decreased initially during regeneration, reached its lowest level at 7 days post-injury (7 dpi), and then increased from 9 dpi, returning to normal levels by 15 dpi (Fig. 1e, f). Furthermore, we quantified the number of nephrons in the Tg(gtshβ:GFP)45 kidneys, which involved labeling of PTs with GFP, at 0, 3, 5, 7, 9, and 11 days after injury (Supplementary Fig. S1a). By quantification, we found that the number of PTs (corresponding to the number of nephrons) exhibited a pattern of change similar to that of the expression of penka (Supplementary Fig. S1b). Based on this correspondence, we speculate that changes in penka expression may play a crucial role in kidney regeneration.
Deficiency of PENK-A accelerates kidney regeneration
To investigate the impact of penka on kidney regeneration, two penka mutants were generated using CRISPR/Cas9-induced gene knockout. The two mutation types identified were penka−1+24 (a 1-bp deletion and a 24-bp insertion) and penka−8 (an 8-bp deletion) (Supplementary Fig. S2a). Since there was no significant disparity in the regeneration phenotype observed between the two mutants, we primarily employed the penka−1+24 mutant for subsequent experiments. Regarding the embryonic and adult stages, including the size and structure of the adult kidneys, there were no discernible differences between the penka−1+24 mutants and the wild type (WT) (Supplementary Fig. S2b, c). In addition, an off-target analysis was conducted on this strain. We utilized CRISPRScan for off-target prediction at the target site and selected the top five potential off-target sites for sequencing analysis46,47. No off-target effects were observed at these five sites in the mutants compared to the WT (Supplementary Fig. S2d, e). Regarding the expression of lhx1a, a marker gene of RPCAs22,25, the penka−1+24 mutant kidneys exhibited high expression levels at 5 dpi, as shown by RT-PCR analyses, whereas the control kidneys reached this level at 7 dpi (Fig. 2a, b). Furthermore, whole-mount in situ hybridization (WISH) of lhx1a revealed that, compared to that in the WT counterparts, the number of RPCAs in the mutant adult kidneys significantly increased at 5 dpi, while it decreased at 9 dpi (Fig. 2c, d). These findings indicate that loss of the penka gene can accelerate kidney regeneration. In addition, we performed knockdown experiments using penka vivo-morpholino (MO) (100 μM, 10 μL per fish), which effectively interferes with the processing of penka mRNA (Supplementary Fig. S3a, b). After intraperitoneal injection of penka vivo-MO, we observed a significant increase in the number of RPCAs at 5 dpi and 7 dpi, as determined by qRT-PCR and WISH analysis of lhx1a expression (Supplementary Fig. S3c‒e). Subsequently, we intraperitoneally injected the PENK-A antagonist naloxone methiodide48 (NAL-M, 2.0 µM, 10 µL per fish) at 2, 4, 6, and 8 dpi. RT-PCR and WISH analysis of the kidneys treated with NAL-M revealed a significant increase in the number of lhx1a+ RPCAs compared to that in the control group at 5 dpi. At 7 dpi, there was a slight increase in the number of RPCAs, but interestingly, at 9 dpi, the number decreased (Fig. 2e‒g). These findings indicate that penka acts as a negative regulator of kidney regeneration and that deletion of penka accelerates kidney regeneration.
a RT-PCR analysis of lhx1a expression in WT and penka−/− zebrafish kidneys during Gent-induced AKI. b The band intensities in a are normalized to that of the loading control, β-actin, and the relative expression levels of lhx1a were quantified (n = 3 biological replications per group). The data are presented as the fold change relative to the 0 dpi WT groups. c WISH analysis of lhx1a was performed in WT and penka−/− zebrafish kidneys without injury (Un-Inj) and at 5, 7, and 9 dpi. d Quantitation of lhx1a+ RPCAs (blue points) per kidney (n = 5) for each condition in (c). e, f RT-PCR (e) and WISH (f) analyses of lhx1a in Un-Inj, 5, 7, and 9 dpi kidneys after administration of NAL-M or DMSO (n = 3 in e, n = 4 in f). g The lhx1a+ RPCAs per kidney (n = 4) were quantified for each condition in (f). The data in (b), (d), and (g) were analyzed by two-sided t-test and are presented as mean values ± SD. Scale bars in (c) and (f), 600 μm. Source data are provided as a Source data file.
PENK-A acts as a brake in kidney regeneration
When the PENK-A agonist Met-ENK49 (100 µM, 10 µL per fish) and tramadol50 (TRAM, 16 µM, 10 µL per fish) were administered via intraperitoneal injection at 2, 4, and 6 dpi, a noticeable decrease in the expression of lhx1a and a reduction in the number of RPCAs were observed in the treated group at 7 dpi through RT-PCR, qRT-PCR, and WISH (Fig. 3a–f). A dose-dependency assay using TRAM and Met-ENK was performed, which demonstrated that higher concentrations (100 µM Met-ENK, 16 µM TRAM, 10 µL per fish) exerted stronger inhibitory effects on the expression of lhx1a and resulted in greater reductions in the number of RPCAs than lower concentrations (50 µM Met-ENK, 10 µM TRAM, 10 µL per fish), as observed through RT-PCR, qRT-PCR, and WISH (Fig. 3a–f). To further investigate the role of PENK-A activation, we generated a transgenic line, Tg(hsp70l:penka), with endogenous overexpression of penka following heat shock (Fig. 3g). A previous study has demonstrated that heat shock accelerates the kidney regeneration response23. Upon heat shock at 2, 4, and 6 dpi, we found significant increases in the expression of lhx1a and the number of RPCAs in injured WT kidneys, whereas no such increases were observed in injured Tg(hsp70l:penka) kidneys at 7 dpi, as observed through RT-PCR and WISH (Fig. 3g–i). These findings suggest that PTECs can finely regulate the number of neonatal nephrons by modulating penka expression.
a‒e RT-PCR (a, c), qRT-PCR (b, d), and WISH (e) analyses of lhx1a were performed on kidneys administered with Met-ENK (a, b) or TRAM (c, d) at various doses at 7 dpi after AKI (n = 3 in b and d). Met-ENK 50, 50 μM Met-ENK; Met-ENK 100, 100 μM Met-ENK; TRAM 10, 10 μM TRAM; and TRAM 16, 16 μM TRAM. f The lhx1a+ RPCAs per kidney (n = 5) were quantified for each condition in (e). The data in (b) and (d) are presented as the fold change relative to the Un-Inj group. g RT-PCR analysis of penka and lhx1a in WT and Tg(hsp70l:penka) zebrafish kidneys with heat shock (HS) or without HS (Un-HS) in the Un-Inj group or at 7 dpi (n = 3). h WISH analysis of lhx1a in WT and Tg(hsp70l:penka) zebrafish kidneys with HS (n = 6 in WT group, n = 10 in Tg(hsp70l:penka) group) or Un-HS (n = 9 in WT group, n = 6 in Tg(hsp70l:penka) group) in the Un-Inj group or at 7 dpi. i Quantitation of lhx1a+ RPCAs per kidney for each condition in (h). j‒l RT-PCR (j), qRT-PCR (k), and WISH (l) analyses of lhx1a at 7 dpi after administration (at 4 and 6 dpi) of Met-ENK 100 (100 μM Met-ENK, 10 μL per fish) or TRAM 16 (16 μM TRAM, 10 μL per fish) following AKI. The data in (k) (n = 3) are presented as the fold change relative to the Un-Inj group. m The lhx1a+ RPCAs per kidney (n = 5) were quantified for each condition in (l). n Confocal images showing adult Tg(lhx1a:DsRed) kidneys at 5 dpi after administration (at 2 and 4 dpi) of Met-ENK or DMSO following AKI (n = 9). o Quantitation of the individual RPCs (iRPCs, arrowhead) and RPCAs in (n). Data in (b), (d), (f), (i), (k), (m), and (o) were analyzed by two-sided t-test and are presented as mean values ± SD. Scale bars in (e), (h), and (l), 600 μm; (n) 50 μm. Source data are provided as a Source data file.
The expression of penka increased beginning on 9 dpi and returned to normal levels at 15 dpi, indicating its association with the termination stage of kidney regeneration. To investigate this relationship, we intraperitoneally injected TRAM (16 µM, 10 µL per fish) and Met-ENK (100 µM, 10 µL per fish) at 4 and 6 dpi, which correspond to the peak period of kidney regeneration. Consequently, the expression of lhx1a and the number of RPCAs significantly decreased, as observed through RT-PCR and WISH (Fig. 3j–m). To further support our findings, we employed a recently developed transgenic line, Tg(lhx1a:DsRed), in which individual RPCs and RPCAs are labeled25. We examined the injured kidneys of adult Tg(lhx1a:DsRed) fish after treatment with Met-ENK (100 µM, 10 µL per fish), and observed a decrease in the number of lhx1a+ RPCAs and a significant increase in the number of individual RPCs at 5 dpi (Fig. 3n, o). This experiment confirmed that early activation of PENK-A can prematurely terminate kidney regeneration.
PENK-A affects kidney regeneration by regulating H2O2 production
To elucidate the mechanisms underlying PENK-A’s regulation of kidney regeneration, we analyzed the expression of PENK-A receptors. Upon analyzing the scRNA-seq data of adult zebrafish kidneys38, we observed low expression levels of genes encoding canonical PENK-A receptors, such as δ-opioid receptors and μ-opioid receptors. However, genes encoding noncanonical opioid receptors in the opioid growth factor receptor (ogfr) family51, including ogfr, ogfrl1, and ogfrl2, exhibited high expression levels (Fig. 4a). Notably, these genes were predominantly expressed in renal medullary cells, such as macrophages, neutrophils, T cells and neurons (Fig. 4a). In our previous studies, we established that ROS, particularly H2O2, serve as the initial signals for kidney regeneration29. H2O2 production primarily occurs in renal medullary cells. We observed a gradual increase in the expression of the H2O2 synthase gene, duox1, and the production of H2O2 labeled with pentafluorobenzenesulfonyl fluorescein (PBSF) in the injured adult kidneys, which peaked at 5 dpi. Inhibition of H2O2 production impairs the formation of new nephrons29. To further confirm the location of H2O2 production, we utilized injured adult kidneys from the Tg(cdh17:DsRed) transgenic line, in which renal tubules are specifically marked25,52. We performed PBSF staining and observed that after 3 dpi, the majority of H2O2 was generated in the renal medulla (Fig. 4b). Interestingly, previous studies have demonstrated that exogenous opioids can hinder tissue regeneration by suppressing ROS production in mice, as exemplified by the regeneration of the inguinal fat pad53. Thus, based on these findings, we hypothesized that endogenous enkephalin PENK-A may exert its effects through a similar mechanism.
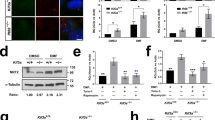
a Gene expression of PENK-A receptors (ogfr, ogfrl1, and ogfrl2) in kidney cells. VECs (vascular endothelial cells), DTECs (distal tubular epithelial cells), Mφ (macrophages), HSCs (hematopoietic stem cells), MSCs (mucin-secreting cells), and RICs (renal interstitial cells). b Confocal images of H2O2 signal in adult Tg(cdh17:DsRed) kidneys after Gent-induced AKI, detected using the PBSF fluorescence probe. Scale bar, 50 μm. c Relative H2O2 concentration in kidneys after NAL-M or DMSO administration following AKI, presented as fold change relative to the DMSO-treated Un-Inj group (n = 3). d Relative H2O2 concentrations in the kidneys of WT, penka+/−, and penka−/− zebrafish following AKI, presented as fold change relative to the Un-Inj WT group (n = 3). e H2O2 concentration in kidneys after TRAM 10 (10 μM TRAM, 10 μL per fish), TRAM 16 (16 μM TRAM, 10 μL per fish), or DMSO administration following AKI, presented as fold change relative to the DMSO-treated Un-Inj groups (n = 3). f H2O2 concentration in kidneys after Met-ENK 50 (50 μM Met-ENK, 10 μL per fish), Met-ENK 100 (100 μM Met-ENK, 10 μL per fish), or DMSO administration following AKI, presented as fold change relative to the DMSO-treated Un-Inj groups (n = 3). g H2O2 concentrations in the kidneys of WT and Tg(hsp70l:penka) with HS or without HS (Un-HS) following AKI, presented as fold change relative to the Un-HS and Un-Inj WT groups (n = 3). No significant differences between Un-HS WT, HS WT, and Un-HS Tg(hsp70l:penka) zebrafish were found using two-tailed t-test. h, i RT-PCR (h) and WISH (i) analyses of lhx1a in 5 dpi penka−/− kidneys after administration (at 2 and 4 dpi) of VAS2870, duox1 vivo-MO, Con vivo-MO, or DMSO after AKI, Scale bar, 600 μm. j Quantification of lhx1a+ RPCAs per kidney (n = 5) for each condition in (i). Data in (c), (d), (e), (f), (g), and (i) were analyzed by two-sided t-test and are presented as mean values ± SD. p values are listed. Source data are provided as a Source data file.
To investigate whether PTECs regulate the production of H2O2 through penka, we utilized a fluorimetric hydrogen peroxide assay kit to accurately measure the H2O2 levels in the kidneys. We injected NAL-M and observed that the H2O2 levels in the treatment group were higher than those in the control group in all stages (Fig. 4c). In addition, we assessed the H2O2 levels in adult penka−1+24 mutant kidneys. Interestingly, we observed significantly elevated H2O2 levels in the injured kidneys of homozygous penka−1+24 mutants throughout all stages of regeneration (Fig. 4d), while the H2O2 levels in injured WT kidneys remained lower. The injured kidneys of the heterozygous penka−1+24 mutants exhibited intermediate H2O2 levels (Fig. 4d). These results indicate a correlation between the penka expression level and H2O2 production.
To investigate the impact of PENK-A pathway reactivation on H2O2 production during regeneration, we examined regenerating adult kidneys treated with TRAM and Met-ENK or kidneys of Tg(hsp70l:penka) fish subjected to heat shock in at 2, 4, and 6 dpi. The results revealed that the levels of H2O2 were lower than those in the control group at all stages (Fig. 4e‒g). In addition, a higher concentration of the agonist led to more significant inhibition of H2O2 production (Fig. 4e, f), demonstrating a dose-dependent relationship between PENK-A and H2O2. Our previous studies have demonstrated that injection of the NADPH oxidase antagonist VAS2870 or the H2O2 synthase duox1 vivo-MO effectively inhibits H2O2 production in injured kidneys29. To confirm that penka deficiency accelerates kidney regeneration through H2O2, we intraperitoneally injected VAS2870 or duox1 vivo-MO into penka−1+24 mutants. The rapid regeneration observed at 5 dpi in the mutants was inhibited (Fig. 4h‒j), providing evidence that PENK-A regulates kidney regeneration through H2O2. Overall, the findings indicate that during the initial stage of regeneration, decreased penka expression promotes H2O2 production, thereby accelerating kidney regeneration. However, in the late stage, as the number of neonatal nephrons increases, the expression of PENK-A also rises. This increase in PENK-A can inhibit the production of H2O2, thereby contributing to the termination of the kidney regeneration process.
The PENK-A–H2O2 pathway regulates kidney regeneration by affecting the remodeling of global H3K4me3 in the kidneys
H2O2 is a relatively stable ROS, that can freely diffuse among cells54. Previous studies have indicated that H2O2 can induce downregulation of H3K4me3, that occurs at the promoter region and is associated with the activation of nearby gene expression, in Caenorhabditis elegans and HeLa cells55. This suggests that H2O2 may play a role in gene switching and regulation of gene expression through histone modification. In light of this, we sought to investigate whether H2O2-induced changes have similar effects in the zebrafish kidneys. To address this, we investigated the changes in H3K4me3 levels during kidney regeneration in WT and penka−1+24 mutant kidneys at 3, 5, and 7 dpi. Remarkably, among WT and penka−1+24 mutant kidneys, our Western blot analyses revealed significantly lower H3K4me3 levels at 3 dpi in injured kidneys than in uninjured kidneys (Fig. 5a, b). Furthermore, in WT kidneys, a subsequent increase in H3K4me3 levels was observed at 5 dpi, and by 7 dpi, the difference from the levels in the uninjured group was not statistically significant (Fig. 5a, b). However, at 5 and 7 dpi, the H3K4me3 levels in penka−1+24 mutant kidneys were lower than those in WT kidneys (Fig. 5a, b). These findings align with the temporal pattern of H2O2 generation and persistence. Consequently, we propose that the burst of H2O2 may induce remodeling of H3K4me3 in renal cells.
a Western blot analysis of H3K4me3 levels in WT and penka−/− zebrafish kidneys following AKI. b The protein band intensities in a were normalized to the loading control, Histone 3 (H3), and the relative expression levels of H3K4me3 were quantified (n = 3 biological replications per group). The data are presented as the fold change relative to the Un-Inj WT group. c Western blot analysis of H3K4me3 levels in WT and penka−/− zebrafish kidneys at 3 dpi after administration (at 2 dpi) of duox1 vivo-MO, VAS2870, or DMSO following AKI. d The protein band intensities in (c) were normalized to the loading control, H3, and the relative expression levels of H3K4me3 were quantified (n = 3 biological replications per group). The data are presented as the fold change relative to the Un-Inj WT group. e Western blot analysis of H3K4me3 levels in the kidneys at 3 dpi after administration (at 2 dpi) of CPI-455 or DMSO following AKI. f The protein band intensities in e were normalized to the loading control, H3, and the relative expression levels of H3K4me3 were quantified (n = 3). The data are presented as the fold change relative to the Un-Inj groups. g, h RT-PCR (g) and WISH (h) analyses of lhx1a at 7 dpi after administration (at 2, 4, and 6 dpi) of CPI-455 or DMSO following AKI (n = 3). i The lhx1a+ RPCAs per kidney (n = 5) were quantified for each condition in (h). j Confocal images showing adult Tg(lhx1a:DsRed) kidneys at 5 dpi after administration (at 2 and 4 dpi) of duox1 vivo-MO (n = 4), CPI-455 (n = 7), or DMSO (n = 8) following AKI. Scale bar, 100 μm. k Quantitation of individual RPCs (iRPCs, arrowheads) and RPCAs in (j). The data in (b), (d), (f), (i), and (k) were analyzed by two-sided t-test and are presented as mean values ± SD. Source data are provided as a Source data file.
To investigate the relationship between H2O2 and H3K4me3 in the injured kidney, we examined the changes in H3K4me3 levels in samples where H2O2 production was inhibited. Inhibition of H2O2 production using VAS2870 or duox1 vivo-MO in WT and penka−1+24 mutant kidneys prevented the decrease in H3K4me3 levels observed at 3 dpi in Western blot analyses (Fig. 5c, d). These findings indicate that H2O2 generation is necessary for the remodeling of H3K4me3. We also injected the H3K4me3 demethylase KDM5 inhibitor CPI-45556 (80 µM, 10 µL per fish) intraperitoneally in WT zebrafish and found that the level of H3K4me3 in the treatment group was significantly higher than that in the control group at 3 dpi (Fig. 5e, f). In addition, the number of RPCAs was significantly lower in the treatment group compared to the control group at 7 dpi (Fig. 5g–i). These results suggest that the initial burst of H2O2 at 3 dpi triggers remodeling of H3K4me3 in kidney cells, which is crucial for kidney regeneration. In addition, they imply that penka regulates the remodeling of H3K4me3 by manipulating the production of H2O2.
Remodeling of H3K4me3 promotes RPCA formation
During kidney regeneration, H2O2 is generated in cells localized in the renal medulla, where lhx1a+ RPCs are also found22,29. The aforementioned data reveal an increase in the number of individual RPCs and a decrease in the number of RPCAs in PENK-A agonist-treated regenerating kidneys (Fig. 3n, o). Therefore, we hypothesized that inhibiting H2O2 would yield similar effects. Upon knockdown of the H2O2 synthase duox1 using duox1 vivo-MO at 2 and 4 dpi in Tg(lhx1a:DsRed) kidneys, we observed a significantly lower number of lhx1a+ RPCAs in the treated group than in the control group at 5 dpi, while the number of individual lhx1a+ RPCs was higher in the treated group (Fig. 5j, k). These findings suggest that H2O2 influences the process of RPCs aggregation to form RPCAs. In addition, we examined the effect of CPI-455 on RPCA formation and found that CPI-455 injection produced a phenotype similar to that elicited by duox1 vivo-MO (Fig. 5j, k). These data indicate that H2O2 affects the aggregation of RPCs to generate RPCAs. RPCs undergo mesenchymal–epithelial transition (MET) during the aggregation process and subsequently differentiate into mature nephrons25,26. We speculate that H2O2 may activate the expression of genes crucial for the RPC MET process by increasing H3K4me3 levels in the promoter regions of these genes, ultimately leading to the promotion of RPCA formation.
The PENK-A–H2O2 pathway controls kidney regeneration by regulating tcf21
To identify direct targets of H3K4me3 remodeling during kidney regeneration, we performed chromatin immunoprecipitation sequencing (ChIP-seq) to examine changes in H3K4me3 levels in all adult kidney cells at 0, 3, and 5 dpi. ChIP-seq analysis revealed significant changes in the H3K4me3 levels of genes associated with apoptosis and the response to stimuli during kidney regeneration (Supplementary Fig. S4a–c). In addition, we observed significant alterations in the promoter region of tcf21 (Fig. 6a), a gene known to play a crucial role in MET57. Specifically, the H3K4me3 levels upstream of the tcf21 ATG start codon exhibited a significant decrease at 3 dpi followed by an increase at 5 dpi. Based on these findings, we hypothesize that tcf21 may be closely linked to the regulatory effects of PENK-A on kidney regeneration.
a ChIP-seq analysis of the H3K4me3 pattern in the promoter region of tcf21. The H3K4me3 level upstream of the ATG start codon (red box) was decreased significantly at 3 dpi and increased at 5 dpi. b qRT-PCR analysis of tcf21 in zebrafish kidneys during AKI. The data were presented as the fold change relative to the 0 dpi group (n = 3 biological replications per group). c FACS coupled with RT-PCR analysis of tcf21 expression in lhx1a:DsRed-labeled RPCs at 5 dpi after AKI. Water was used as the RT-PCR negative control. Mk, Marker. d Confocal images revealed that the combination of tcf21 FISH with Pax2a immunofluorescence showed high expression of tcf21 in Pax2a+ RPCAs at 5 dpi after AKI. Scale bar, 50 μm. e qRT-PCR analysis of tcf21 in WT and penka−/− kidneys following AKI (n = 3 biological replications per group). The data were presented as the fold change relative to the 0 dpi WT group. f qRT-PCR analysis of tcf21 in 7 dpi WT kidneys after administration (at 2, 4, and 6 dpi) of Met-ENK, Con vivo-MO, duox1 vivo-MO, VAS2870, CPI-455, and DMSO following AKI (n = 3 biological replications per group). The data were presented as the fold change relative to the 7 dpi DMSO-treated group. The data in (b), (e), and (f) were analyzed by two-sided t-test and are presented as mean values ± SD. Source data are provided as a Source data file.
TCF21 is a helix-loop-helix domain-containing transcription factor that participates in regulating cell differentiation and cell fate transformation through MET57. We investigated the expression of the tcf21 gene at different time points during kidney regeneration using qRT-PCR and observed that tcf21 expression began to increase at 5 dpi, which coincided with the formation of RPCAs (Fig. 6b). To confirm whether tcf21 is expressed in RPCs, we sorted lhx1a+ RPCs at 5 dpi using fluorescence-activated cell sorting (FACS) and performed RT-PCR using Tg(lhx1a:DsRed)-injured kidneys. The results demonstrated high expression of tcf21 in the sorted lhx1a+ cells (Fig. 6c). In addition, we employed a combined approach with tcf21 FISH and Pax2a (a marker for RPCAs22,25,29) immunofluorescence in injured kidneys and observed high expression of tcf21 in Pax2a+ RPCAs (Fig. 6d). We also examined the expression of tcf21 in penka−1+24 mutant kidneys through qRT-PCR analysis. In comparison to WT, the penka−1+24 mutant kidneys displayed an elevated tcf21 expression at 5 and 7 dpi (Fig. 6e). However, in the Met-ENK treated WT group, the qRT-PCR analysis results indicated a significant decrease in the expression of tcf21 at 7 dpi (Fig. 6f). The intraperitoneal injection of VAS2870 or duox1 vivo-MO into WT fish also resulted in the suppression of the upregulation of tcf21 expression (Fig. 6f). Furthermore, in the CPI-455 treated group, the expression of tcf21 did not show an increase when H3K4me3 remodeling was inhibited (Fig. 6f), which corresponded to the change in lhx1a expression (Fig. 5g). These findings suggest that PENK-A influences the formation of RPCAs through its effects on tcf21.
Due to the inability of zebrafish tcf21 mutants to survive to adulthood, we suppressed tcf21 expression through intraperitoneal injection of tcf21 vivo-MO58 (25 μM, 10 µL per fish) (Supplementary Fig. S5a, b). Treatment with tcf21 vivo-MO significantly reduced the number of RPCAs, while increasing the number of individual RPCs in Tg(lhx1a:DsRed) kidneys (Fig. 7a, b). WISH and RT-PCR analyses also revealed a significant decrease in the expression of lhx1a and a reduction in the number of RPCAs in the tcf21 vivo-MO group compared to the control group (Fig. 7c‒e). Moreover, the results of lhx1a RT-PCR and WISH in penka−1+24 mutant kidneys injected with tcf21 vivo-MO also indicated a reduction in RPCAs formation (Fig. 7c‒e). In addition, we constructed a transgenic line, Tg(hsp70l:tcf21), that can effectively overexpress tcf21 after heat shock (Fig. 7h). Heat shock was conducted at 2 and 4 dpi, and we found significantly more RPCAs in the injured kidneys of the Tg(hsp70l:tcf21;lhx1a:DsRed) fish than in those of control fish (Fig. 7f). RT-PCR and WISH also revealed an increase in lhx1a expression and a significant increase in RPCAs (Fig. 7h–j). Furthermore, we observed that activating the PENK-A signaling pathway through intraperitoneal injection of Met-ENK (100 µM, 10 µL per fish) restored the number of RPCAs in the injured kidneys of heat-shocked Tg(hsp70l:tcf21;lhx1a:DsRed) fish (Fig. 7f). lhx1a RT-PCR and WISH confirmed these findings (Fig. 7h‒j). When treated with CPI-455 (80 µM, 10 µL per fish) in heat-shocked Tg(hsp70l:tcf21;lhx1a:DsRed) fish, it was found that the overexpression of tcf21 could rescue the inhibition of RPC aggregation by CPI-455 (Fig. 7f) and reactivate kidney regeneration (Fig. 7h‒j). These results provide further evidence that PENK-A regulates kidney regeneration through its effects on tcf21.
a Confocal images showing adult Tg(lhx1a:DsRed) kidneys at 5 dpi afer administration (at 2 and 4 dpi) of tcf21 vivo-MO, or Con vivo-MO following AKI (n = 5 biological replications per group). b Quantitation of individual RPCs (iRPCs, arrowhead) and RPCAs in a. c, d RT-PCR (n = 3) (c) and WISH (d) analyses of lhx1a in 7 dpi WT and penka−/− kidneys with administration (at 2, 4, and 6 dpi) of tcf21 vivo-MO (n = 5 in WT group, n = 8 in penka−/− group) or Con vivo-MO ((n = 5 in WT group, n = 4 in penka−/− group) following AKI. e The lhx1a+ RPCAs per kidney were quantified for each condition in d. f Confocal images showing 5 dpi Tg(lhx1a:DsRed;hsp70l:tcf21) kidneys with HS (at 2 and 4 dpi) or Un-HS after administration (at 2 and 4 dpi) of Met-ENK (n = 9 in Un-HS group, n = 5 in HS group), CPI-455 (n = 9 in Un-HS group, n = 6 in HS group), or DMSO (n = 8) following AKI. g Quantitation of individual RPCs (iRPCs, arrowheads) and RPCAs in (f). h RT-PCR analysis of tcf21 and lhx1a in 7 dpi WT and Tg(hsp70l:tcf21) kidneys with HS (at 2, 4, and 6 dpi) or Un-HS after administration (at 2, 4, and 6 dpi) of Met-ENK, CPI-455 or DMSO following AKI (n = 3). i WISH analysis of lhx1a in 7 dpi WT and Tg(hsp70l:tcf21) kidneys with HS (at 2 and 4 dpi) or Un-HS after administration (at 2, 4, and 6 dpi) of Met-ENK (n = 5), CPI-455 (n = 5) or DMSO (n = 5 in Un-HS group, n = 4 in HS group) following AKI. j Quantitation of lhx1a+ RPCAs per kidney was performed for each condition in (i). Data in (b), (e), (g), and (j) were analyzed by two-sided t-test and are presented as mean values ± SD. Scale bars in (a) and (f), 50 μm; (d) and (i), 600 μm. Source data are provided as a Source data file.
In general, the results indicate that a sudden loss of PENK-A in PTECs can trigger a burst of H2O2 production within the kidney. The burst of H2O2 can modify the H3K4me3 levels at the tcf21 promoter region, altering the activity of the tcf21 promoter and promoting the expression of tcf21, thereby facilitating the expansion of RPCAs. During the later stages of regeneration, as PTECs recover, the increased expression of penka inhibits H2O2 production, thereby promoting the termination of kidney regeneration (Fig. 8). Thus, PTECs can regulate their own regeneration through a negative feedback mechanism mediated by the PENK-A signaling pathway.
PTEC-expressed PENK-A levels decreased with the loss of PTs after AKI. This decrease triggered an increase in H2O2 production, resulting in the expansion of RPC aggregation. This process occurred by upregulating tcf21 expression through remodeling of H3K4me3 in the tcf21 promoter. In the later stage of regeneration, as PTECs recovered, elevated penka expression suppressed the production of H2O2, facilitating the termination of kidney regeneration.
Discussion
In this study, we investigated the involvement of endogenous opioids in zebrafish kidney regeneration using genetic approaches. Our findings demonstrate that penka expression is responsive to tissue damage during zebrafish kidney regeneration. A decrease in penka expression creates a conducive environment for the production of substantial amounts of H2O2 within the kidneys. Through analysis of a penka mutant, we observed increased H2O2 production in the kidneys of homozygous mutant adult zebrafish accompanied by early activation of kidney regeneration. The H2O2 levels in heterozygous mutants fell between those of WT and homozygous mutants, indicating a dependency on penka expression levels for H2O2 production. Given the combined results of pharmacological treatments and penka overexpression experiments, we conclude that PTECs can finely regulate H2O2 production by precisely controlling penka expression, thereby influencing kidney regeneration.
Clinical studies have provided evidence of a positive correlation between increased PENK and exacerbation of AKI. Higher concentrations of PENK may hinder AKI repair process32,33,35. In zebrafish, we observed a significant decrease in PENK-A expression prior to kidney tissue regeneration, suggesting that PENK-A could be an essential molecular factor in achieving rapid tissue repair following kidney injury in zebrafish. The role of ROS in the pathogenesis of kidney diseases has been well-documented. Elevated ROS levels contribute to renal inflammation, renal tubular injury, and renal fibrosis30,31. Consequently, screening for drugs that can inhibit excessive ROS production holds promise for the treatment of kidney diseases. Our study reveals the significant regulatory role of endogenous PENK-A in renal H2O2 production, positioning PENK as a potential drug target for kidney disease treatment. Currently, perioperative pain relief protocols routinely involve the use of opioid analogs as exogenous agents59. Further investigation of the relationship between PENK and the development and progression of kidney disease could enable these drugs to be utilized in the clinical management of kidney disease.
In a previous study on an AKI model induced by lipopolysaccharide (LPS), the overall level of H3K4me3 was observed to be significantly lower than that in compared to the control group60. However, the underlying reason for this decline remains unclear. Our investigation revealed a direct relationship between the H2O2 burst at 3 dpi and the decrease in H3K4me3 level at 3 dpi in the zebrafish kidneys. Blocking H2O2 production effectively inhibited the alterations in H3K4me3 levels. Notably, we observed that the levels of H3K4me3 began to rise at 5 and 7 dpi, indicating that the abrupt change in H2O2 production profoundly induced H3K4me3 remodeling in the kidney. Previous studies have also demonstrated that transient ROS exposure can modulate the global levels of H3K4me3 in both Caenorhabditis elegans and mammalian cells, leading to enhanced stress resistance55. Consequently, we propose that H3K4me3 remodeling can reshape the gene expression pattern of kidney cells and drive their participation in the process of kidney regeneration. By screening genes associated with alterations in renal H3K4me3 levels, we identified tcf21 as a direct influencer of the regeneration process in RPCs. Knockdown of tcf21 expression effectively hindered the formation of RPCAs. However, we acknowledge that tcf21 may not be the sole factor influenced by H3K4me3 remodeling, and further investigations are warranted to elucidate the involved signaling pathways.
In this study, we made the significant discovery that PENK-A expressed by PTECs plays a pivotal role in determining the pace of kidney regeneration through its control over H2O2 production. A burst in H2O2 triggers remodeling of H3K4me3 in kidney cells. We further demonstrated that this remodeling affects the expression of tcf21 in RPCs, which is a critical gene involved in the formation of RPCAs. Collectively, our findings unveil the inhibitory role of PENK-A in kidney regeneration. Leveraging these insights will enable the design of safe and effective clinical interventions for the treatment of renal diseases.
Methods
Zebrafish husbandry and lines used
Zebrafish were produced, grown, and maintained according to standard protocols61. The following transgenic lines were used in this study: Tg(lhx1a:DsRed)25, Tg(cdh17:DsRed)25,52, Tg(gtshβ:GFP)45, Tg(hsp70l:penka), and Tg(hsp70l:tcf21). Adult zebrafish aged between 3 to 12 months were used for the experiments, and approximately equal sex ratios were employed. Anesthesia was administered using 0.0168% buffered tricaine methanesulfonate (MS-222, Sigma). The AB strain of zebrafish served as the WT control for this study. Animal care and use protocol was approved by the Institutional Animal Care and Use Committee of the Army Medical University, China (SYXK-PLA-2007035).
Single-cell gene expression profiling
For analysis of the expression of penka and its receptors in zebrafish kidneys, we utilized a portion of recently published scRNA-seq data (GSE100910)38. The Seurat package (version 4.2.0) was employed to perform data normalization, dimensionality reduction, clustering, and differential expression analysis.
Human renal biopsy samples
Renal biopsies were performed as part of routine clinical diagnostic investigations. Three patients with AKI and three patients with no detectable lesions verified by renal biopsy were enrolled in this study from the Department of Nephrology, Xinqiao Hospital, Chongqing, China. Patients with inflammatory and autoimmune-associated diseases, diabetes, polycystic kidney disease, and pregnancy were excluded from the study. Kidney biopsies were obtained from these patients for FISH analysis. The human studies conducted in this research were approved by the Ethics Committee of Xinqiao Hospital, Army Medical University (No. 2023-YAN-143-01). The study design and conduct adhered to all applicable regulations concerning the use of human participants and were in accordance with the principles set forth in the Declaration of Helsinki. Informed written consent was provided by all participants. Furthermore, the study complies with the guidelines provided by the Ministry of Science and Technology (MOST) for the Review and Approval of Human Genetic Resources.
FISH of human renal biopsy samples
The paraffin-embedded renal biopsy samples were sectioned to a thickness of 3 μm. These sections were permeabilized with proteinase K (10 μg/mL, Roche) in PBS with 0.1% Tween-20 (PBT) for 10 min with gentle rocking. Digoxigenin-labeled riboprobes were synthesized from cDNA fragments containing the sequence of human PENK (Supplementary Table S1). To detect the probes, we utilized an anti-digoxigenin-peroxidase antibody (Roche, 11207733910) and a TSA Plus Fluorescein system (PerkinElmer, NEL741001KT). Following FISH, the sections were stained with LTL (10 μg/mL, Vector Laboratories, FL−1321-2). Finally, images were captured using a Nikon A1 confocal microscope. Detailed antibody information is listed in Supplementary Table S2.
Zebrafish AKI model
Intraperitoneal injection of Gent was employed to induce AKI in adult zebrafish following a previously described method25. In brief, Gent (2.7 μg/μL, 20 μL per fish), diluted in water, was intraperitoneally injected into the WT line or other zebrafish lines. Each injected zebrafish was then placed into an individual container. Zebrafish exhibiting proteinuria at 1 dpi were selected for subsequent experiments.
Nephron counting
Each nephron contained only one segment of gtshβ:GFP-labeled PTs45. Therefore, the number of gtshβ:GFP-labeled PTs segment was used to counting the number of nephrons. To perform the counting, kidneys were carefully removed from the adult Tg(gtshβ:GFP) fish and imaged in a 1.5 mm by 1.5 mm area using a Nikon A1 confocal microscope. The total number of GFP positive segments was determined using ImageJ. Before counting, all the pictures were relabeled, and the person conducting the analysis had no knowledge of the treatment conditions for each sample. This step was taken to minimize subjective bias and maintain objectivity and accuracy in the analysis. To assess the variation in the number of nephrons following Gent-induced AKI, the baseline number of nephrons in the uninjured kidneys was considered 100%. Subsequently, the percentage change in nephrons after AKI compared to the uninjured condition was utilized to depict the extent of kidney damage.
CRISPR‒Cas9 mutagenesis
The CRISPR/Cas9 target site in penka was selected using the CRISPR design tool46 (https://www.crisprscan.org/). The guide RNA targeted the sequence 5’-GGCTTCATGAAGCGTTACGGCGG-3’ (target underlined) in penka exon 2. To construct the gRNA, the T7 promoter and gRNA scaffold were cloned into the pMD 19-T vector. The DNA template for gRNA synthesis was obtained by PCR amplification. Subsequently, the gRNA was synthesized using a Hiscribe T7 High Yield RNA Synthesis Kit (NEB, E2040S). For CRISPR/Cas9 mutagenesis, a mixed solution containing 1 nL of Cas9 protein (0.7 μM, NEB, M0646T) and gRNA (75 ng/μL) was injected into one-cell stage WT embryos. After the injection, genomic DNA of the embryos was obtained at 24 h post-fertilization (hpf) to check for mutagenesis at the target site. The primers used to amplify the penka gene are listed in Supplementary Table S1. The PCR products were then subjected to Sanger sequencing to confirm mutagenesis. Once the mutagenesis was confirmed, other injected embryos were raised to adulthood as F0 fish. The F1 generation were obtained by mating F0 fish with WT zebrafish and raising them to adulthood. Genomic DNA was extracted from tail tissue of the F1 fish and analyzed using methods mentioned above. Two penka mutant alleles (penka−1+24 and penka−8) were identified through PCR and DNA sequencing. These alleles were subsequently chosen for further propagation, leading to the establishment of stable penka mutant lines to facilitate subsequent analyses. To assess off-target effects, CRISPRScan was used to predict potential off-target sites46,47, and the top five sites were selected for sequencing analysis. Primers and sequencing data were listed in Source data file.
WISH
WISH was conducted following previously described methods25,29. Briefly, after removing the internal organs, except for the kidneys, zebrafish were fixed overnight with 4% paraformaldehyde (PFA). After three washes with PBT, the fixed kidneys were dissected from the body and permeabilized with proteinase K (10 μg/mL, Roche, 3508838) in PBT with rocking for 1 h. Digoxigenin-labeled riboprobes were generated from cDNA fragments containing the sequences of zebrafish lhx1a25,29. An anti-digoxigenin-alkaline phosphatase antibody (Roche, 11093274910) and NBT/BCIP substrate (Roche, 11681451001) were used to detect the probe. Images were captured using a BX3-CBH microscope. Detailed antibody information is listed in Supplementary Table S2.
Combined FISH and immunofluorescence in zebrafish kidneys
Combined FISH and immunofluorescence were performed following previously described methods25,62. Briefly, kidneys were harvested and fixed in 4% PFA overnight at 4 °C, and then frozen sections were created at 100-μm thickness. The sections were permeabilized with proteinase K (10 μg/mL, Roche) in PBT for 20 min with rocking. Digoxigenin-labeled riboprobes were generated from cDNA fragments comprising the sequences of zebrafish tcf21, slc20a1a, trpm7, and penka (Supplementary Table S1). An anti-digoxigenin-peroxidase antibody (Roche, 11207733910) and a TSA Plus Fluorescein system (PerkinElmer, NEL741001KT) were used to detect the probes. After FISH, the sections were stained for immunofluorescence. The primary antibodies used were anti-Met-ENK (Abcam, ab22620) and anti-Pax2a (Abcam, ab229318). The secondary antibody goat anti-rabbit IgG H&L Alexa Fluor 633 (Invitrogen, A21070) was used at a dilution of 1:500. Images were captured using a Nikon A1 confocal microscope. Detailed antibody information is listed in Supplementary Table S2.
RT-PCR and qRT-PCR
RNA was extracted from kidney tissues using TRIzol reagent (Invitrogen, 15596018). A Prime Script II 1st strand cDNA Synthesis Kit (Takara, 9767) was used to synthesize cDNA, which was then subjected to PCR using Taq Master Mix (Vazyme, p112-01) for RT-PCR or TB Green Premix EX Taq II (Takara, RR820A) for qRT-PCR. All primers for lhx1a25, penka, tcf21, and β-actin25 are listed in Supplementary Table S1. Gene expression was normalized to β-actin mRNA expression. Full scan blots are provided as a Source data file.
Western blotting
Zebrafish kidneys were obtained and homogenized with a 1 mL syringe and needle in cell lysis buffer (50 mM PBS, pH 7.4, 1% SDS, and 0.5% Triton X-100) containing protease inhibitor (Beyotime, P1005). The lysate was centrifuged at 12,000 × g for 20 min at 4 °C, and the resulting supernatant was used as the protein sample. The protein concentration was determined using a bicinchoninic acid protein assay kit (CWBIO, CW0014S). Western blotting was carried out following standard protocols. The levels of total H3K4me3 and histone H3 were detected using the enhanced chemiluminescence (ECL) method with the antibodies listed in Supplementary Table S2. Full scan blots are provided as a Source data file.
Pharmaceutical treatment
During kidney regeneration, TRAM (10 or 16 μM, 10 μL per fish), NAL-M (2.0 μM, 10 μL per fish), Met-ENK (50 or 100 μM, 10 μL per fish), VAS2870 (8 μM, 10 μL per fish), CPI-455 (80 μM, 10 μL per fish), or 0.1% DMSO (10 μL per fish) was intraperitoneally injected individually every other day starting from 2 dpi until the kidneys were collected for further analysis. To test the termination signal of kidney regeneration, TRAM, Met-ENK, or 0.1% DMSO (10 μL per fish) was intraperitoneally injected into zebrafish individually at 4 and 6 dpi. Kidneys were collected at 7 dpi for subsequent experiments. Detailed information about these inhibitors is listed in Supplementary Table S2.
Vivo-MO
The duox1 vivo-MO29, tcf21 vivo-MO58, penka vivo-MO, and control vivo-MO29 were designed as previously described and are listed in Supplementary Table S1. To verify the efficiency of tcf21 vivo-MO and penka vivo-MO (two splice blocking morpholinos) in adult kidneys, tcf21 vivo-MO (25 μM, 10 μL per fish) or penka vivo-MO (100 μM, 10 μL per fish) was intraperitoneally injected into zebrafish at 2, 4, and 6 dpi, and kidneys were collected at 7 dpi for RNA extraction and RT-PCR using the identification primers (Supplementary Table S1). During kidney regeneration, the vivo-MOs were intraperitoneally injected every other day beginning on 2 dpi until the kidneys were collected for further analysis.
Generation of transgenic zebrafish lines
The coding sequence of penka or tcf21 was amplified using PCR. The resulting fragments were then directionally cloned into the SalI/NotI site of the hsp70l-loxP-mCherry-STOP-loxP-H2B-GFP_cryaa-cerulean/pI-SceI plasmid, leading to the generation of the hsp70l:penka_cryaa-cerulean/pI-SceI or hsp70l:tcf21_cryaa-cerulean/pI-SceI plasmid. For the transgenesis process, 1 nL of the injection mix containing 30 pg of each plasmid DNA and I-SceI restriction enzyme (2 U/μL, NEB, R0694S) was injected into one-cell-stage embryos. At ~96 h post-fertilization, we employed fluorescence microscopy to screen embryos displaying robust expression of blue fluorescent protein (cerulean) in their eyes. These embryos were then raised to generate F0 transgenic fish. The F1 generation was obtained by mating F0 transgenic fish with WT zebrafish. Based on whether they expressed the blue fluorescent protein cerulean in their eyes, four F1 fish were identified for each transgenic line. Subsequently, these F1 fish were crossed with WT zebrafish, and the resulting offspring were subjected to heat-shock on the third day after birth using the method described below. Total RNA was extracted from these heat-shocked embryos and converted into cDNA. Subsequently, the expression levels of penka or tcf21 were analyzed using RT-PCR in these heat-shocked embryos. The fish line with the highest expression level of penka or tcf21 was selected for further propagation, establishing the stable Tg(hsp70l:penka) or Tg(hsp70l:tcf21) lines for further analyses.
Heat shock treatment
For heat shock treatment, zebrafish were transferred to preheated system water at 39 °C and kept at 39 °C for 30 min. After heat shock, the zebrafish were returned to the 28.5 °C system water. Zebrafish were heat shocked every other day after 2 dpi until the kidneys were collected for subsequent experiments.
Imaging and quantification of lhx1a + individual RPCs and RPCAs
Kidneys were carefully removed from the adult Tg(lhx1a:DsRed) zebrafish and imaged using a Nikon A1 confocal microscope. The individual DsRed+ cells and DsRed+ RPCAs were counted using ImageJ. Before counting, all the pictures were relabeled to ensure that the analyst conducting the analysis remained blind to the treatment conditions of each sample. This precautionary measure was implemented to minimize subjective bias and maintain objectivity and accuracy during the analysis.
in situ H2O2 imaging
For in situ H2O2 imaging, adult Tg(cdh17:DsRed) zebrafish were intraperitoneally injected with a PBSF solution (Santa Cruz, SC-205429A; 100 μM, 10 μL per fish) to detect the location and level of H2O2. Three hours post-injection, the fish were anesthetized with 0.016% MS-222, sacrificed, and dissected to obtain the kidneys. The kidneys were then photographed using a Nikon A1 confocal microscope.
H2O2 level measurement
H2O2 levels in adult zebrafish kidneys were assessed using a fluorimetric hydrogen peroxide assay kit (Sigma, MAK166) following the manufacturer’s instructions. Briefly, kidney protein lysates were diluted to 1 mg/mL in NP-40 lysis buffer without protease inhibitors to avoid interference with the assay. In 96-well plates, 50 μL of each sample and standards of 0, 0.1, 0.3, 1, 3, and 10 μM H2O2 were added. Each reaction was then mixed with 50 μL of the reaction mixture, which consisted of assay buffer with 20 units/mL horseradish peroxidase and 1% infrared fluorometric peroxidase substrate. The plates were incubated at room temperature for 10 min and subsequently read fluorometrically at 640 nm excitation and 680 nm emission using a SpectraMAX M3 (Molecular Devices).
FACS
To obtain lhx1a:DsRed-labeled cells, kidneys from ten Tg(lhx1a:DsRed) zebrafish were manually dissected in PBS and 0.05% trypsin-EDTA solution at 5 dpi. DsRed positive cells were sorted using a MoFlo XDP flow cytometer (Beckman) and collected for RNA extraction.
ChIP-seq
Total DNA from uninjured zebrafish kidneys and injured zebrafish kidneys at 3 and 5 dpi was used for ChIP assays. For each assay, ten kidneys were utilized for total DNA extraction. The extracted DNA was then sheared into fragments ranging from 100 to 500 base pairs using an ultrasonic crusher. In the ChIP pull-down assay, 25 µg of chromatin samples (in equal volume) were incubated with 2.5 µg of specific antibodies (anti-H3K4me3 from CST, Cat# 9751) or rabbit IgG (as a negative control, from CST, Cat# 66326) for 24 h at 4 °C on a vertical rotor. The antibody–DNA‒protein complexes were subsequently immunoprecipitated using a CUT&RUN Assay Kit (CST, Cat# 86652) following an agarose bead wash and incubation of the complex with the beads for 3 h at 4 °C. After coincubation, the beads were washed three times using incubation buffer. The DNA fragments were then extracted using the phenolchloroform method to construct the Illumina sequencing library. Library construction was performed by Novogene Corporation (Beijing, China). Subsequently, pair-end sequencing of the samples was conducted on the Illumina platform. The quality of the sequencing library was assessed using the Agilent Bioanalyzer 2100 system.
ChIP-seq data analysis
The sequencing reads for ChIP and input DNA were aligned to the zebrafish reference genome files (GRCz11). The GRCz11 index was built using Burrows–Wheeler Aligner (BWA, v0.7.12), and clean reads were then mapped to the reference genome using BWA-MEM (v0.7.12). To build an enrichment model and predict fragment size, a specific number of windows were used as samples. Subsequently, peak calling analysis was performed based on the predicted fragment size. ChIPseeker was employed to retrieve the nearest genes surrounding the identified peaks and annotate the genomic regions associated with each peak. The ChIPseeker tool was utilized to confirm peak-related genes, and Gene Ontology (GO) enrichment analysis was conducted to identify enriched functions. In addition, KOBAS software was used to test the statistical enrichment of peak-related genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Differential peak analyses were based on the fold enrichment of peaks observed in different experiments. A peak was considered differential when the odds ratio between two groups was greater than 2. Utilizing the same methodology, genes associated with differential peaks were identified, and GO and KEGG enrichment analyses were performed on these genes as well.
Statistics
Unless otherwise stated, all experiments were conducted with at least three independent replicates. The results are presented as the means ± standard deviations (SDs). Statistical analysis was carried out using Excel (Microsoft Office Home and Student 2019 version) and GraphPad Prism (version 8.02) for Microsoft Windows. The data were analyzed by two-sided t-test and are presented as mean values ± SD.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All datasets generated in this study have been deposited in the Gene Expression Omnibus repository under the series number GSE217831. Source data are provided with this paper.
References
Rayego-Mateos, S. et al. Molecular mechanisms of kidney injury and repair. Int. J. Mol. Sci. 23, 1542 (2022).
Michalopoulos, G. K. & Bhushan, B. Liver regeneration: biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 18, 40–55 (2021).
Liu, Y., Lou, W. P. & Fei, J. F. The engine initiating tissue regeneration: does a common mechanism exist during evolution? Cell Regen. 10, 12 (2021).
Gomes, R. N., Manuel, F. & Nascimento, D. S. The bright side of fibroblasts: molecular signature and regenerative cues in major organs. NPJ Regen. Med. 6, 43 (2021).
Ghosh-Roy, A., Wu, Z., Goncharov, A., Jin, Y. & Chisholm, A. D. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J. Neurosci. 30, 3175–3183 (2010).
Yan, D. & Jin, Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron 76, 534–548 (2012).
Love, N. R. et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 15, 222–228 (2013).
Romero, M. M. G., McCathie, G., Jankun, P. & Roehl, H. H. Damage-induced reactive oxygen species enable zebrafish tail regeneration by repositioning of Hedgehog expressing cells. Nat. Commun. 9, 4010 (2018).
Santabarbara-Ruiz, P. et al. Ask1 and Akt act synergistically to promote ROS-dependent regeneration in Drosophila. PLoS Genet. 15, e1007926 (2019).
Bangsgaard, N. et al. Contact allergy to epoxy resin: risk occupations and consequences. Contact Dermat. 67, 73–77 (2012).
Godwin, J. W., Pinto, A. R. & Rosenthal, N. A. Macrophages are required for adult salamander limb regeneration. Proc. Natl Acad. Sci. USA 110, 9415–9420 (2013).
Boulter, L. et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 18, 572–579 (2012).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Satoh, A., Graham, G. M., Bryant, S. V. & Gardiner, D. M. Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum). Dev. Biol. 319, 321–335 (2008).
Kumar, A., Godwin, J. W., Gates, P. B., Garza-Garcia, A. A. & Brockes, J. P. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 (2007).
Cigliola, V., Ghila, L., Chera, S. & Herrera, P. L. Tissue repair brakes: a common paradigm in the biology of regeneration. Stem Cells 38, 330–339 (2020).
Apte, U. et al. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology 50, 844–851 (2009).
Liu, B. et al. Investigation of the role of glypican 3 in liver regeneration and hepatocyte proliferation. Am. J. Pathol. 175, 717–724 (2009).
Jin, J. et al. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 61, 315–325 (2015).
Huck, I., Gunewardena, S., Espanol-Suner, R., Willenbring, H. & Apte, U. Hepatocyte nuclear factor 4 alpha activation is essential for termination of liver regeneration in mice. Hepatology 70, 666–681 (2019).
Heallen, T. et al. Hippo signaling impedes adult heart regeneration. Development 140, 4683–4690 (2013).
Diep, C. Q. et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature 470, 95–100 (2011).
Kamei, C. N., Liu, Y. & Drummond, I. A. Kidney regeneration in adult zebrafish by gentamicin induced injury. J. Vis. Exp. 3, e51912 (2015).
Augusto, J., Smith, J., Smith, S., Robertson, S. & Reimschuessel, R. Gentamicin-induced nephrotoxicity and nephroneogenesis in Oreochromis nilotica, a tilapian fish. Dis. Aquat. Org. 26, 49–58 (1996).
Liu, X. et al. Renal interstitial cells promote nephron regeneration by secreting prostaglandin E2. Elife 12, e81438 (2023).
Kamei, C. N., Gallegos, T. F., Liu, Y., Hukriede, N. & Drummond, I. A. Wnt signaling mediates new nephron formation during zebrafish kidney regeneration. Development 146, dev168294 (2019).
Gallegos, T. F., Kamei, C. N., Rohly, M. & Drummond, I. A. Fibroblast growth factor signaling mediates progenitor cell aggregation and nephron regeneration in the adult zebrafish kidney. Dev. Biol. 454, 44–51 (2019).
McCampbell, K. K., Springer, K. N. & Wingert, R. A. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015, 547636 (2015).
Chen, J. et al. Dual roles of hydrogen peroxide in promoting zebrafish renal repair and regeneration. Biochem. Biophys. Res. Commun. 516, 680–685 (2019).
Aranda-Rivera, A. K., Cruz-Gregorio, A., Aparicio-Trejo, O. E. & Pedraza-Chaverri, J. Mitochondrial redox signaling and oxidative stress in kidney diseases. Biomolecules 11, 1144 (2021).
Nie, J. & Hou, F. F. Role of reactive oxygen species in the renal fibrosis. Chin. Med. J. 125, 2598–2602 (2012).
Beunders, R. et al. Proenkephalin (PENK) as a novel biomarker for kidney function. J. Appl. Lab. Med. 2, 400–412 (2017).
Breidthardt, T. et al. Proenkephalin for the early detection of acute kidney injury in hospitalized patients with chronic kidney disease. Eur. J. Clin. Invest. 48, e12999 (2018).
Rosen, H., Polakiewicz, R. D., Benzakine, S. & Bar-Shavit, Z. Proenkephalin A in bone-derived cells. Proc. Natl Acad. Sci. USA 88, 3705–3709 (1991).
Jantti, T. et al. Predictive value of plasma proenkephalin and neutrophil gelatinase-associated lipocalin in acute kidney injury and mortality in cardiogenic shock. Ann. Intensive Care 11, 25 (2021).
Denning, G. M. et al. Proenkephalin expression and enkephalin release are widely observed in non-neuronal tissues. Peptides 29, 83–92 (2008).
Thadhani, R., Pascual, M. & Bonventre, J. V. Acute renal failure. N. Engl. J. Med. 334, 1448–1460 (1996).
Tang, Q. et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 214, 2875–2887 (2017).
Gonzalez Nunez, V., Gonzalez Sarmiento, R. & Rodriguez, R. E. Characterization of zebrafish proenkephalin reveals novel opioid sequences. Brain Res. Mol. Brain Res. 114, 31–39 (2003).
Majumdar, A., Lun, K., Brand, M. & Drummond, I. A. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development 127, 2089–2098 (2000).
Sander, V. et al. Transcriptional profiling of the zebrafish proximal tubule. Am. J. Physiol. Ren. Physiol. 317, F478–F488 (2019).
Poureetezadi, S. J., Cheng, C. N., Chambers, J. M., Drummond, B. E. & Wingert, R. A. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. Elife 5, e17551 (2016).
Fang, Y. et al. Metformin effectively treats Tsc1 deletion-caused kidney pathology by upregulating AMPK phosphorylation. Cell Death Discov. 6, 52 (2020).
Giurgea-Marion, L., Toubeau, G., Laurent, G., Heuson-Stiennon, J. A. & Tulkens, P. M. Impairment of lysosome-pinocytic vesicle fusion in rat kidney proximal tubules after treatment with gentamicin at low doses. Toxicol. Appl. Pharm. 86, 271–285 (1986).
Wang, Y., Sun, Z. H., Zhou, L., Li, Z. & Gui, J. F. Grouper tshbeta promoter-driven transgenic zebrafish marks proximal kidney tubule development. PLoS ONE 9, e97806 (2014).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015).
Uribe-Salazar, J. M. et al. Evaluation of CRISPR gene-editing tools in zebrafish. BMC Genom. 23, 12 (2022).
Lewanowitsch, T. & Irvine, R. J. Naloxone and its quaternary derivative, naloxone methiodide, have differing affinities for mu, delta, and kappa opioid receptors in mouse brain homogenates. Brain Res. 964, 302–305 (2003).
Liu, Z. et al. Met-enkephalin inhibits ROS production through Wnt/beta-catenin signaling in the ZF4 cells of zebrafish. Fish. Shellfish Immunol. 88, 432–440 (2019).
Kathwate, G. H. & Karuppayil, S. M. Tramadol, an opioid receptor agonist: an inhibitor of growth, morphogenesis, and biofilm formation in the human pathogen, Candida albicans. Assay. Drug Dev. Technol. 14, 567–572 (2016).
Kareem, Z. Y., McLaughlin, P. J. & Kumari, R. Opioid growth factor receptor: anatomical distribution and receptor colocalization in neurons of the adult mouse brain. Neuropeptides 99, 102325 (2023).
Liao, S. et al. Establishment of a drug screening model for cardiac complications of acute renal failure. Biomolecules 11, 1370 (2021).
Labit, E. et al. Opioids prevent regeneration in adult mammals through inhibition of ROS production. Sci. Rep. 8, 12170 (2018).
Groeger, G., Quiney, C. & Cotter, T. G. Hydrogen peroxide as a cell-survival signaling molecule. Antioxid. Redox Signal 11, 2655–2671 (2009).
Bazopoulou, D. et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 576, 301–305 (2019).
Vinogradova, M. et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12, 531–538 (2016).
Jiang, X. & Yang, Z. Multiple biological functions of transcription factor 21 in the development of various cancers. Onco Targets Ther. 11, 3533–3539 (2018).
Lee, G. H., Chang, M. Y., Hsu, C. H. & Chen, Y. H. Essential roles of basic helix-loop-helix transcription factors, Capsulin and Musculin, during craniofacial myogenesis of zebrafish. Cell Mol. Life Sci. 68, 4065–4078 (2011).
Hyland, S. J. et al. Perioperative pain management and opioid stewardship: a practical guide. Healthcare 9, 333 (2021).
Liu, Y., Yu, Y., Zhang, J. & Wang, C. The therapeutic effect of dexmedetomidine on protection from renal failure via inhibiting KDM5A in lipopolysaccharide-induced sepsis of mice. Life Sci. 239, 116868 (2019).
Westerfield, M. The zebrafish book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th ed. (Univ. of Oregon Press, 2000).
He, J., Mo, D., Chen, J. & Luo, L. Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nat. Protoc. 15, 3361–3379 (2020).
Acknowledgements
This work was funded by The National Key Research and Development Program of China (2017YFA0106600), the National Natural Science Foundation of China (No. 32070822, 82030023, 82322012, 31771609), The Key support object training project of Army Medical University (No. 2019R025), Young Doctoral Training Program of the Second Affiliated Hospital of Army Medical University (No. 2022YQB013 and 2022YQB060).
Author information
Authors and Affiliations
Contributions
C.L., J.Zhao, and Y.H. conceived the study; C.L. and X.L. designed the study; X.L., Z.H., J.Zhang, X.T., T.Y., Y.Z., S.L., F.L., Z.X., L.D., and W.Y. performed all the experiment; X.L., C.L., and J.Zhang carried out microscopy; C.L., Z.H., and X.L. prepared the manuscript; all authors approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Camila Lima and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, C., Liu, X., He, Z. et al. Proenkephalin-A secreted by renal proximal tubules functions as a brake in kidney regeneration. Nat Commun 14, 7167 (2023). https://doi.org/10.1038/s41467-023-42929-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-42929-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.