Abstract
Advances in cryogenic transmission electron microscopy have revolutionised the determination of many macromolecular structures at atomic or near-atomic resolution. This method is based on conventional defocused phase contrast imaging. However, it has limitations of weaker contrast for small biological molecules embedded in vitreous ice, in comparison with cryo-ptychography, which shows increased contrast. Here we report a single-particle analysis based on the use of ptychographic reconstruction data, demonstrating that three dimensional reconstructions with a wide information transfer bandwidth can be recovered by Fourier domain synthesis. Our work suggests future applications in otherwise challenging single particle analyses, including small macromolecules and heterogeneous or flexible particles. In addition structure determination in situ within cells without the requirement for protein purification and expression may be possible.
Similar content being viewed by others
Introduction
Cryo-electron microscopy1,2,3 (cryo-EM), is a powerful method for visualising a wide range of biological macromolecules at near-atomic resolution and is particularly powerful for studying the structure of biologically significant protein complexes which are often difficult to solve using X-ray or nuclear magnetic resonance (NMR) methods4. Numerous technical developments including optimized specimen preparation5,6, improved image reconstruction and processing algorithms7,8,9, new electron sources and energy filters10 and a generation of direct electron detectors have allowed the structures of many biomolecules with weights greater than 100 kDa to be solved at close to atomic resolution10, using single particle analysis (SPA)11.
However, despite these advances, SPA has been almost exclusively based on the use of phase contrast images recorded at high defocus. A major limitation of this approach is that the low-frequency information from weak-phase objects such as most macromolecules12, is attenuated and high frequency information suffers from rapid reversals in the phase contrast transfer function (PCTF). This can cause difficulties in the accurate identification, classification and alignment of particles to a common reference13. To overcome the low contrast and signal-to-noise ratio (SNR) in defocused cryo-EM images that has often precluded high resolution 3D reconstructions using the SPA method, it is therefore important to develop high-contrast phase-sensitive imaging modes. Phase plates have been successfully used to increase low-frequency contrast and SNR for in-focus images13,14,15,16. However, the routine use of phase plates is often restricted by signal attenuation at high frequencies and inconsistent fabrication giving rise to unstable phase shifts16.
An alternative strategy is based on the use of phase information recovered using electron ptychography17, which has been recently demonstrated under low dose conditions for biological samples embedded in vitreous ice. Ptychography is a lensless imaging approach originally proposed by Hoppe18 where a sample is scanned by a suitably conditioned probe in a 2D array. This provides an array of diffraction patterns recorded in the far field as a function of probe position forming a four-dimensional (4D) dataset. Using this 4D dataset, quantitative phase data with high spatial resolution can be recovered commonly using one of several iterative phase retrieval algorithms19,20. This method has been used with a variety of radiations including light, X-rays, and electrons, in wide ranging applications in both the physical and life sciences17,21,22,23,24. Using electrons, ptychography has generated considerable interest given its potential for super-resolution imaging24,25, high-contrast light-element detection26,27,28, 3D optical sectioning28,29,30 and potential coupling to spectroscopic data acquisition31. Moreover, as ptychography utilizes the full diffraction pattern, it is dose-efficient17,32,33 particularly when data is recorded using direct electron detectors34,35 which record data with high signal-to-noise at low electron dose. This approach has recently been demonstrated for micrometer wide phase reconstruction of an unstained virus-infected cell at a dose of 27 e/Å2 17.
Here we describe a standardised protocol for 3D SPA using ptychographic data recorded under cryogenic conditions (cryo-EPty SPA), and experimentally demonstrate that this can restore 3D information across a wide bandwidth of spatial frequencies by adjusting the convergence semi angle (CSA) of the electron probe. A 3D map with a wide bandwidth in the Fourier domain has been synthesised by combining data for different CSAs that shows better contrast than conventional cryo-EM SPA. We also show, using simulated data that cryo-EPty SPA has future potential for structural studies of small molecules.
Results
Protocol for ptychographic SPA
4D data collection: 4D datasets were acquired in a scanning diffraction configuration shown schematically in Fig. 1a, whereby a defocused probe with a known CSA, α was raster scanned over a cryo-sample for a field of view (FOV) that can reach micrometer scale17. The defocus value of the probe and the scanning step-size were adjusted such that the overlap ratio between each adjacent illuminated area (Fig. 1b) was maintained in the range between 95%33 and 60%36 which is essential to ensure computational convergence of the iterative algorithm used36. We note that this parameter affects the total dose32. A fast direct electron detector34,35 that was synchronised with the scanning system of the microscope was used to collect the 4D diffraction datasets (Fig. 1c). Moving from one FOV to the next was achieved by shifting the scanned illumination area, although mechanical movement of the stage could also be used.
Schematic diagrams of the optical configuration a and data acquisition for ptychography b. c Typical diffraction patterns and corresponding probe positions used for ptychography (blue). Ptychographic reconstructed amplitude d and phase e of an object function. Instances of amplitude f and phase g of particles picked from d and e, respectively. h Schematic diagram of the optical configuration for TEM data acquisition. i Movie frames collected in TEM mode (left) and translations calculated for individual frames (right). j Motion-corrected image from a movie. k CTF correction by fitting the amplitude spectrum of j. l Picked particles instances from j.
Post-acquisition scanning alignment and focusing: The probe positions and defocus values used for data collection are accurately known and considered as prior knowledge in the ptychographic reconstruction used here. However, due to unavoidable instabilities in the microscope37 such as scan distortions including jitter, stretching and warping, together with possible mechanical stage drift and defocus drift, the actual values can deviate from their nominal settings as shown in Fig. 1c (right). To overcome this a starting estimate of probe positions can be calculated using cross-correlation38 and position refinement algorithms based on conjugate gradient descent methods39, annealing algorithms40 or serial cross-correlation41 can then be included in the iterative reconstruction to eliminate these positional errors.
As the defocus value directly determines the probe size at the sample plane, an accurate initial estimate can speed up the convergence of the reconstruction, even though the probe function can ultimately be retrieved within certain reconstruction algorithms42 including that used in this work. The initial defocus value can be accurately estimated from the Ronchigram43 or by using a computational multislice algorithm28,29,30 as used here.
Iterative ptychographic reconstruction: Given optimised probe positions and defoci, the object and probe complex functions can be simultaneously retrieved from the 4D datasets using one of several alternative reconstruction methods including iterative methods such as the extended ptychographic iterative engine (ePIE)20 or difference map (DM)22 or an analytical Wigner distribution deconvolution (WDD)19. Here ePIE was used. For thick biological samples, ptychography can also retrieve 3D structural information by computational optical sectioning28,29, which has further potential for use in imaging large volumes of biological material. Viral particles have been experimentally reconstructed in 2D using ePIE17, demonstrating that both the amplitude and phase of the sample wavefunction can be recovered at low dose for cryogenic samples, as shown in Fig. 1d, e.
Particle picking: From the phase and amplitude of the specimen wavefunction, particle-picking procedures44,45,46 that have been developed for cryo-EM SPA can be directly applied to both. Using both enables a cross-check and can also provide a coordinate reference if one signal is weak. Multiple individual particles can then be sequentially picked from the phase and amplitude and formed into two positionally coordinated stacks of particle phases and amplitudes, as shown in Fig. 1f, g. For the higher CSA data (4.83 mrad), the amplitude was used to aid particle picking in the phase as shown in Supplementary Fig. 1c, f.
3D SPA reconstruction: An overall workflow as shown in Fig. 2, similar to that used in SPA can be applied to either the reconstructed phase or amplitude stacks using standard software packages such as Relion7 or EMAN8. It is important to note that contrast flipping to correct for the contrast transfer function (CTF) is not required as the ptychographic transfer function has no reversals17.
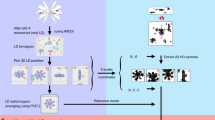
a–c 3D electron density maps for low, medium, and high CSAs respectively. d–f Corresponding bandpass filtered maps of a–c, using the selected bandpass ranges where the information transfer is strongest. g Ultrawide bandwidth map obtained by Multi-band Fourier Synthesis (h) Band-limit map reconstructed by conventional TEM-SPA.
Multi-band Fourier synthesis: The bandwidth of information transfer for ptychography can be varied in the Fourier domain by changing the CSA17. Hence, multiple SPA 3D maps of the same sample with different spatial frequency bandwidths (low, medium and high frequencies as shown in Fig. 2a–c) can be obtained for different values of α. Furthermore, by combining the spatial frequencies most strongly transferred (Fig. 2d–f) in each SPA 3D map in the Fourier domain it is possible to generate a 3D map that contains a wider bandwidth of information transfer than that of any individual map as shown in Fig. 2g. We term this procedure Multi-band Fourier Synthesis (details are given in Methods).
Cryo-ptychography with varying CSAs
Following the protocol illustrated in Fig. 1, experimental cryo-EPty SPA datasets were recorded in different areas for vitrified samples of rotavirus double-layered viral particles (DLPs) using different values of the CSA, α. Single particle reconstructed phases for α = 1.03 mrad, 3.26 mrad, and 4.83 mrad (Fig. 3a–c) (corresponding amplitude data shown in Supplementary Fig. 2) were extracted from the ptychographic reconstructions using the ePIE20 algorithm (Supplementary Fig. 1) at doses of 22.6, 24.1, 24.6 e/Å2, respectively. For comparison, a previously published cryo-EM image of rotavirus DLPs recorded at a similar dose of 15 ~ 20 e/Å2 47 is shown in Fig. 3e. At α = 1.03 mrad, the phase (Fig. 3a) shows strong low frequency transfer in the range from 0.014 to 0.116 nm−1 (8.6–72 nm) which defines the overall shape of the viral capsid proteins, consistent with our previous work17. As the CSA increased to 4.83 mrad (Fig. 3c), higher resoluion features of the viral capsid proteins are resolved.
a–c Representative experimental ptychographic phase of a virus particle with CSAs, α = 1.03, 3.26 and 4.83 mrad from sets of typically 257, 443 and 498 reconstructed particles, respectively. d Representative simulated ptychographic phase of a particle for α = 4.83 mrad from a set of typically 305 simulated particles. e Representative TEM particle image from a set of typically 378 selected particles in ref. 47. f–j 3D maps refined with 232, 318, 241, 292 and 269 particles, respectively, corresponding to the particle instances (a–e). k–o Representative VP6 trimers selected from a total of 260 VP6 trimes in the outer shell of a rotavirus DLP and p–t central slices (the 125th slice from 248 slices in z-direction) extracted from the 3D maps in (f–j), respectively. Scale bars: 25 nm (e, j, t); 2.5 nm (o).
To understand how frequency transfer is affected by α, it is necessary to examine the information transfer as a function of spatial frequency for ptychography. This determines how strongly each spatial frequency in the object is transferred, and hence the strength of the corresponding component in the final reconstruction. The transfer function calculated in previous work17,48 shows that for a given value of α the transfer function behaves as a bandpass filter that transfers components in a specified band of frequencies strongly. By varying α, this bandpass filter can be tuned to select either high or low periodicity features with no contrast reversals. This allows the reconstruction of multiple 3D maps from SPA data recorded with different values of the CSA which give rise to signal transfer in different bandwidths. These can subsequently be combined in the Fourier domain into a 3D map with a broad band of information transfer (See Methods). Details of the experimental settings for ptychographic data acquisition are provided in Supplementary Table 1 and Supplementary Text 1. Ptychographic reconstruction details are provided in Supplementary Text 2.
Multi-band SPA reconstructions
Using the SPA pipeline (Fig. 2), three 3D density maps of rotavirus DLPs (Fig. 3f–h) were reconstructed from ptychographic phase data for different values of the CSA (calculation details are given in Supplementary Table 2 and Text 3), using the Relion3.17 software package. The number of viral particles used for each map varied between 232 to 318, significantly less than that used for many conventional SPA reconstructions, although we note that icosahedral symmetry was imposed on the data. The low-angle 3D maps reconstructed from data at α = 1.03 mrad (Fig. 3f) and 3.26 mrad (Fig. 3g) clearly show the overall features of the outer VP6 capsid layer including 260 VP6 trimers (a typical VP6 trimer at 1.03 mrad and 3.26 mrad is shown in Fig. 3k and Fig. 3l). These are arranged in a T = 13 icosahedral lattice47, with the channels surrounded by the trimers. For α = 4.83 mrad, higher spatial frequency information is transferred17 such that finer details of the outer VP6 layer are visible in the 3D map (Fig. 3h). The central channels and three individual monomers can be directly visualised in the 3D volume of a trimer (Fig. 3m) extracted from the 3D map (Fig. 3h). It is also evident that the VP6 trimers are broader at the base than at the tip, and that each VP6 monomer follows an approximately helical path from the base inside the DLP to the tip of the trimer (Fig. 3m). These results are consistent with those from the cryo-EM SPA 3D map (Fig. 3j, o) and is further confirmed by line-profiles (Supplementary Fig. 3) extracted across a single VP6 trimer in the central slices of the 3D maps (Fig. 3p–t). The sharp valley in the blue line (Supplementary Fig. 3) indicates that the resolution at α=4.83 mrad is sufficient to resolve the channel in the trimer.
The resolution of the 3D maps (Fig. 3f–h, j) are estimated as 3.72 nm, 3.29 nm, 1.86 nm, and 2.09 nm, respectively, using a Fourier Shell Correlation49 (FSC = 0.143, gold-standard). For simplicity, procedures50 to correct the effects of the CTF (for the TEM data) and MTF were not implemented in these reconstructions and we note that phase flipping is not required for the ptychographic phase. The 3D map calculated from the TEM data shows an oscillation in the FSC curve with a deep valley close to zero at 0.5 nm−1 (Fig. 4a), which is attributed to the zero-crossings and contrast reversal in the TEM CTF. These could however, be compensated in the 3D maps by implementation of CTF correction11,51, use of a wide range of defocus values52 and collection of data from a sufficiently large number of particles with varying defocus values. However, the FSC curve of the 3D maps using cryo-EPty SPA shows a gradual decrease with spatial frequency with no oscillations, due to the continuous positive nature of the ptychographic transfer function over its entire spatial frequency range17.
a Fourier shell correlation curves of the 3D maps in Fig. 3f–j. b Radially averaged amplitude spectra calculated from the 3D maps in Fig. 3f–h, j and Fig. 4(c). For each spectrum for a particular CSA value a bold line segment indicates the selected bandwidth within which the information transfer is strongest at that value of the CSA. For α = 1.03 mrad, the low-frequency bandwidth from 0~0.16 nm−1 (area 1 below the bold line segment in black) is selected. For α = 3.26 and 4.83 mrad, the selected bandwidths are at medium frequency from 0.17~0.23 nm−1 (area 2 below the bold segment in red) and high frequency from 0.23~0.76 nm−1 (area 3 below the bold segment in blue), respectively. c Ultrawide bandwidth 3D map obtained by multi-band Fourier synthesis from the 3D maps in Fig. 3f–h with α = 1.03 mrad (232 particles), 3.26 mrad (318 particles) and 4.83 mrad (241 particles), respectively. Scale bars: 25 nm (c).
To validate this interpretation of our experimental results, cryo-EPty SPA simulations using the multislice method53,54 were carried out for a Rotavirus DLP model (3KZ4) using a software package described previously55 (for details of the calculations see Supplementary Table 3 and Text 4). When imaging frozen hydrated biological samples, a major resolution and contrast limiting factor is image blurring due to beam induced sample motion56,57, which in many instances cannot be prevented58. To model these effects, a range of empirical isotropic motion factors from 0-0.5 nm57 were included in the ptychographic simulations (see Supplementary Text 4). The simulated results (Supplementary Fig. 4) clearly show that the resolution (Supplementary Table 4) of the 3D maps for different CSAs were degraded as expected with increased motion factors.
For comparison with the experiment results (Fig. 3f–h), simulations (Supplementary Fig. 4) with various motion factors were calculated and the degree of fit between simulation and experiment was evaluated by normalized cross-correlation using the CHIMERA59 software (details are described in Supplementary Text 5). This fitting suggests that the motion factors present in the experimental data are in the range of 0.3–0.7 nm57 for the current experiments (Supplementary Fig. 5). Using α = 4.83 mrad as an example (Fig. 3i and Supplementary Fig. 6c), the VP6 trimers (Fig. 3n) extracted from the 3D map with an average motion factor of 0.5 ± 0.1 nm show similar features to those present in the experimental reconstruction (Fig. 3m). Line profiles (Supplementary Fig. 3) extracted from the central slices of the 3D map also show agreement between simulation and experiment. Furthermore, the FSC curves (Fig. 4a) indicate that the resolution of the simulated and experimental reconstructions are almost identical (1.83 nm vs 1.86 nm). Therefore, we conclude that the loss of resolution in the experiment with α = 4.83 mrad is equivalent to the effect of a 0.5 ± 0.1 nm beam-induced motion during acquisition as shown in Supplementary Table 4.
In conventional cryo-EM, the effects of beam-induced motion can be reduced or eliminated by optimizing the hole size of the perforated carbon films used for cryo-sample preparation57 or by using UltrAufoil grids60 or HexAu grids61 and recording movies of TEM images followed by drift correction to restore high-resolution information62 (as shown in Fig. 1i). Similarly, in ptychography, multi-frame diffraction patterns32 could be collected at each scanning position (Fig. 1c) followed by alignment of the patterns for motion correction. In future this approach will allow cryo-EPty SPA to achieve higher resolution.
To further explore the full potential of cryo-EPty SPA, a simulation with α = 15 mrad was calculated for a model of apoferritin (PDB-7A6A) without beam-induced motion. Supplementary Fig. 7a shows the resultant ptychographic phase of apoferritin and the corresponding 2D classifications, from which a 3D density map of apoferritin (Supplementary Fig. 7b) was reconstructed at a global resolution of 0.22 nm (Supplementary Fig. 7c) from 2826 particles at dose of 73.24 e/Å2, for a B-factor of −0.655556 nm2. An enlarged image (Supplementary Fig. 7d) extracted from the 3D map reveals backbone carbonyl groups, and distinct side-chain structural details including aromatic rings. This indicates that cryo-EPty SPA is capable of solving atomic structures of proteins and their ligand-bound complexes if larger values of α are used with sample motion correction and we note that the values of α required are easily obtained with Cs corrected optics.
Wide bandwidth 3D map synthesis
Radial averages of 3D amplitude spectra were calculated (Fig. 4b) to directly compare the 3D information transfer as a function of spatial frequency for various values of the CSA. By tracing the intensity envelope of the amplitude spectra for the three values of the CSA used, the strongest information transfer bandwidth can be identified for each. For example, the 3D map for α = 1.03 mrad has strongest transfer in a low-frequency bandwidth (area 1 below the bold line segment in black as shown in Fig. 4b). The bandwidths with the strongest information transfer at different values of the CSA are identified in Fig. 4b and their ranges are given in Supplementary Table 5. Using this information, the 3D maps from datasets with different values of the CSA were then band-pass filtered using the corresponding selected bandwidths, giving band-pass filtered 3D maps (Supplementary Fig. 8), each of which, individually gives the best information transfer within a given spatial frequency bandwidth. In summary our approach is to choose, for any given spatial frequency, the data from the CSA that has the strongest signal for that frequency and to discard the weaker signals from other CSA data. Although this is not an optimally efficient reconstruction strategy since some data from any one experiment is discarded, for the case of SPA reconstruction adding additional data does not increase the dose of any one particle and each dataset is recorded from a different set of particles.
By combining these filtered 3D maps (Supplementary Fig. 8), a wide bandwidth 3D map (Fig. 4c) can be synthesised using the procedure illustrated in Fig. 2. Compared to the 3D maps using a dataset with a single value of α (Fig. 3f–h), the wide bandwidth 3D map (Fig. 4c) contains strong information transfer across a wider range of spatial frequencies (shaded beige in Fig. 4b) and hence provides improved transfer of both low and high frequency information (Supplementary Table 5). This conclusion is further supported by simulations (Supplementary Fig. 9).
Discussion and conclusions
We have demonstrated cryo-EPty SPA for reconstruction of rotavirus particles both experimentally and with simulations using the protocol shown in Figs. 1 and 2. It is therefore useful to discuss the differences in SPA reconstructions using different input data, specifically comparing the use of defocused phase contrast TEM images and reconstructed ptychographic phases.
In terms of the respective optical configurations, the major difference is that TEM uses static plane wave illumination (Fig. 1h), whereas a convergent beam is scanned across the sample during ptychographic data acquisition (Fig. 1a). Moreover as ptychography records diffraction patterns, the objective lens aberrations are not important in determining the information transfer. Ultimately, for this reason, ptychography can theoretically reach a resolution set by the diffraction limit as has been demonstrated for radiation resistant samples24,63.
For TEM data (Fig. 1i), the FOV and the pixel sampling in an image must be balanced due to the finite pixel size and array size of imaging detectors (typically a maximum of ca. 4096 × 4096 pixels) and consequently there is a trade-off between resolution and FOV (e.g. a sampling of 0.1 nm per pixel leads to approximately a 400 nm FOV for a typical 4k × 4k detector). For a given detector size, finer sampling must be used, at higher magnifications, resulting in a reduction in the number of particles imaged in each frame for a smaller FOV64. In contrast, for ptychography the sampling of the image is limited by the highest angle collected by the detector. Therefore, even when using smaller detectors (256 × 256 Merlin Medipix334 and 128 × 128 EMPAD35) ptychography can still obtain an unlimited FOV at high resolution if sufficient probe positions are recorded.
Cryo-EM SPA can be used to fine tune the low order aberrations before image acquisition (Fig. 1h), usually in an adjacent area to the target area. In contrast ptychographic reconstruction algorithms can reconstruct both probe and object functions20 and compensate for residual low order aberrations after data acquisition28,29. This is particularly important at low dose where accurate determnination of the aberrations during acquisition is challenging due to a low SNR.
The ptychographic transfer function leads to reconstructions that are free of contrast reversals over the entire spatial frequency range recovered. As a result, there is no need for phase reversal correction during SPA reconstruction as is routinely used in cryo-EM SPA (Fig. 1j–k). Furthermore, as we have shown, the ptychographic transfer function can also be adjusted by changing the CSA of the probe17, giving multiple reconstructions with different optimal contrast transfer bandwidths. Subsequently, using a multi-band Fourier synthesis as described here, a 3D map with high signal transfer is obtained over a wide spatial frequency range. Similarly, TEM SPA can also combine multiple defocus values to provide sufficient contrast for selection (high defocus) and sufficient resolution (low defocus). Although, as already described some data is discarded in the process described in the case of SPA reconstructions this is acceptable as more images can be added without increasing the dose of any one particle.
Furthermore, we emphasise that the complex wavefunction (both phase and amplitude) of an object is recovered using ptychography, which provides additional information for particle picking or model initialization in SPA. Finally, ptychography using an inverse multi-slice method29 can computationally section z-slices of interest from thicker biological samples, where the projection approximation breaks down as a consequence of significant multiple scattering within the sample.
However, there are still experimental challenges, mainly related to beam-induced motion which limits the achievable resolution for ptychographic SPA reconstruction of biological samples embedded in vitrified ice. The first is to eliminate or reduce the sample drift. Considering the various possible mechanisms of beam-induced motion, such as a build-up of charge65, radiolysis of the specimen and mechanical stress in the ice layer and carbon support66,67, methods have been proposed to overcome these and to mitigate the effects of motion57. An alternative is to implement drift correction in the ptychographic data acquisition. For this, a movie of diffraction patterns needs to be recorded at each position and a drift-correction algorithm, adapted to ptychographic iterative reconstruction will be required to reduce the effect of the motion blur. The third possibility is to outrun the sample motion by recording data at high spatio-temporal resolution. For example, an ultra-fast detector with a much high frame rate, such as 10,000 fps68 (1000 fps used in our work) has been developed. This together with a pulsed electron source, potentially allows the data acquisition to outrun the sample motion and avoiding blurring effects69.
In summary, we have described a workflow for high contrast, wide spatial frequency-band SPA reconstructions which could be extended to higher resolution. With improved microscope stability, improved reconstruction algorithms (machine learning) specifically optimized for low signal data70, ultrafast detectors, and pulsed electron sources, ptychography has potential application for structure determination with some advantages over the use of conventional phase contrast images.
Methods
Samples
A suspension of Rotavirus DLPs (strain SA11) was prepared using a previously described method17. The DLPs/AuNPs suspension (4 μl) was placed onto holey carbon EM grids (Quantifoil). Each grid was blotted for 5 s and subsequently plunged into a liquid ethane/propane mixture cooled by liquid nitrogen at 80% humidity. The grids were then transferred and stored under liquid nitrogen. Further details of the sample preparation are provided in Supplementary Text 1.
Cryo-ptychography experiment setup
The cryo-EPty experiments were performed in a scanning diffraction mode on a JEOL ARM 300CF operated at 300 kV with data recorded on a pixelated Merlin Medipix3 detector34. The vitrified EM samples were transferred into a Gatan 698 Elsa cryo-transfer holder to ensure that the specimen temperature was maintained at −176 ± 2 °C. Three probe CSAs of 1.03 mrad, 3.26 mrad, and 4.83 mrad were used to record ptychographic datasets. Detailed additional experimental parameters for these values of the CSA are given in Supplementary Table 1 and Supplementary Text 1.
ePIE reconstructions
Ptychographic reconstructions were carried out using the ePIE20 algorithm run for 100 iterations. Detailed reconstruction parameters for each value of α are given in Supplementary Table 1 and Supplementary Text 2. The degree of redundancy was estimated as 217, 336, and 155 for the datasets at α values of 1.03, 3.26 and 4.83 mrad, respectively. Details of the redundancy calculation used are provided in Supplementary Text 6. The computational time cost for ptychographic data collection and processing is described in Supplementary Text 7.
Single particle reconstruction
Using the e2box.py program in EMAN2.28, 257, 443, 498, and 378 particles were picked for α = 1.03 mrad, 3.26 mrad, and 4.83 mrad and TEM, respectively. Subsequently, the 3D reconstruction workflow as described was implemented using the software package Relion 3.17. Further details are provided in Supplementary Text 3.
Ptychographic simulations
Multislice simulations of cryo-EPty SPA reconstructions using models of rotavirus DLPs and apoferritin used were carried out using the multislice algorithm53,54 implemented in the Matlab code InSilicoTEM55. The atomic potential maps of rotavirus DLPs and apoferritin were built from the 3KZ4 and 7A6A models in the protein data bank. Detailed simulation parameters and related information for various CSA values are given in Supplementary Table 3 and Supplementary Text 4.
Multi-band SPA reconstructions
We define \({\{{\alpha }_{i}\}}_{i=1}^{n}=\{{\alpha }_{1},{\alpha }_{2},{\alpha }_{3},\ldots,{\alpha }_{n}\}\) as a set of n different values of α for which a corresponding set of 3D electron density maps \(\{{V}_{{\alpha }_{i}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }})\}_{i=1}^{n}=\{{V}_{{\alpha }_{1}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }}),\,{V}_{{\alpha }_{2}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }}),{V}_{{\alpha }_{3}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }}),\ldots,{V}_{{\alpha }_{n}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }})\}\) were reconstructed, with \(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }}\) a 3D vector in real space. The amplitude spectra in 3D are given as the modulus of the Fourier transforms of the 3D electron density maps \({W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})=ft({V}_{{\alpha }_{i}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }}))\) forming a set of \(\{{W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\}_{i=1}^{n}=\{{W}_{{\alpha }_{1}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}),\,{W}_{{\alpha }_{2}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}),{W}_{{\alpha }_{3}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}),\ldots,\,{W}_{{\alpha }_{n}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\}\), with \(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}\) a 3D vector in Fourier space.
To evaluate the information transfer of \({W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\), we calculate one-dimensional radially averaged amplitude spectra \({l}_{{\alpha }_{i}}(q)\) for each αi as a function of a radius, q in 3D Fourier space as shown in Fig. 4b. The amplitude spectra are expressed as:
where \({\sum }_{q-\frac{b}{2} < |\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}|\le q+\frac{b}{2}}|{W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})|\) is the sum of the modulus of \({W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\) with a bin width, b at a radius, q and \({\sum }_{q-\frac{b}{2} < |\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}|\le q+\frac{b}{2}}1\) is the total number of voxels within this bin width. From this, a set of radially averaged amplitude spectra \(\{{l}_{{\alpha }_{i}}(q)\}_{i=1}^{n}=\{{l}_{{\alpha }_{1}}\left(q\right),{l}_{{\alpha }_{2}}\left(q\right),{l}_{{\alpha }_{3}}\left(q\right),\ldots,{l}_{{\alpha }_{n}}\left(q\right)\}\) can be obtained.
Each spectrum corresponding to a particular value, αi has a bandwidth \({B}_{{\alpha }_{i}}\) (a segment in q space as shown in Fig. 4b), within which the strength of the information transfer \({l}_{{\alpha }_{i}}(q)\) is strongest. The bandwidth for αi is given as:
which forms a set of bandwidths \({\{{B}_{{\alpha }_{i}}\}}_{i=1}^{n}=\{{B}_{{\alpha }_{1}},\,{B}_{{\alpha }_{2}},\,{B}_{{\alpha }_{3}},\ldots,\,{B}_{{\alpha }_{n}}\}\).
To carry out the multi-band Fourier synthesis, the signals most strongly transferred in each element of the Fourier amplitude spectrum set \(\{{W}_{{\alpha }_{i}}(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\}_{i=1}^{n}\) are assigned to \({W}_{\max }(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\) subject to the condition that \(|\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }}|\) is within the corresponding element of the bandwidth set \({\{{B}_{{\alpha }_{i}}\}}_{i=1}^{n}\) given by:
The multi-band Fourier synthesis 3D map \({V}_{\max }(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }})\) can then be generated by 3D inverse Fourier transforming \({W}_{\max }(\mathop{{{{\bf{q}}}}}\limits^{{\rightharpoonup }})\) to give:
which contains a much wider bandwidth and stronger information transfer than those of any individual element in the 3D map set \(\{{V}_{{\alpha }_{i}}(\mathop{{{{\bf{r}}}}}\limits^{{\rightharpoonup }})\}_{i=1}^{n}\).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. The raw data (diffraction datasets) in this study have been deposited in the Zenodo database [https://doi.org/10.5281/zenodo.7701710]. The cryo-EPty maps have been deposited in the Electron Microscopy Data Base (EMDB) under accession codes EMD-35828 (CSA 1.03 mrad), EMD-35916 (CSA 3.26 mrad) and EMD-35917 (CSA 4.83 mrad). The reconstructed phase image data are available as EMPIAR datasets under accession codes EMPIAR-11500 (CSA 1.03 mrad), EMPIAR-11503 (CSA 3.26 mrad), and EMPIAR-11504 (CSA 4.83 mrad). Previously published structural models can be accessed using PDB accession codes 3KZ4 and 7A6A.
Code availability
Distributions of ePIE code are subject to an agreed licence agreement with Phase Focus Ltd. and the authors of this paper are not permitted to distribute the ePIE code used under the terms of that agreement. The EMAN2 code is available as open source under a General Public License (GPL) version v2.0 from the authors [https://github.com/cryoem/eman2]. The Relion3 code is available as open source under a GNU General Public licence v2.0 [https://github.com/3dem/relion].
References
Dubochet, J., Adrian, M., Chang, J.-J., Lepault, J. & McDowall, A. W. in Cryotechniques in Biological Electron Microscopy (eds R. A. Steinbrecht & K. Zierold) 114–131 (Springer Berlin Heidelberg, 1987).
Frank, J. In Single-Particle Cryo-Electron Microscopy Vol. Volume 10 Series in Structural Biology 69–72 (World Scientific, 2017).
Henderson, R. & Unwin, P. N. T. Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975).
Cheng, Y. Membrane protein structural biology in the era of single particle cryo-EM. Curr. Opin. Struct. Biol. 52, 58–63 (2018).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
Glaeser, R. M. Proteins, interfaces, and cryo-EM grids. Curr. Opin. Colloid Interface Sci. 34, 1–8 (2018).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Yip, K. M., Fischer, N., Paknia, E., Chari, A. & Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 (2020).
Cheng, Y., Grigorieff, N., Penczek, Pawel, A. & Walz, T. A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Danev, R. & Nagayama, K. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy 88, 243–252 (2001).
Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl Acad. Sci. USA 111, 15635–15640 (2014).
Danev, R. & Baumeister, W. Cryo-EM single particle analysis with the Volta phase plate. eLife 5, e13046 (2016).
Danev, R. & Baumeister, W. Expanding the boundaries of cryo-EM with phase plates. Curr. Opin. Struct. Biol. 46, 87–94 (2017).
Zhou, L. et al. Low-dose phase retrieval of biological specimens using cryo-electron ptychography. Nat. Commun. 11, 2773 (2020).
Hoppe, W. Diffraction in inhomogeneous primary wave fields 1. Principle of phase determination from electron diffraction interference. Acta Crystallogr. A-Crystal Phys. Diffraction Theor. Gen. Crystallogr. A 25, 495–501 (1969).
Rodenburg, J. & Bates, R. The theory of super-resolution electron microscopy via Wigner-distribution deconvolution. Phil. Trans. R. Soc. Lond. A 339, 521–553 (1992).
Maiden, A. M. & Rodenburg, J. M. An improved ptychographical phase retrieval algorithm for diffractive imaging. Ultramicroscopy 109, 1256–1262 (2009).
Rodenburg, J. M. et al. Hard-X-ray lensless imaging of extended objects. Phys. Rev. Lett. 98, 034801 (2007).
Thibault, P. et al. High-resolution scanning X-ray diffraction microscopy. Science 321, 379 (2008).
Dierolf, M. et al. Ptychographic X-ray computed tomography at the nanoscale. Nature 467, 436–439 (2010).
Jiang, Y. et al. Electron ptychography of 2D materials to deep sub-angstrom resolution. Nature 559, 343–349 (2018).
Nellist, P. D., McCallum, B. C. & Rodenburg, J. M. Resolution beyond the ‘information limit’ in transmission electron microscopy. Nature 374, 630–632 (1995).
Wang, P., Zhang, F., Gao, S., Zhang, M. & Kirkland, A. I. Electron ptychographic diffractive imaging of boron atoms in LaB6 crystals. Sci. Rep. 7, 2857 (2017).
Yang, H. et al. Electron ptychographic phase imaging of light elements in crystalline materials using Wigner distribution deconvolution. Ultramicroscopy 180, 173–179 (2017).
Yang, H. et al. Simultaneous atomic-resolution electron ptychography and Z-contrast imaging of light and heavy elements in complex nanostructures. Nat. Commun. 7, 12532 (2016).
Gao, S. et al. Electron ptychographic microscopy for three-dimensional imaging. Nat. Commun. 8, 163 (2017).
Chen, Z. et al. Electron ptychography achieves atomic-resolution limits set by lattice vibrations. Science 372, 826–831 (2021).
Song, B. et al. Hollow electron ptychographic diffractive imaging. Phys. Rev. Lett. 121, 146101 (2018).
Song, J. et al. Atomic resolution defocused electron ptychography at low dose with a fast, direct electron detector. Sci. Rep. 9, 3919 (2019).
Chen, Z. et al. Mixed-state electron ptychography enables sub-angstrom resolution imaging with picometer precision at low dose. Nat. Commun. 11, 2994 (2020).
Ballabriga, R. et al. The Medipix3RX: a high resolution, zero dead-time pixel detector readout chip allowing spectroscopic imaging. J. Instrum. 8, C02016 (2013).
Tate, M. W. et al. High dynamic range pixel array detector for scanning transmission electron microscopy. Microsc. Microanal. 22, 237–249 (2016).
Bunk, O. et al. Influence of the overlap parameter on the convergence of the ptychographical iterative engine. Ultramicroscopy 108, 481–487 (2008).
Braidy, N., Le Bouar, Y., Lazar, S. & Ricolleau, C. Correcting scanning instabilities from images of periodic structures. Ultramicroscopy 118, 67–76 (2012).
Hurst, A. C., Edo, T. B., Walther, T., Sweeney, F. & Rodenburg, J. M. Probe position recovery for ptychographical imaging. J. Phys.: Conference Series 241, 012004 (2010).
Tripathi, A., McNulty, I. & Shpyrko, O. G. Ptychographic overlap constraint errors and the limits of their numerical recovery using conjugate gradient descent methods. Opt. Express 22, 1452–1466 (2014).
Maiden, A. M., Humphry, M. J., Sarahan, M. C., Kraus, B. & Rodenburg, J. M. An annealing algorithm to correct positioning errors in ptychography. Ultramicroscopy 120, 64–72 (2012).
Zhang, F. et al. Translation position determination in ptychographic coherent diffraction imaging. Opt. Express 21, 13592–13606 (2013).
Hue, F., Rodenburg, J. M., Maiden, A. M. & Midgley, P. A. Extended ptychography in the transmission electron microscope: possibilities and limitations. Ultramicroscopy 111, 1117–1123 (2011).
Lupini, A. R., Wang, P., Nellist, P. D., Kirkland, A. I. & Pennycook, S. J. Aberration measurement using the Ronchigram contrast transfer function. Ultramicroscopy 110, 891–898 (2010).
Sigworth, F. J. Classical detection theory and the cryo-EM particle selection problem. J. Struct. Biol. 145, 111–122 (2004).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Bell, J. M., Chen, M., Durmaz, T., Fluty, A. C. & Ludtke, S. J. New software tools in EMAN2 inspired by EMDatabank map challenge. J. Struct. Biol. 204, 283–290 (2018).
Zhang, X. et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl Acad. Sci. USA 105, 1867 (2008).
O’Leary, C. M. et al. Contrast transfer and noise considerations in focused-probe electron ptychography. Ultramicroscopy 221, 113189 (2021).
Van Heel, M. & Schatz, M. Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 (2005).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Danev, R., Iijima, H., Matsuzaki, M. & Motoki, S. Fast and accurate defocus modulation for improved tunability of cryo-EM experiments. IUCrJ 7, 566–574 (2020).
Cowley, J. M. & Moodie, A. F. The scattering of electrons by atoms and crystals. I. A new theoretical approach. Acta Crystallogr 10, 609–619 (1957).
Goodman, P. & Moodie, A. F. Numerical evaluations of N-beam wave functions in electron scattering by the multi-slice method. Acta Crystallogr. A 30, 280–290 (1974).
Vulovic, M. et al. Image formation modeling in cryo-electron microscopy. J. Struct. Biol. 183, 19–32 (2013).
Henderson, R. & Glaeser, R. M. Quantitative analysis of image contrast in electron micrographs of beam-sensitive crystals. Ultramicroscopy 16, 139–150 (1985).
Brilot, A. F. et al. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 177, 630–637 (2012).
Glaeser, R. M., McMullan, G., Faruqi, A. R. & Henderson, R. Images of paraffin monolayer crystals with perfect contrast: minimization of beam-induced specimen motion. Ultramicroscopy 111, 90–100 (2011).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Russo, C. J. & Passmore, L. A. Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Naydenova, K., Jia, P. & Russo, C. J. Cryo-EM with sub–1 Å specimen movement. Science 370, 223–226 (2020).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Rodenburg, J. M. In Advances in Imaging and Electron Physics Vol. 150 (ed Hawkes) 87–184 (Elsevier, 2008).
Feathers, J. R., Spoth, K. A. & Fromme, J. C. Experimental evaluation of super-resolution imaging and magnification choice in single-particle cryo-EM. J. Struct. Biol.: X 5, 100047 (2021).
Brink, J., Sherman, M. B., Berriman, J. & Chiu, W. Evaluation of charging on macromolecules in electron cryomicroscopy. Ultramicroscopy 72, 41–52 (1998).
Glaeser, R. M. Retrospective: radiation damage and its associated “information limitations”. J. Struct. Biol. 163, 271–276 (2008).
Glaeser, R. M. & Taylor, K. A. Radiation damage relative to transmission electron microscopy of biological specimens at low temperature: a review. J. Microsc 112, 127–138 (1978).
Philipp, H. T. et al. Very-High Dynamic Range, 10,000 Frames/Second pixel array detector for electron microscopy. Microsc. Microanal. 28, 425–440 (2022).
Evans, J. E. & Browning, N. D. Enabling direct nanoscale observations of biological reactions with dynamic TEM. Microscopy (Oxf) 62, 147–156 (2013).
Pelz, P. M., Qiu, W. X., Bücker, R., Kassier, G. & Miller, R. J. D. Low-dose cryo electron ptychography via non-convex Bayesian optimization. Sci. Rep. 7, 9883 (2017).
Acknowledgements
P.W. acknowledges funding from the National Natural Science Foundation of China (11874199) and the University of Warwick Research Development Fund (RDF) 2021-22 Science Development Award. L.Q.Z. acknowledges the fellowship of China National Postdoctoral Program for Innovative Talents (No. BX2021119) and the Project funded by China Postdoctoral Science Foundation (No. 2022M711564). C.H., J.S.K., E.L., and A.I.K. acknowledge The Rosalind Franklin Institute, funded by the UK Research and Innovation, Engineering and Physical Sciences Research Council. D.I.S. is supported by the UK Medical Research Council (MR/N00065X/1). P.Z. is supported by the Wellcome Trust (206422/Z/17/Z) and the European Research Council Advanced Grant (101021133). We thank Diamond Light Source for access and support in the use of the electron Physical Science Imaging Centre (Instrument E02, EM17918) that contributed to the results presented. For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence (where permitted by UKRI, ‘Open Government Licence’ or ‘Creative Commons Attribution No-derivatives (CC BY-ND) licence’ may be stated instead) to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
P.W. and A.I.K. conceived the overall project. X.D.P. and L.Q.Z. carried out the ptychographic reconstructions, multi-band 3D SPA analysis and relevant simulations supervised by P.W. P.W., L.Q.Z., C.H., J.S.K., and E.L. acquired the ptychographical data. T.S. provided technical assistance with the data acquisition. M.B., D.I.S., and P.Z. prepared the various cryo-samples and provided cryo-EM biological structural analysis. X.D.P., L.Q.Z., C.H., J.S.K., E.L., Y.M.H., P.D.N., A.I.K., and P.W. discussed ptychographical data analysis. P.W., X.D.P., L.Q.Z., J.S.K., and A.I.K. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Richard Leapman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pei, X., Zhou, L., Huang, C. et al. Cryogenic electron ptychographic single particle analysis with wide bandwidth information transfer. Nat Commun 14, 3027 (2023). https://doi.org/10.1038/s41467-023-38268-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-38268-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.