Abstract
Trace metals have been an important ingredient for life throughout Earth’s history. Here, we describe the genome-guided cultivation of a member of the elusive archaeal lineage Caldarchaeales (syn. Aigarchaeota), Wolframiiraptor gerlachensis, and its growth dependence on tungsten. A metagenome-assembled genome (MAG) of W. gerlachensis encodes putative tungsten membrane transport systems, as well as pathways for anaerobic oxidation of sugars probably mediated by tungsten-dependent ferredoxin oxidoreductases that are expressed during growth. Catalyzed reporter deposition-fluorescence in-situ hybridization (CARD-FISH) and nanoscale secondary ion mass spectrometry (nanoSIMS) show that W. gerlachensis preferentially assimilates xylose. Phylogenetic analyses of 78 high-quality Wolframiiraptoraceae MAGs from terrestrial and marine hydrothermal systems suggest that tungsten-associated enzymes were present in the last common ancestor of extant Wolframiiraptoraceae. Our observations imply a crucial role for tungsten-dependent metabolism in the origin and evolution of this lineage, and hint at a relic metabolic dependence on this trace metal in early anaerobic thermophiles.
Similar content being viewed by others
Introduction
Evidence is mounting that some of the most ancient life forms inhabited terrestrial hot springs present on the anoxic early Earth1,2,3,4. Such habitats could have mobilized and concentrated nutrients including trace metals essential to thermophiles, but that differ from the metals essential to extant aerobic mesophiles4. Contemporary terrestrial hot springs are reasonable analogs for those on early Earth, often exhibiting low reduction potentials and sulfur-rich anoxic conditions5. In the presence of sulfide, several trace metals, including molybdenum (Mo), copper (Cu), nickel (Ni), and cobalt (Co) react with sulfide and precipitate as solid phases6, rendering them inaccessible to microbial cells. This, in turn, could have represented a strong selective pressure to evolve metalloenzymes that were dependent on less thiophilic trace metals, such as manganese (Mn) and tungsten (W).
In modern hot springs, most microbial lineages lack a cultured representative7,8, making the assessment of nutrient requirements difficult. Although cultivation-independent “omics” techniques hold tremendous promise in assessing the functional potential and activity of communities in their natural state, results from such studies are limited by what is known of characterized proteins from cultured organisms. Yet, for uncultivated lineages, many of the proteins are poorly annotated, often leading to an incomplete picture of their ecology and physiology. As such, there is a great need to bring these organisms into the culture so that the traits that permit the habitation of terrestrial hot springs, both today and in the geological past, can be examined.
Candidatus Caldarchaeales (syn. Aigarchaeota) is a yet-uncultivated lineage detected in high-temperature environments that was first described based on a genomic assembly from a thermal stream in a subsurface gold mine in Japan9,10. Although originally designated as candidate phylum Aigarchaeota, this lineage has been reassigned to the order Ca. Caldarchaeales within the phylum Thermoproteota and class Nitrososphaeria11. For simplicity, we use this latter taxonomy without the Candidatus designation for uncultivated taxa. Caldarchaeales are commonly found in hydrothermal and geothermal ecosystems8, where they can be abundant12,13,14. Metagenomics and 16S rRNA gene amplicon surveys have previously shown a high abundance of Caldarchaeales in sediments of Great Boiling Spring (GBS, Nevada, USA)15. In particular, members of the putative genus “Aigarchaeota Group 4” (AigG4)8 were abundant in in-situ GBS enrichments cultivated on lignocellulosic substrates such as corn stover16,17. AigG4 species from GBS and from hot springs in Tengchong, China (GMQ_bin_10 and JZ_bin_10 in ref. 12) have been predicted to be anaerobic heterotrophs that degrade polysaccharides and peptides13,14. However, these predictions have yet to be experimentally validated and other physiological traits of AigG4 remain unknown.
Here, we report the enrichment and stable laboratory growth of an AigG4 species from GBS we designate Wolframiiraptor gerlachensis. After initial unsuccessful attempts to grow the species without amending growth media with spring water, analysis of a metagenome-assembled genome (MAG) of W. gerlachensis suggested possible dependence on the biologically rare trace metal W, as evidenced by six annotated W-dependent ferredoxin oxidoreductases that could play central roles in anaerobic carbohydrate degradation. Indeed, stable cultivation of W. gerlachensis in a synthetic medium containing corn stover or a monosaccharide mix required the addition of W. NanoSIMS analysis demonstrated that xylose is the carbohydrate monomer preferred by W. gerlachensis in in-situ corn stover-degrading enrichments. Based on these experimental results and the genomic potential of the MAG, we propose that W is essential for carbohydrate metabolism and plays a critical role in one or more of the annotated W-dependent ferredoxin oxidoreductases, which could replenish reducing equivalents for a type-4 [NiFe] membrane-bound hydrogenase, producing a proton motive force. Using environmental metagenomes from three continents and a marine hydrothermal vent, we present evidence that both putative tungstate (WO42–) transporters and tungstoenzymes were ancestral in this lineage, herein designated the family Wolframiiraptoraceae, and trace their evolution among 78 high-quality MAGs representing four genera and 11 species. While it is known that W is important for several archaeal groups18,19,20,21, including methanogens, representatives of the Thermococcales and Thermoproteales are the only known archaea that require W for growth18. Our investigation is unique in showing long-term W-dependent growth dynamics in a mixed culture, identifying W-requiring archaea outside of the Methanobacteriota (formerly Euryarchaeota), and demonstrating large-scale expansions of W-dependent enzymes in a microbial lineage.
Results
A stable enrichment culture of an anaerobic member of the Caldarchaeales
Because W. gerlachensis was previously enriched on corn stover incubated in GBS17, we deployed new corn stover in-situ enrichments and incubated them for 6 months (Supplementary Fig. 1). The water above the incubation site was 86.5 °C, with a pH of 7.31 at the time the enrichment was deployed. After incubation, W. gerlachensis was present in the in-situ enrichment at ~3% of the total community based on 16S rRNA gene tag analysis (Fig. 1a), similar to previous results17, along with other taxa previously observed in enrichments or GBS sediments16,17. Based on the reproducible enrichment of W. gerlachensis on corn stover, and the genome-predicted anaerobic lifestyle13,14, we used the in-situ enrichment to inoculate 13 different anaerobic media that contained corn stover and/or xyloglucan and keratin as carbon sources. Some enrichments also contained previously sampled GBS spring water that was filtered (0.2 µm) and autoclaved at a 1:1 ratio with synthetic medium (Supplementary Table 1) to test if unknown components of the spring water were required for growth. The abundance of W. gerlachensis under culture conditions without added spring water dropped below the level of detection. However, W. gerlachensis was maintained after multiple transfers in two cultures with added spring water (A8 and A10) at concentrations of ~0.25–1.0 × 106 16S rRNA gene copies mL–1 culture, with a relative abundance of ~0.5–7% (Fig. 1b).
a Taxonomic composition of a sediment in-situ enrichment from Great Boiling Spring inferred from 16S rRNA gene tag sequencing. b Wolframiiraptor (W. gerlachensis) and total microbial abundances within enrichment cultures, as determined by qPCR. The absolute abundance and proportion of W. gerlachensis was stable after three transfers. Curves marked by dark coloration represent 16S rRNA gene quantities of the total community and curves marked by light coloration show 16S rRNA gene quantities belonging to W. gerlachensis in cultures A8 and A10 (all other cultures have no colored fields). Source data are provided as a Source Data file.
The two culture conditions that maintained stable populations of W. gerlachensis contained GBS spring water but were otherwise identical to synthetic media enrichments where W. gerlachensis was not maintained (compare A10 with A1, and A8 with A3; Supplementary Table 1). Additional experiments determined that nitrate (0.1 mM) was inhibitory to W. gerlachensis (explaining the lack of growth in culture A9) and that robust growth was dependent on anoxic conditions and on the presence of corn stover rather than other substrates (Supplementary Figs. 1 and 2 and Supplementary Table 1). As a result, most subsequent experiments were performed using a variation of the A10 medium (A10X2) with corn stover as the main carbon source under anoxia. Under these conditions, W. gerlachensis has been maintained at an abundance of 106–107 16S rRNA gene copies mL–1 culture for over 4 years. To our knowledge, these represent only the second reported laboratory cultures containing members of the Caldarchaeales22.
W is essential to maintain the growth of W. gerlachensis
Since GBS spring water was necessary for the maintenance of W. gerlachensis cultures (Fig. 1b and Supplementary Table 1), we compared the composition of the synthetic medium to GBS spring water using inductively coupled plasma mass spectrometry (ICP-MS) (Supplementary Data 1). Biologically relevant trace elements in the GBS spring water that were absent in the synthetic medium trace metal solution (Supplementary Data 2) included Ni (2.2 nM), which was however in the complete corn stover medium (19–26 nM), and W (263 nM)23. Only W was present in the spring water and not elsewhere, such as in the substrate (~0.11 nM) or media components (<0.05 nM, Supplementary Data 2). Furthermore, preliminary analysis of a W. gerlachensis MAG obtained from the laboratory cultures indicated the presence of multiple W-associated enzymes and WO42– ABC transporters. We therefore hypothesized that W was the key component in the GBS spring water allowing maintenance of W. gerlachensis.
The utilization of W was conceivable given its previous recognition as an essential element for diverse ferredoxin oxidoreductases in the marine heterotrophic hyperthermophile Pyrococcus furiosus (Thermococcales)24,25,26,27,28. Nevertheless, W biochemistry is rare in microorganisms, although the low redox potential of W cofactors makes them especially suitable for anaerobes29. For instance, the low redox potential of the W6+/W4+ couple makes it preferable to Mo, which is more bioavailable in oxic environments and serves equivalent cellular functions in formaldehyde oxidoreductases and benzoyl CoA reductases30,31. In acetylene hydratase, W enables sharing of electrons of the outer orbital shell with the substrate and provides electrostatic stabilization to transition states32. In GBS, W is likely derived from minerals of the Scheelite (CaWO4) and Wolframite groups ([Fe, Mn]WO4), within W-rich deposits overlaying Triassic clays that can be extensive in the Great Basin33.
We confirmed that W. gerlachensis is dependent on W using a synthetic medium with discrete additions of WO42– (Fig. 2). After two transfers in a synthetic medium without W amendment, W. gerlachensis fell to very low or undetectable levels based on amplicon sequence variant (ASV) counts, whereas addition of 0.1 or 2 µM WO42– alleviated this effect (Fig. 2a). Importantly, the addition of W to the synthetic medium led to W. gerlachensis abundances indistinguishable from those in the original spring water-containing medium (ANOVA, p > 0.05). These results were also reflected in W. gerlachensis absolute abundance as determined by qPCR. The omission of W induced a drop of W. gerlachensis 16S rRNA genes by 2–3 orders of magnitude (Fig. 2b). Tungsten starvation induced loss of W. gerlachensis earlier (after the second transfer) in corn stover relative to sugar mix cultures (Fig. 2c and Supplementary Fig. 3), with orders-of-magnitude differences in W. gerlachensis 16S rRNA genes with varying W concentrations. Concentrations of 20 nM WO42– yielded higher W. gerlachensis 16S rRNA gene numbers than 5 nM WO42– (corn stover) and 1 nM WO42– (sugar mix, ANOVA, p < 0.05 for both), suggesting 20 nM WO42– was the critical threshold to maintain W. gerlachensis over at least three transfers in synthetic medium (Fig. 2c). We should note that, given ~0.8 mM sulfide added to the medium as reducing agent, the dominant W species is likely WS42–, which microbial cells would be exposed to. Regardless of the W speciation, low W levels decreased the total culture biomass as assessed by DNA concentrations in extracts (Supplementary Fig. 3), suggesting that microorganisms requiring or using W may play important roles in community productivity, despite the relative abundances of only a few community members being directly affected by the absence of W. The critical W concentration to maintain W. gerlachensis in culture is higher than that required by P. furiosus (at least 1.5 nM but less than 15 nM)34. Nevertheless, P. furiosus growth dynamics were not determined over multiple transfers. There may be differences in the W-requiring biochemistry between these two organisms, while the comparison between mixed and pure cultures may also be obscured by inter-species W cycling.
a ASV abundances based on 16S rRNA gene tags from synthetic medium with and without added spring water or W. The 12 most abundant taxa are shown, which consistently made up >98% of the total ASVs. Caldimicrobium and W. gerlachensis ASV counts from triplicate cultures were statistically different between synthetic medium with and without spring water (without W addition) as denoted by the asterisk (ANOVA, *p = 0.07, **p = 0.03, n = 3). ASV counts are reported as raw reads; see Supplementary Data 9 including normalized ASV counts that show identical trends. b Absolute abundance of W. gerlachensis based on 16S rRNA gene qPCR on corn stover incubations. Central marks in boxes indicate the median, with the bottom and top edges as the 25th and 75th percentiles. Error bars denote SD of biological replicates (n = 3). c Abundance of W. gerlachensis 16S rRNA genes measured after each transfer and the resulting growth. Cultures were grown in corn stover and sugar mix in synthetic medium amended with discrete W concentrations (indicated by black arrows). 16S rRNA gene tags retained at the third transfer indicated no ASVs affiliated with W. gerlachensis in all replicates without W (Supplementary Fig. 4). Source data are provided as a Source Data file.
Because the abundance of Caldimicrobium was also affected by W limitation (Fig. 2a and Supplementary Fig. 4), we were unable to exclude the possibility that W. gerlachensis was affected indirectly via Caldimicrobium, potentially due to a mutualistic relationship between the two. Nevertheless, the presence of multiple W-associated genes in the W. gerlachensis MAG, discussed below, suggested that the requirement for W is direct. To probe the genomic potential for W. gerlachensis anaerobic growth on carbohydrates, we annotated a high-quality MAG from a hybrid assembly of short- and long-read sequence data derived from the laboratory cultures (N50 = 86,582 bp, estimated 98.06% completeness and 0.49% contamination, containing both 16S and 23S rRNA genes). This MAG encoded six W-dependent ferredoxin oxidoreductases as well as a complete WO42– transmembrane ABC transport system of the Tup family, which corroborated our initial hypothesis that W is required for growth in synthetic medium.
W-dependent ferredoxin oxidoreductases are extremely oxygen-sensitive cytoplasmic tungstoenzymes that are involved in sugar and peptide catabolism in a variety of thermophiles. They oxidize various aldehydes, derived from amino acids or sugars, to corresponding carboxylic acids with ferredoxin as the electron acceptor35. These enzymes can generally be separated into aldehyde:ferredoxin oxidoreductases (AOR)24, glyceraldehyde-3-phosphate:ferredoxin oxidoreductases (GAPOR)25,36,37, and formaldehyde:ferredoxin oxidoreductases (FOR)26, in addition to tungstoenzymes with unknown function (WOR4)27 or broad substrate specificity (WOR5)28. All characterized enzymes in this family require incorporation of W, which is coordinated by a pyranopterin cofactor, similar to molybdoenzymes38,39. Dissolved W is mainly available as the tetrahedral oxyanion WO42– in reduced spring waters40. Tungstate is taken up by ABC-type transporters containing highly selective binding proteins (TupA, WtpA). Molybdate is transported by other distinct transmembrane proteins (ModABC). Although we found several annotated MoO42– cofactor biosynthesis protein-encoding genes within the W. gerlachensis MAG, no Mod genes were present, supporting that multiple cellular processes in W. gerlachensis are dependent on W and not Mo.
To test if the identified genes encoding tungstoenzymes and W transporters were expressed, we performed reverse transcriptase-polymerase chain reaction (RT-PCR) assays specific for these W. gerlachensis transcripts in community RNA. While several of the oxidoreductase genes were transcribed, WO42– ABC-type transporters were only weakly transcribed (Supplementary Fig. 5). Transcripts for the WO42–-binding portion of the ABC complex (MCF3653660.1) were detected in the sugar mix culture (irrespective of reverse transcriptase), and the transmembrane subunit transcripts were detected in the corn stover culture. Increased synthesis rates of the periplasmic binding proteins over other subunits have previously been observed for ABC-type transporters in Escherichia coli, including MoO42– transporters, where the synthesis rate of the periplasmic binding protein exceeded that of the transmembrane portion by at least 100-fold41. For the putative W-dependent ferredoxin oxidoreductases, the gene encoding protein MCF3653435.1 was transcribed in corn stover and sugar mix cultures, whereas those encoding MCF3653440.1 and MCF3653608.1 were only transcribed in corn stover cultures (Supplementary Fig. 5). The apparent higher expression of tungstoenzymes in corn stover over sugar mix cultures is consistent with higher sensitivities to W starvation (Fig. 2c). Thus, the expression of W-dependent enzymes varied in enrichments provided with different primary carbon substrates, pointing to the direct involvement of tungstoenzymes in the catabolism of complex carbohydrates in corn stover.
Xylose is the preferred carbohydrate monomer for W. gerlachensis
To investigate carbohydrate metabolism in W. gerlachensis in the absence of an axenic culture, we used a combined CARD-FISH-nanoSIMS approach42 that allowed us to directly interrogate the metabolism of single cells in the complex community of the enrichment. We amended GBS in situ corn stover enrichments with different 13C-labeled substrates (amino acids, starch, glucose, xylose, and ribose) to track carbohydrate metabolism, and with 15N-labeled ammonium to track overall assimilatory activity. Cells from non-FISH-labeled incubations collected after 24 h were significantly enriched in both 15N and 13C compared to unlabeled controls (Kruskal–Wallis test, p < 0.001; Fig. 3 and Supplementary Fig. 6), indicating active metabolism. Carbon and nitrogen enrichment were positively correlated (p = 0.003, Supplementary Fig. 6), but the 13C/15N enrichment varied, suggesting that some cells were metabolically active (labeled with 15N ammonium) but did not incorporate any of the 13C-labeled substrates.
a Isotopic enrichments in CARD-FISH-identified W. gerlachensis (white data points) and other (black data points) cells. Central marks in boxes indicate the median, while the bottom and top edges are the 25th and 75th percentiles. Each point reflects the enrichment of a single cell. b Fluorescent (CARD-FISH) and nanoSIMS ion and ion ratio images reflecting 13C and 15N incorporation in a representative W. gerlachensis cell identified by CARD-FISH in xylose treatments (Cy3, magenta arrow). An adjacent cell (no Cy3 label, white arrow) does not exhibit 13C and 15N enrichment. The higher relative enrichment of 13C and 15N in W. gerlachensis corresponds to higher single-cell anabolic activity compared to other surrounding cells. The minimum isotope abundance ratios are set to natural abundance ratios. Source data are provided as a Source Data file.
The W. gerlachensis population identified with CARD-FISH and subsequently analyzed by nanoSIMS was significantly enriched with 13C and 15N in all except the ribose-amended incubations (Fig. 3 and Supplementary Fig. 6; Kruskal–Wallis test, p < 0.001). W. gerlachensis cells were the most active (15N enriched) in the xylose treatment. Xylose incubations also contained W. gerlachensis cells most enriched in 13C (median of 15 atom% excess and 100% of cells enriched), consistent with direct assimilation of xylose and a preference for it over the other substrates tested. Median 13C enrichment of W. gerlachensis cells incubated with labeled glucose, amino acids or starch was less than 1 atom% excess (potentially due to cross-feeding on metabolites derived from these substrates), and no isotope enrichment was detected in 13C ribose incubations.
A hot spring sediment community composed of 4% Caldarchaeales degraded the xylose polymer xylan and was used to purify a thermostable xylanase enzyme (>70% activity at 70 °C43), indicating the participation of this lineage in polysaccharide-degrading consortia. We also found xylose isomerase (EC 5.3.1.5) and multiple xylulose kinase (EC 2.7.1.17) encoding genes in the W. gerlachensis MAG. In light of our NanoSIMS results, these data suggest W. gerlachensis is an important contributor to the anaerobic degradation of lignocellulose by the microbial community at GBS, likely as members of a cellulolytic consortium17. The precursor of xylose is likely hemicellulose in corn stover, but other sources might be present in natural hot spring environments, including both allochthonous (e.g., nearby plants) and autochthonous (e.g., biofilm) materials.
W-dependent ferredoxin oxidoreductases putatively drive aldehyde metabolism
Next, we identified the potential substrates of the six W-requiring ferredoxin oxidoreductases encoded by the W. gerlachensis MAG, and likely to be compromised by W limitation, employing primary sequence and structural analyses. Over 2600 sequences of known or annotated AOR, FOR, GAPOR, and WOR homologs served as reference sequences to interrogate the W-associated ferredoxin oxidoreductase homologs encoded in the W. gerlachensis MAG. Based on phylogenetic analysis, three homologs were distantly related to well-characterized AORs, herein referred to as AOR-like 1, AOR-like 2, and AOR-like 3; one homolog was distantly related to characterized FORs, referred to herein as FOR-like; and one homolog was distantly related to characterized GAPORs, here termed GAPOR-like (Fig. 4 and Supplementary Figs. 7 and 8). To gain insight into the putative functions of these oxidoreductases, we modeled the protein structure of MAG-encoded oxidoreductase sequences belonging to the genus Wolframiiraptor, along with selected reference sequences, and applied multidimensional scaling on the predicted structures (Fig. 4b). Structural similarities to a characterized AOR from P. furiosus, as quantified based on a sum-of-pairs score using a protein database reference, emerged in the three AOR-like lineages (Cluster I and II, Fig. 4b), supporting the phylogenetic analysis. The other oxidoreductases were structurally related to the GAPOR from Caldicellulosiruptor bescii (Clostridia)37 (Cluster III, Fig. 4b) and diverse FORs (Cluster IV, Fig. 4b), again congruent with the phylogenetic analysis. With the genomic assessments, coupled with the protein phylogenetic-structural analyses, we further looked for evidence of physiological dependence on W.
a Maximum-likelihood phylogenetic tree of W-dependent ferredoxin oxidoreductase protein sequences retrieved from MAGs belonging to Wolframiiraptoraceae, and reference sequences. Illustrated is the masked alignment of the W-dependent ferredoxin oxidoreductase sequences, excluding all alignment positions with sequence information in less than 75% of the taxa analyzed. Branch support was inferred with SH-aLRT and Ultrafast bootstrapping and support at deeper nodes among the lineages in the phylogeny is indicated. W-dependent ferredoxin oxidoreductase lineages were inferred based on the topologies of the masked and full alignments (Supplementary Fig. 7 and Supplementary Data 7). The scale bar indicates the number of amino acid changes per site. b Multidimensional scaling of modeled protein structures. Data points with the most similar structural neighborhoods are positioned near each other. Protein models are based on the amino acid sequences aligned in a. Reference sequences are color-coded. K-means clustering was applied to identify significant data clusters (colored ellipses) based on minimum cluster r2. Clusters included AORs from both references and those from representative members of the genus Wolframiiraptor. One sequence (MCF3653595.1) was excluded from the analysis (<300 amino acids). Dotted lines are used to indicate putative groups identified in a and b. All AOR-like lineages grouped together in Clusters I and II, while GAPOR-like sequences grouped in Cluster III and FOR-like sequences grouped in Cluster IV. c Pyruvate, derived from xylose via the pentose phosphate pathway (PPP) and the Embden–Meyerhof pathway (EMP), is stepwise converted to organic acids. Within this conversion, AOR and GAPOR couple aldehyde oxidation with ferredoxin reduction. Electrons from ferredoxin are used by a membrane-bound proton-translocating [NiFe]-hydrogenase (Mbh) to generate a proton motive force. Besides this main pathway, aldehydes could additionally be derived from amino acid metabolism and ferredoxin could also be recycled by an H2-consuming sulfide dehydrogenase89 after H2 activation by Mbh (dashed lines). W uptake protein (subunits A-C), Tup; Oligopeptide transporter, Opp; Aromatic aminotransferase or branched-chain amino acid aminotransferase, Ar/Bcat; 2-Ketoisovalerate ferredoxin:oxidoreductase, VOR; Maltose/maltodextrin transporter, Mal; Xylose isomerase, Xyl; Xylulose kinase, Xk; Ribose 5-phosphate isomerase, Rpi; Transketolase, Tk; Coenzyme A (-acyl), CoASH; Aldehyde:ferredoxin oxidoreductase, AOR; Ion translocator, Mrp; sulfide dehydrogenase, SuDH; F0F1(V)-type ATP synthase, ATPase.
W-requiring ferredoxin oxidoreductases are found at the heart of sugar and peptide metabolisms in thermophilic anaerobes44,45. While glycolysis in thermophilic bacteria is catalyzed by the conventional glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase, glucose catabolism in several hyperthermophilic archaea differs from that canonical pathway and appears to proceed via a modified Embden–Meyerhof (EM) pathway that involves novel W-dependent enzymes46. On one hand, phylogenetic analysis and structural modeling of the homologs represented by MCF3653440.1 in W. gerlachensis predicts an encoded GAPOR (Fig. 4, GAPOR-like lineage). On the other hand, the W. gerlachensis MAG also encodes an annotated glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) and a phosphoglycerate kinase (EC 2.7.2.3) and we thus cannot determine with certainty if pyruvate is originating via the canonical or modified (archaea-typical) EM pathway. Similar to the amino acid catabolism in P. furiosus, W. gerlachensis likely decarboxylates 2-keto acids, such as pyruvate, to aldehydes that are then oxidized by W-dependent ferredoxin oxidoreductases to form carboxylic acids (Fig. 4c)47,48,49. The pivotal step of the metabolic inhibition occurs when W-requiring ferredoxin oxidoreductases become insufficiently supplied with the W-cofactor tungstopterin and ferredoxin reduction halts. A ferredoxin-dependent, membrane-bound proton-translocating [NiFe]-hydrogenase (Mbh) encoded by the W. gerlachensis genome would then increasingly lack reduced ferredoxin to trigger tunneling of H+ ions across the cell membrane, thereby leading to a decreased transmembrane H+ gradient and retarded ATP formation (Fig. 4).
Inactivation of W-dependent ferredoxin oxidoreductases would be highly effective at depriving the cell of energy because (i) the encoded ATPase is predicted to be proton-dependent, based on alignment with V-type ATPases50, inducing a proton gradient of 3 ion equivalents across the membrane and (ii) these oxidoreductases can make up a significant proportion of cell biomass. For instance, in Thermococcus strain ES1, these oxidoreductases constitute 1% of the total protein mass51 and the intracellular W-dependent ferredoxin oxidoreductase concentration in P. furiosus reached 110 µM44. Interestingly, the W. gerlachensis MAG revealed a putative ferredoxin-dependent, membrane-bound, and proton-translocating [NiFe] hydrogenase complex of 10 subunits (Mbh-Mrp) including conserved cysteine pairs in the N- and C-terminus of the catalytic subunit MbhL (Supplementary Fig. 9) that ligate the [NiFe] metallocluster52, along with several Hyp-type [NiFe] metallocluster assembly proteins. We were not able to find rnf genes53 in the W. gerlachensis MAG and we hence suspect ferredoxin (as the reducing equivalents produced by AOR) would predominantly be recycled by the Mbh-Mrp. A similar Mbh-Mrp complex has previously been described for P. furiosus54,55 and is a type-4 [NiFe] energy-conserving hydrogenase56 that was also detected in other Caldarchaeales genomes8,12. W addition into culture media of the marine thermophile Thermotoga maritima resulted in stimulation of hydrogenase activity57. The carbon and energy metabolism in that bacterium are similar to that of W. gerlachensis58: Formation of pyruvate via the EM pathway, production of organic acids and H2 and therefore coupling of W enzymatic activity to a hydrogenase and alternative reduction of S0 to H2S by H2. Across domains and comprising terrestrial as well as marine habitats, W-associated anaerobic heterotrophic processes are therefore important for thermophilic life.
Distribution of tungstoenzymes across the family Wolframiiraptoraceae
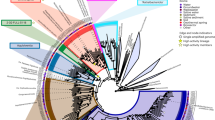
In evaluating the scope of tungstoenzymes in the Caldarchaeales, we recruited and annotated 77 additional high-quality MAGs related to W. gerlachensis that were extracted from metagenomes from geothermal springs in Tengchong, China; Yellowstone National Park, United States; and Taupo, New Zealand, along with a deep-sea hydrothermal mineral deposit from the Eastern Lau Spreading Center in the Southern Pacific. The assignment of these MAGs to the family Wolframiiraptoraceae was defined based on monophyly using conserved marker gene sets and relative evolutionary divergence values. The MAGs could be assigned to four genus-level lineages [including previously recognized AigG4 (syn. EX4484-121, herein Wolframiiraptor), AigG5 (syn. JGI-000106-J15, herein Geocrenenecus), and AigG7 (syn. NZ13-MG1, herein Terraquivivens)8], and 11 species (Fig. 5 and Supplementary Fig. 10), based on relative evolutionary divergence (Supplementary Data 3), average amino acid identity, and average nucleotide identity (Supplementary Data 4).
a Average amino acid identity (AAI) among designated type genomes for species-level groups within the family Wolframiiraptoraceae. The same color scheme used in b is used for taxa indicated with numbers. For full ANI and AAI matrices of the respective genera, see Supplementary Data 4. b A maximum-likelihood phylogeny was inferred from the concatenated, partitioned sequence alignment of 122 conserved archaeal marker sequences (ar122). Appropriate evolutionary models were applied for each partition. Branch support for the phylogeny was inferred from 1000 pseudoreplicates, using traditional bootstrapping and supported branches are indicated with filled circles at nodes. Species-level groups, as supported by phylogenetics, average nucleotide identity (ANI), and relative evolutionary divergence are indicated with different colors, with species belonging to the same genus indicated in different shades of the same color. Taxon names in bold and indicated with a star are the proposed nomenclatural types for the taxa. Gray taxon names represent medium-quality MAGs that are publicly available. c Summary of gene tree/species tree reconciliation analyses with predicted gains, transfers, and losses, for W-associated enzymes (ABC-type transporter A subunits and W-dependent oxidoreductases) within the family. Hypothesized evolutionary paths for the genes of interest are indicated in the colors used in the dot matrix on the right of the cladogram and numbered according to the dot matrix order. The consensus presence or absence profiles for species with multiple genome sequences available were determined by data as indicated for the majority of genomes in the species. Where genes encoding these proteins are present in some genomes within a taxon, colored dots are partially filled, while fully filled dots in the dot matrix indicate the presence in the majority of the genomes belonging to that taxon. For full ABC-type transporter phylogeny, including KOs K02020, K05772, and K15495, see Supplementary Figs. 10 and Fig. 5, and Supplementary Fig. 7 for phylogenies and structural modeling of W-dependent ferredoxin oxidoreductases.
To assess the potential importance of W within the family Wolframiiraptoraceae, we interrogated the presence of tungstoenzymes within the genomes of these taxa. The three AOR-like, FOR-like, and GAPOR-like enzymes were widely distributed within the family, each present in more than half of the Wolframiiraptoraceae genomes (Fig. 4 and Supplementary Figs. 7 and 8), sparse in other Caldarchaeales, and absent from close relatives in the Nitrososphaeria (Supplementary Fig. 11). In addition, three divergent putative tungstoenzymes were identified in the Geocrenenecus and Terraquivivens MAGs.
All protein-coding sequences annotated as the substrate-binding subunits of the Mod (K02020), Tup (K05772), or Wtp (K15495) ABC transporter systems within the family were analyzed to determine the putative substrate specificity of these transporters (Supplementary Fig. 12a, b). All species of the genera Wolframiiraptor, Terraquivivens, and the sole representative of Benthortus encode homologs of the TupABC transporter. Notably, only a single genome of Geocrenenecus and members of the species T. tengchongensis encoded novel TupA- and/or WtpA-like proteins. The WtpA-like protein has been identified before as a ModA-like homolog in Pyrobaculum species14,59. The consensus lack of a MoO42–/WO42– transport system in the Geocrenenecus species is not unique, as several completely sequenced genomes from organisms known to utilize MoO42–/WO42– lack any of these annotated ABC transporters59.
Scaling up the distribution analysis of MoO42–/WO42– transport proteins across entire thermophilic community metagenomes indicated unequal modA, tupA, and wtpA gene frequencies among microbial communities inhabiting springs that span a spectrum of geochemical conditions (Supplementary Figs. 13 and 14). While the WO42– transporters show a high presence (~70%) in spring waters with temperatures >60 °C, wtpA is generally more abundant than tupA and is the most abundant MoO42–/WO42– transporter among microbial communities of low-pH (<4) springs.
To better understand how Wolframiiraptoraceae acquired W-dependent enzymes, we inferred associated evolutionary events through phylogenetic analyses, ancestral character state reconstructions (Supplementary File 2), and gene tree/species tree reconciliation analyses based on the W-dependent ferredoxin oxidoreductases and MoO42–/WO42– transporters (Fig. 5c). These analyses revealed a complex evolutionary history characterized by rampant lateral transfer from other Thermoproteota to various Wolframiiraptoraceae lineages. Although it remains difficult to distinguish between phylogenetic noise and true signal for lateral acquisitions in single-gene trees, the presence of one tungstoenzyme (AOR-like 1) in the ancestral node to the Wolframiiraptoraceae was well supported by all analyses. This ancestral enzyme, combined with extensive capture of W-associated enzymes from Thermoproteota and other sources (Supplementary Figs. 7, 8, and 10 and Supplementary Dataset 2), and multiple gene family expansions and losses throughout the evolution of the Wolframiiraptoraceae, implies an important role for W in the diversification of the family. Given the close phylogenetic grouping of the Wolframiiraptoraceae TupA transporter subunit, AOR-like 3, FOR-like, and GAPOR-like sequences with those from diverse Thermoproteota (Supplementary Figs. 7, 8, and 10 and Supplementary File 3), these enzymes may have been inherited through separate horizontal acquisitions. However, the notable progressive increases in the number and diversity of W-dependent enzymes within Caldarchaeales, and in particular Wolframiiraptoraceae, suggest an early emergence of W utilization within the Thermoproteota and a critical role for W metabolism in the evolution of the Wolframiiraptoraceae.
Discussion
In the present study, we demonstrate that W is required to culture the anaerobic species W. gerlachensis, the first physiologically characterized cultured member of the Caldarchaeales. The basis of this requirement, as evidenced by poor biomass yields in cultures in synthetic medium without amended W, is most likely W-dependent ferredoxin oxidoreductases involved in the degradation of xylose-derived pyruvate, which becomes non-functional without W. This dependence on W is seemingly a characteristic of several close relatives of W. gerlachensis, with W utilization feasibly representing an ancestral trait within the family and important for the diversification of the Wolframiiraptoraceae. The levels of W required by W. gerlachensis (≥20 nM) are below those that are typically inhibitory to the growth of other microorganisms where W inhibition has been demonstrated18. The standard inclusion of this trace element may aid in the cultivation of additional members of the Wolframiiraptoraceae and other novel thermophiles.
It has been suggested that molybdoenzymes did not evolve prior to the widespread oxygenation of the Earth, which allowed for oxidative weathering of sulfide minerals that hosted Mo60. In contrast to Mo, W is less affinitive to sulfide complexation and it is possible that the apparent favorability of W over Mo in the Wolframiiraptoraceae is a relic from adaptations to early microbial habitats where W was more bioavailable than Mo. On the early Earth, such potential habitats were more common and extended beyond hot springs into the marine realm. About 2.3 billion years ago, increasingly sulfidic bottom waters of the oceans underwent a strong geochemical shift from ferruginous to euxinic55,56,57. Investigation of contemporary euxinic sediments in the Black Sea revealed a critical concentration of ~11 µM aqueous sulfide to cause complete Mo fixation from the water column61 due to reduction to insoluble Mo-sulfide species or transformation to particle-reactive thiomolybdate (MoS42–)62. Such sulfide levels based on dissimilatory sulfite/sulfate reduction63 or volcanically derived sources63,64 likely prevailed both in terrestrial hot springs and marine basins. The availability of W theoretically could have constituted a regulatory switch on the fermentative degradation of organic matter and could have influenced fluxes of organic acids, which, in turn, were crucial substrates for sulfur reduction and methanogenesis. The selective pressure for W utilization likely dropped with the oxygenation of the Earth and the dispersion of soluble Mo, but the remnants of ancient W-based pathways likely perpetuated in the carbohydrate metabolisms of anaerobic, thermophilic archaea.
Methods
In-situ enrichments and sampling
GBS water used in the preparation of cultivation medium was sampled in 2013, 2016, 2017, and 2018, passed through a 0.2 µm filter, and stored in 20 L plastic containers in the laboratory without temperature control. In situ enrichments on ammonia-fiber explosion-treated corn stover (kindly provided by Bruce Dale, Michigan State University) in GBS were performed as described in Peacock et al.17. Samples were taken after approximately six months of incubation on March 29, 2016. At that time, the temperature of the water was 86.0 °C and the pH was 7.15, typical for the main source pool of GBS16. For sampling, a nylon bag containing corn stover was removed from the conical tube, the bag was cut open with a sterile scissors, and approximately half of the contents was immediately transferred to 10 mL of 0.2 µm filtered GBS water (previously sampled, sparged with N2, reduced by the addition of cysteine to 0.05 g/L, and sterilized by autoclaving) in a 25 mL Balch tube under a stream of N2 gas applied through a 0.2 µm filter. The tube was sealed with a butyl rubber stopper, the headspace was flushed for 3 min with N2 gas, and the tube was then shaken to disperse the corn stover. Previously prepared anaerobic media (50 mL in 160 mL serum vials sealed with butyl rubber stoppers; see below) were inoculated with 0.2 mL of the corn stover slurry using a sterile, N2-flushed needle and syringe. The bottles were transported back to the laboratory at ambient temperature and were transferred to high-temperature incubators within 24 h of sample collection. The remaining in-situ corn stover enrichment was placed in 1.5 mL centrifuge tubes, frozen immediately, and transported back to the laboratory on dry ice, and then stored at –80 °C for DNA extraction.
Laboratory culturing
The 13 different media and incubation conditions used are shown in Supplementary Table 1. All media were based on GBS salts synthetic medium, which contained per liter of 18.2 MΩ cm water: 3 g NaCl, 0.3 g Na2SO4, 0.15 g KCl, 0.015 g CaCl2·2H2O, 0.123 g MgSO4·7H2O, and 5 mL of a trace mineral solution64. The trace mineral solution, based on ref. 65, contained per liter: 3.7 g Na2-EDTA, 1.1 g FeSO4·7H2O, 0.028 g ZnSO4·7H2O, 0.098 g MnCl2·4H2O, 0.031 g H3BO3, 0.024 g CoCl2·6H2O, 0.017 g CuCl2·2H2O, and 0.024 g NaMoO4·2H2O. The “spring water medium” consisted of a 1:1 mix of the above synthetic medium and water collected from GBS as described above. All media contained 0.107 g/L NH4Cl. The medium was then sparged with N2 gas and transferred into serum vials in an anaerobic chamber. The vials were sealed with butyl rubber stoppers and aluminum crimps, the headspace was exchanged with N2 by three cycles of vacuuming to 585 mm Hg and repressurizing to 15 psi with N2, and the vials were sterilized by autoclaving64. Prior to inoculation, the following were added to all media at the indicated final concentrations from concentrated, anoxic stocks: 0.025% keratin, 0.002% xyloglucan, 0.02% Na2S, 0.02% cysteine, 0.002% yeast extract, 0.0014% NaH2PO4·H2O, and 1.6 mL/L of a vitamin solution66. All stocks were sterilized by autoclaving except yeast extract and vitamins, which were passaged via a 0.2 µm filter. Additions for different media conditions in the initial enrichments were made from sterile, anoxic stocks to final concentrations shown in Supplementary Table 1, except that corn stover (0.02% m/v) was added to individual serum vials in the anaerobic chamber prior to sealing. Follow-up experiments used the same base synthetic or spring water media with modifications of the specific substrates that were excluded or added anoxically. After stable laboratory cultures were obtained, the culture conditions for routine transfer and maintenance used the base spring water medium with 2 mM sodium bicarbonate and either corn stover (0.02% m/v) or a “sugar mix” (0.01% m/v each of glucose, xylose, mannose, and D- and L-arabinose) for major carbon sources. Laboratory cultures were kept at 80 °C and initially transferred to fresh media (1/100 volume) every 2 weeks for the first two transfers after inoculation with in situ enrichments and then every 3 weeks thereafter.
Quantification of W in spring water and media by ICP-MS and ICP-OES
W levels in laboratory media and filtered GBS spring water were measured using ICP-MS after the addition of nitric acid (OPTIMA grade, Fisher Chemical, Fair Lawn, NJ, USA) to 1% final concentration at Huffman Hazen Laboratories (Golden, CO, USA). W levels in filtered GBS spring water were also measured using inductively coupled plasma optical emission spectroscopy (ICP-OES) with an Agilent 5110 ICP-OES (Santa Clara, CA, USA) and dilutions of a W calibration standard (PLW9-2Y, SPEX CertiPrep, Metuchen, NJ, USA). Nitric acid was added to samples and standards to a final concentration of 1% before analysis, and optical emission at 207.91 nm was used for W quantification. Broad trace element analysis by ICP-MS on GBS spring water and media was performed on samples prepared as described above at the Analytical Chemistry Laboratory of New Mexico Bureau of Geology and Mineral Resources, New Mexico Institute of Mining and Technology.
16S rRNA gene tag sequencing
16S rRNA gene tag amplification and sequencing on initial enrichment cultures was performed essentially as described67. A modified forward primer 515 (5′ GTGYCAGCMGCCGCGGTAA) with a Y instead of a C at the fourth position from the 5′ end was used to increase coverage of Archaea, and a corresponding change was made to the SeqRead1 primer. Sequencing (2 × 250 bp) was performed on an Illumina iSeq sequencer using the V2 reagent kit. Additional 16S rRNA gene tag sequencing on laboratory cultures tested with different levels of W was performed as described68 at Argonne National Laboratories.
Genomic DNA and RNA extraction
For the laboratory culture, biomass (from 10 to 40 mL culture) was harvested by centrifugation at 10 min at 9300 g and 4 °C in 50 mL conical tubes. Resulting pellets were resuspended in 1 mL of supernatant, transferred to 1.5 mL tubes, and centrifuged at 16,100 g for 5 min at 4 °C. The supernatant was removed and the pellets were stored at –80 °C until nucleic acid extraction. DNA was extracted using the Fast DNA SPIN Kit for Soil (MP Biomedicals) as described following the manufacturer’s recommendations, except that homogenization by bead-beating was performed twice for 30 s at a speed setting of 4.5 m/s. DNA was eluted in 75–100 µL of DES solution and further diluted 10-fold in 10 mM trishydroxyaminomethane pH 8. To obtain long fragments of DNA for Oxford Nanopore sequencing, DNA was extracted using the cetyltrimethyl ammonium bromide protocol of the Joint Genome Institute66. RNA was extracted using the FastRNA Spin Kit for Soil (MP Biomedicals) and the High Pure RNA Isolation Kit (Roche) as described in Hamilton et al.69, followed by an additional DNase treatment using the TURBO DNase kit (Ambion) according to the manufacturer’s instructions. For all other information regarding metagenomic DNA extraction, please refer to Supplementary Note 1.
Quantitative PCR
Assays were performed in 96-well plates (Applied Biosystems) in a StepOnePlus Quantitative Real-Time PCR machine (Applied Biosystems) using PowerUp SYBR Green Master Mix (Applied Biosystems). Cycling conditions for AigG4-specific primers (Supplementary Table 2) were as follows: initial denaturation for 15 min at 95 °C followed by 45 cycles of denaturation (20 s at 95 °C), annealing (25 s at 64 °C), and extension (50 s at 72 °C). Specificity of qPCR primers was verified by melt curve analysis. Using a plasmid with a fragment of the AigG4 16S SSU rDNA sequence as a standard (SSW_L4_C03; ref. 23), an AigG4 standard curve was generated over 6 orders of magnitude from 5.0 × 102 to 5.0 × 108 (r2 = 0.98–1.00). The reported template abundances are the average and standard deviation of qPCR assays performed in triplicate. Quantification of total bacterial and archaeal 16S rRNA genes was performed similarly using primers 806r and 515f with the modification described above for 16S rRNA gene tag sequencing (Supplementary Table 2), and with an annealing temperature of 55 °C.
Reverse transcriptase PCR
RNA was converted to cDNA with random hexamer primers using the Verso cDNA kit (ThermoScientific), including control reactions without added reverse transcriptase. cDNA was amplified with primers targeting AigG4 predicted W-associated genes and AigG4 16S rRNA (Supplementary Table 3) in the Axygen® Maxygene™ II Thermal Cycler as follows: at 95 °C for 3 min followed by 35 cycles of denaturation (30 s at 95 °C), annealing (30 s at annealing temperature appropriate for specific primer set), and extension (72 °C for 5 min) and finally, 72 °C for 1 min. Primers targeting AigG4 predicted W-associated enzymes were designed using Primer365. DNA was quantified using the Qubit 4.0 with HS DNA reagent kit.
Stable isotope labeling
Stable isotope labeling was performed on-site at GBS in July 2017 using in situ corn stover enrichments prepared and incubated as described above but using 5 g packets of corn stover incubated at the GBS “C” site (outflow)16 for 34 days. Two packets (approximately 10 g) of freshly harvested corn stover enrichment were added to 180 mL of anaerobic, autoclaved GBS water in a sealed 500 mL bottle with an N2 headspace and shaken to form a slurry. Samples of this slurry (2 mL) were then added via syringe to sealed 60 mL amber serum vials containing 8 mL of anaerobic, autoclaved GBS water with an N2 headspace, generating 10 mL microcosms. Various individual 13C-labeled substrates (Cambridge Isotope Laboratories) were then added from concentrated, anaerobic stocks to the microcosms at 0.001% (mass/volume) final concentration: algal amino acids, algal starch, glucose, xylose, ribose, or an equal-mass mixture of acetate, propionate, and butyrate; a control with no label added was also performed and did not yield enriched cells. In addition, 0.5 mM 15N-NH4Cl was added to all microcosms. Preparation of microcosms was done in a hot water bath at 75 °C to prevent excessive cooling. Unlabeled controls were processed as described below immediately after preparation. Sealed microcosms with labeled substrates were incubated at 75 °C for 4 or 24 h. After incubation, microcosm vials were immersed in ice water to cool, shaken vigorously for 30 s, opened, and decanted into 15 mL conical tubes. Tubes were centrifuged for 30 s at 11 g in a clinical centrifuge to pellet large fragments of corn stover. Supernatant (4.5 mL) was then split between three 1.5 mL tubes and centrifuged for 5 min at 16,100 g to pellet cells. Pellets from the three tubes were pooled in 0.25 mL of 1×PBS, and 0.5 mL of 3% paraformaldehyde (PFA) was added and mixed. Samples were fixed for 1 h on ice. After fixation, tubes were centrifuged for 5 min at 16,100 g. Resulting pellets were washed twice with 1×PBS, resuspended in 200 µL of 50% ethanol, and stored on ice (in the field) and then at –20 °C in the laboratory. Prior to performing FISH, Nycodenz density cushion centrifugation70 was used to separate cells from higher density particles in the samples. Cells harvested from the aqueous-Nycodenz interface were pelleted by centrifugation, washed twice in water, and resuspended in 50% ethanol.
CARD-FISH
Detection of W. gerlachensis (AigG4) cells was performed using CARD-FISH using a horseradish peroxidase-labeled probe 5′ CRGAAAGGCCTTCAACCTGT (Biomers.net, Germany). This probe is specific to Aigarchaeota Group 4 and Group 5, and was designed based on multiple sequence alignment of 16S rRNA genes of Aigarchaeota and other Archaea and checked using the Probe Match function of the Ribosomal Database Project16. Optimization of formamide concentrations used during hybridization was performed using the Clone-FISH technique70 with the AigG4 16S rRNA gene sequence SSW_L4_C03 (EU635908)23 expressed from the T7 promoter on plasmid pET23(+) in E. coli strain JM109(DE3). Formamide concentrations from 0 to 50% were tested in 10% increments. The highest formamide concentration that gave strong signal (20%) was then used to test for signal in the E. coli strain without induction of expression of the AigG4 16S rRNA as well as Staphylococcus aureus and Bacillus subtilis as negative controls. CARD-FISH was performed as previously described71 with minor modifications. Briefly, cells were fixed using a 1% PFA solution for 1 h, followed by storage at –20 °C until use. The protocol was performed on fixed cells spotted and air dried on custom synthesized 14-well indium tin oxide (ITO)-coated slides rather than using filters. Cells were permeabilized by treatment with 10 mg/mL lysozyme (for Clone-FISH in E. coli and negative controls) or 1 µg/mL proteinase K (for samples with AigG4) in 50 mM EDTA and 100 mM Tris (pH 8) for 60 min at 37 °C, endogenous peroxidases were inactivated by treatment with 0.01 M HCl for 15 min at ambient temperature (~25 °C), and dehydrated using 96% EtOH. Hybridization was performed at 20% formamide for 16 h at 46 °C, followed by washing at 48 °C, CARD using tyramide labeled with Alexafluor 555 (Invitrogen), and counterstaining by incubation in ice-cold 1 ug/mL DAPI for 10 s. Cells were visualized with a Leica DM5500B microscope with a ×100 magnification dry immersion objective. Fluorescence and brightfield images were collected at diverse locations with FISH-positive cells and the X-Y location of those images was noted, as well as fiducial locations to enable navigation in the nanoSIMS.
NanoSIMS
ITO-coated slides were cut with a diamond saw in order to fit on metal sample holders and analyzed on a CAMECA nanoSIMS 50 at Lawrence Livermore National Laboratory. The charge-coupled device camera was used to find fiducial locations and locations of the FISH-positive cells using X-Y coordinates and the real-time imaging unit. The primary 133Cs+ ion beam was set to 1.5 pA, corresponding to an approximately 100 nm diameter beam diameter at 16 keV. Rastering was performed over 20 × 20 μm analysis areas with a dwell time of 1 ms pixel–1 for 19–30 scans (cycles) and generated images containing 256 × 256 pixels. Sputtering equilibrium at each analysis area was achieved with an initial beam current of 90 pA to a depth of ~10 nm, thus ensuring analysis of intracellular isotopic material but without sputtering through the ITO coating. After tuning the secondary ion mass spectrometer for mass resolving power of ~7000, secondary electron images and quantitative secondary ion images were simultaneously collected for 12C2–, 13C12C–, 12C14N–, and 12C15N– on individual electron multipliers in pulse counting mode. All nanoSIMS datasets from two independent experimental runs were initially processed using L’Image (http://limagesoftware.net, August 2021) to perform deadtime and image shift correction of ion image data before creating 13C12C/12C2 and 12C14N/12C15N ratio images, which reflected the level of 13C and 15N incorporation into biomass. Regions of interest for isotopic ratio quantification were drawn manually around each cell. A cell was considered isotopically enriched, and therefore anabolically active, if the isotopic composition exceeded three times the standard deviation of the mean isotopic composition of unlabeled (non-AigG4) cells72.
Metagenome sequencing, assembly, and binning
For the AigG4 cultures established from the in-situ enrichment at GBS, extracted DNA were sequenced using the Illumina MiSeq platform (2 × 250 bp), along with Oxford Nanopore sequencing using a MinION device. All extracted metagenomic DNA from geothermal springs from Tengchong, China, and Yellowstone National Park, USA, and the marine hydrothermal vent in the Western Pacific were sequenced on the Illumina NovaSeq, HiSeq3000, or HiSeq4000 platforms, in a paired-end or single-read configuration (Supplementary Note 2). Quality control of sequencing reads are outlined in the Supplementary Notes. Sequencing data were assembled and metagenomes were binned using several software packages (Supplementary Note 2). All MAGs were subjected to quality checks with CheckM v. 1.0.18-1.1.373 and classification with GTDB Toolkit v. 1.4.174. This was followed by gene calling using Prodigal v. 2.6.275 and annotation of all MAGs using eggNOG-mapper v. 276, and gene calling and annotation with Rapid Annotation using Subsystem Technology77 in the case of the hybrid assembly for W. gerlachensis.
Phylogenomic analyses
All MAGs classified as belonging to the family NZ13-MG1 in the order Caldarchaeales were used for the construction of a species tree. As reference sequences, genomes for members of the Nitrososphaerales (syn. Thaumarchaeota), along with publicly available genomes for the Caldarchaeales (syn. Aigarchaeota) were included in the taxon set. The 122 conserved archaeal marker sequences in the ar122 dataset were identified from the genomes of these taxa using the GTDB Toolkit v. 1.4.174. Aligned markers were subjected to testing for appropriate evolutionary models using ProtTest v. 3.478, followed by concatenation and partitioning with FASconCAT-G v. 1.0.479. This partitioned, concatenated matrix was subjected to maximum-likelihood tree reconstruction with RAxML v. 8.2.1280, with the appropriate evolutionary model applied to each partition, and branch support inferred from 1000 bootstrap pseudoreplicates. For description on the phylogenomic analysis for the domain Archaea, please see Supplementary Notes 3–6.
Phylogenetic analyses of genes
For phylogenetic confirmation of function of the predicted genes related to physiology, and specifically tungstoenzymes, all Wolframiiraptoraceae sequences annotated as belonging to the KEGG Orthology (KO) numbers of interest were used to infer phylogenetic trees to place the identified sequences into evolutionary context within the protein families. A detailed description of these analyses is provided in the Supplementary notes (see also Supplementary Data 7 and 8).
Gene tree/species tree reconciliation and ancestral character state reconstruction
For reconciliation analyses, a species tree was inferred from the domain-level phylogenomic tree with Bacteria specified as the outgroup. Similarly, subtrees for all W-associated homologs were inferred from the maximum-likelihood phylogenetic analyses of single-gene trees (see Supplementary materials for full description of single-gene tree construction). The inferred gene trees were reconciled with the species tree for the family ecceTERA v. 1.2.481, with default reconciliation penalties, partially ultrametric trees specified, and the median reconciliation graphs printed as sylvx reconciliations. Resulting sylvx reconciliations were viewed in SylvX v. 4282, and median symmetrical reconciliations were summarized and edited in Inkscape v. 0.92.4.
A cladogram inferred from the species tree constructed from the genomes was used as the basis for performing reconstruction of ancestral character states for the family, with a particular focus on genes relating to physiology and W utilization. As the presence or absence of genes within single MAGs can be unreliable, a majority-rule consensus approach was employed for all genomes of the respective species. This entailed using all eggNOG annotated genomes, and summarizing the genome content based on KO numbers for each genome. The absence (0), presence (1), or presence in multiple copies (≥2) for each species was determined based on all genomes belonging to the species. These consensus presence/absence profiles for the species were then used to reconstruct all potential evolutionary events (gene birth, HGT, duplication, or loss) by optimizing rates and inferring the history of gene families by posterior probabilities, as implemented in COUNT83. The COUNT sessions for the full genome analysis and the tungstoenzyme-specific analysis are available as Supplementary Data 6.
Protein structure modeling
W-dependent oxidoreductase sequences were submitted to the iterative threading assembly refinement (I-TASSER) server84 to generate models of oxidoreductase homologs. Models were selected based on C-score85. All Wolframiiraptor W-dependent oxidoreductases reached at least a C-score of 1.3. Structural comparison of all-against-all was done with DaliLite v5 based on PDB coordinates of the models86,87. The first two eigenvectors of the output were used for the multidimensional-scaling and K-means clustering88 applied in JMP Pro (Version 13.1.0, SAS Institute Inc.).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequence data for nomenclatural types and some non-type genomes were deposited on GenBank under accession numbers listed in Supplementary Data 3, and all other genomes were submitted to eLMSG and/or IMG under under Bioproject nos. PRJNA495050, PRJNA791658, PRJNA807863, PRJNA381623, and PRJNA780009. For nomenclatural types, raw sequence reads were submitted to the Sequence Read Archive (SRA) under the SRA run accessions listed in Supplementary Data 3. All other data are contained within the manuscript and supplements/source data files. Source Data are provided with this paper.
References
Weiss, S. et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681 (2016).
Djokic, T., Kranendonk, M. J. V., Campbell, K. A., Walter, M. R. & Ward, C. R. Earliest signs of life on land preserved in ca. 3.5 Ga hot spring deposits. Nat. Commun. 8, 1–9 (2017).
Damer, B. & Deamer, D. The Hot Spring Hypothesis for an origin of life. Astrobiology 20, 429–452 (2020).
Van Kranendonk, M. J. et al. Elements for the origin of life on land: a deep-time perspective from the Pilbara Craton of Western Australia. Astrobiology 21, 39–59 (2021).
Colman, D. R. et al. Phylogenomic analysis of novel Diaforarchaea is consistent with sulfite but not sulfate reduction in volcanic environments on early Earth. ISME J. 14, 1316–1331 (2020).
Anbar, A. D. & Knoll, A. H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142 (2002).
Lloyd, K. G. et al. Phylogenetically novel uncultured microbial cells dominate Earth microbiomes. mSystems 3, 431 (2018).
Hedlund, B. P. et al. Uncultivated thermophiles: current status and spotlight on ‘Aigarchaeota’. Curr. Opin. Microbiol. 25, 136–145 (2015).
Nunoura, T. et al. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ. Microbiol. 7, 1967–1984 (2005).
Nunoura, T. et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 39, 3204–3223 (2010).
Rinke, C. et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 6, 946–959 (2021).
Hua, Z.-S. et al. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat. Commun. 9, 1–11 (2018).
Takami, H., Arai, W., Takemoto, K., Uchiyama, I. & Taniguchi, T. Functional classification of uncultured ‘Candidatus Caldiarchaeum subterraneum’ using the Maple system. PLoS ONE 10, e0132994 (2015).
Beam, J. P. et al. Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous ‘streamer’ community. ISME J. 10, 210–224 (2016).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Cole, J. K. et al. Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. ISME J. 7, 718–729 (2013).
Peacock, J. P. et al. Pyrosequencing reveals high-temperature cellulolytic microbial consortia in Great Boiling Spring after in situ lignocellulose enrichment. PLoS ONE 8, e59927 (2013).
Kletzin, A. & Adams, M. W. W. Tungsten in biological systems. FEMS Microbiol. Rev. 18, 5–63 (1996).
Hagedoorn, P. L. et al. Purification and characterization of the tungsten enzyme aldehyde:ferredoxin oxidoreductase from the hyperthermophilic denitrifier Pyrobaculum aerophilum. J. Biol. Inorg. Chem. 10, 259–269 (2005).
de Vries, S. et al. Adaptation to a high-tungsten environment: Pyrobaculum aerophilum contains an active tungsten nitrate reductase. Biochemistry 49, 9911–9921 (2010).
Bräsen, C., Esser, D., Rauch, B. & Siebers, B. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175 (2014).
Kato, S. et al. Long-term cultivation and metagenomics reveal ecophysiology of previously uncultivated thermophiles involved in biogeochemical nitrogen cycle. Microbes Environ. 33, 107–110 (2018).
Costa, K. C. et al. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13, 447–459 (2009).
Mukund, S. & Adams, M. W. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J. Biol. Chem. 266, 14208–14216 (1991).
Mukund, S. & Adams, M. W. W. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270, 8389–8392 (1995).
Roy, R. et al. Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: the third of a putative five-member tungstoenzyme family. J. Bacteriol. 181, 1171–1180 (1999).
Roy, R. & Adams, M. W. W. Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 184, 6952–6956 (2002).
Bevers, L. E., Bol, E., Hagedoorn, P.-L. & Hagen, W. R. WOR5, a novel tungsten-containing aldehyde oxidoreductase from Pyrococcus furiosus with a broad substrate specificity. J. Bacteriol. 187, 7056–7061 (2005).
Habib, U. & Hoffman, M. Effect of molybdenum and tungsten on the reduction of nitrate in nitrate reductase, a DFT study. Chem. Cent. J. 11, 1–12 (2017).
Liao, R.-Z. Why is the molybdenum-substituted tungsten-dependent formaldehyde ferredoxin oxidoreductase not active? A quantum chemical study. J. Biol. Inorg. Chem. 18, 175–181 (2013).
Qian, H.-X. & Liao, R.-Z. QM/MM study of tungsten-dependent benzoyl-coenzyme A reductase: rationalization of regioselectivity and predication of W vs Mo selectivity. Inorg. Chem. 57, 10667–10678 (2018).
Liu, Y.-F., Liao, R.-Z., Ding, W.-J., Yu, J.-G. & Liu, R.-Z. Theoretical investigation of the first-shell mechanism of acetylene hydration catalyzed by a biomimetic tungsten complex. JBIC 16, 745–752 (2011).
Kerr, P. F. Tungsten-bearing manganese deposit at Golconda, Nevada. Geol. Soc. Am. Bull. 51, 1359–1390 (1940).
Mukund, S. & Adams, M. W. W. Molybdenum and vanadium do not replace tungsten in the catalytically active forms of the three tungstoenzymes in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178, 163–167 (1996).
Debnar-Daumler, C., Seubert, A., Schmitt, G. & Heider, J. Simultaneous involvement of a tungsten-containing aldehyde:ferredoxin oxidoreductase and a phenylacetaldehyde dehydrogenase in anaerobic phenylalanine metabolism. J. Bacteriol. 196, 483–492 (2014).
Scott, I. M. et al. A new class of tungsten-containing oxidoreductase in Caldicellulosiruptor, a genus of plant biomass-degrading thermophilic bacteria. Appl. Environ. Microbiol. 81, 7339–7347 (2015).
Scott, I. M. et al. The thermophilic biomass-degrading bacterium Caldicellulosiruptor bescii utilizes two enzymes to oxidize glyceraldehyde 3-phosphate during glycolysis. J. Biol. Chem. 294, 9995–10005 (2019).
Johnson, J. L., Rajagopalan, K. V., Mukund, S. & Adams, M. W. Identification of molybdopterin as the organic component of the tungsten cofactor in four enzymes from hyperthermophilic Archaea. J. Biol. Chem. 268, 4848–4852 (1993).
Chan, M. K., Mukund, S., Kletzin, A., Adams, M. W. & Rees, D. C. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 267, 1463–1469 (1995).
Glass, J. B. et al. Geochemical, metagenomic and metaproteomic insights into trace metal utilization by methane‐oxidizing microbial consortia in sulphidic marine sediments. Environ. Microbiol. 16, 1592–1611 (2014).
Li, G.-W., Burkhardt, D., Gross, C. & Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014).
Behrens, S. et al. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol. 74, 3143–3150. https://doi.org/10.1128/AEM.00191-08 (2008).
Knapik, K., Becerra, M. & González-Siso, M.-I. Microbial diversity analysis and screening for novel xylanase enzymes from the sediment of the Lobios Hot Spring in Spain. Sci. Rep. 9, 11195 (2019).
Roy, R., Dhawan, I. K., Johnson, M. K., Rees, D. C. & Adams, M. W. Aldehyde Ferredoxin Oxidoreductase. 266 (American Cancer Society, 2011).
Sevcenco, A.-M. et al. The tungsten metallome of Pyrococcus furiosus. Metallomics 1, 395–402 (2009).
Sakuraba, H. & Ohshima, T. Novel energy metabolism in anaerobic hyperthermophilic archaea: a modified Embden-Meyerhof pathway. J. Biosci. Bioeng. 93, 441–448 (2002).
Ma, K., Hutchins, A., Sung, S.-J. S. & Adams, M. W. W. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl Acad. Sci. USA 94, 9608–9613 (1997).
Mai, X. & Adams, M. W. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178, 5890–5896 (1996).
Adams, M. W. W. & Kletzin, A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archaea. Adv. Prot. Chem. 48, 101–180 (1996).
Mulkidjanian, A. Y., Galperin, M. Y., Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Evolutionary primacy of sodium bioenergetics. Biol. Direct 3, 1–19 (2008).
Heider, J., Ma, K. & Adams, M. W. W. Purification, characterization, and metabolic function of tungsten-containing aldehyde ferredoxin oxidoreductase from the hyperthermophilic and proteolytic archaeon Thermococcus strain ES-1. J. Bacteriol. 177, 4757–4764 (1995).
Schut, G. J. et al. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203 (2013).
Kuhns, M., Trifunović, D., Huber, H. & Müller, V. The Rnf complex is a Na+ coupled respiratory enzyme in a fermenting bacterium, Thermotoga maritima. Commun. Biol. 3, 1–10 (2020).
Sapra, R., Verhagen, M. F. J. M. & Adams, M. W. W. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182, 3423–3428 (2000).
Sapra, R., Bagramyan, K. & Adams, M. W. W. A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc. Natl Acad. Sci. USA 100, 7545–7550 (2003).
Schut, G. J. et al. The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim. Biophys. Acta Bioenerg. 1857, 958–970 (2016).
Juszczak, A., Aono, S. & Adams, M. W. The extremely thermophilic eubacterium, Thermotoga maritima, contains a novel iron-hydrogenase whose cellular activity is dependent upon tungsten. J. Biol. Chem. 266, 13834–13841 (1991).
Selig, M., Xavier, K. B., Santos, H. & Schönheit, P. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167, 217–232 (1997).
Zhang, Y. & Gladyshev, V. N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 379, 881–899 (2008).
Anbar, A. D. et al. A whiff of oxygen before the Great Oxidation Event? Science 317, 1903–1906 (2007).
Neubert, N., Nägler, T. F. & Böttcher, M. E. Sulfidity controls molybdenum isotope fractionation into euxinic sediments: evidence from the modern Black Sea. Geology 36, 775–778 (2008).
Helz, G. R. et al. Mechanism of molybdenum removal from the sea and its concentration in black shales: EXAFS evidence. Geochim. Cosmochim. Acta 60, 3631–3642 (1996).
Shen, Y., Buick, R. & Canfield, D. E. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature 410, 77–81 (2001).
Dodsworth, J. A. et al. Thermoflexus hugenholtzii gen. nov., sp. nov., a thermophilic, microaerophilic, filamentous bacterium representing a novel class in the Chloroflexi, Thermoflexia classis nov., and description of Thermoflexaceae fam. nov. and Thermoflexales ord. nov. Int. J. Sys. Evol. Microbiol. 64, 2119–2127 (2014).
Hanada, S., Hiraishi, A., Shimada, K. & Matsuura, K. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int. J. Sys. Evol. Microbiol. 45, 676–681 (1995).
Murugapiran, S. K. et al. Thermus oshimai JL-2 and T. thermophilus JL-18 genome analysis illuminates pathways for carbon, nitrogen, and sulfur cycling. Stand. Genom. Sci. 7, 449–468 (2013).
Kozich, J. J., Westcott, S. L., Baker, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Friel, A. D. et al. Microbiome shifts associated with the introduction of wild atlantic horseshoe crabs (Limulus polyphemus) into a touch-tank exhibit. Front. Microbiol. 11, 1398 (2020).
Hamilton, T. L., Peters, J. W., Skidmore, M. L. & Boyd, E. S. Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J. 7, 1402–1412 (2013).
Courtois, S. et al. Quantification of bacterial subgroups in soil: comparison of DNA extracted directly from soil or from cells previously released by density gradient centrifugation. Environ. Microbiol. 3, 431–439 (2001).
Pernthaler, A. & Pernthaler, J. In Protocols for Nucleic Acid Analysis by Nonradioactive Probes 353, 153–164 (Humana Press, 2007).
Pett-Ridge, J. & Weber, P. K. In Microbial Systems Biology 91–136 (Humana, New York, NY, 2022). https://doi.org/10.1007/978-1-0716-1585-0_6
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2020).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 1–11 (2010).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Aziz, R. K. et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9, 1–15 (2008).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Kück, P. & Longo, G. C. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 11, 1–8 (2014).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Jacox, E., Chauve, C., Szöllősi, G. J., Ponty, Y. & Scornavacca, C. ecceTERA: comprehensive gene tree-species tree reconciliation using parsimony. Bioinformatics 32, 2056–2058 (2016).
Chevenet, F. et al. SylvX: a viewer for phylogenetic tree reconciliations. Bioinformatics 32, 608–610 (2016).
Csűös, M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26, 1910–1912 (2010).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Wu, S., Skolnick, J. & Zhang, Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 5, 17 (2007).
Holm, L. & Rosenstrïm, P. I. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019).
MacQueen, J. In Some Methods for Classification and Analysis of Multivariate Observations 1, 281–297 (1967).
Ma, K. & Adams, M. W. W. Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J. Bacteriol. 176, 6509–6517 (1994).
Acknowledgements
We thank Jonathan Covington (UNLV) for help assembling a repository of tungstoenzymes, Jim Noblet and Andreas Beyersdorf (CSUSB) for assistance with ICP-OES, Dave and Sandy Jamieson for access to GBS, high school teachers Kathleen Diver, Heather Garcia, Juan Salgado, and Chuong Vu for assistance with fieldwork, and Prof. Aharon Oren for assistance on nomenclature. We acknowledge Tikitere Trust for their enthusiasm for our research and assistance in access and sampling of the Hell’s Gate geothermal features. We extend our gratitude to Mackenzie Lynes and Roland Hatzenpichler for permitting use of metagenome 3300028735 and providing geochemistry information on hot spring LCB-006, as well as to Donald Bryant, Vera Thiel, Mircea Podar, Susannah Tringe, Robert Kelly, Brian Yu, and William Inskeep for sequence data use permissions. We also thank Everett Shock for discussions on W geochemistry in hot springs. Part of this work was carried out at Lawrence Livermore National Laboratory (LLNL) under Contract DE-AC52-07NA2734. Funding was provided by the U.S. National Science Foundation (DEB 1557042 and EAR-1820658), NASA (80NNSC17KO548 and 80NSSC19M0150), and the National Natural Science Foundation of China (No. 91951205).
Author information
Authors and Affiliations
Contributions
J.A.D., B.P.H., A.E.D., S.B. and M.P. conceived of the study. J.D., M.M., C.G., L.G., C.S., C.V., M.Z., N.B., and F.R. conducted cultivation experiments, monitoring, and quantitative PCR. S.B. and J.A.D. analyzed qPCR and 16S data. D.M. performed CARD-FISH. X.M., P.K.W., and J.P.-R. analyzed culture samples with nanoSIMS and S.B. and A.E.D. joined for the nanoSIMS data analysis/interpretation. J.-Y.J., D.R.C., L.M.K., E.S.J., E.S.B., A.D.C., B.S.C., Z.-S.H., W.-J.L., A.-L.R., and M.B.S. were involved in metagenomic sampling, sequencing, assembly, annotation and binning. M.P. performed phylogenomic analyses and ACS reconstructions. S.B. carried out the protein structural modeling. S.B., M.P., and J.A.D. wrote the manuscript and all authors contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buessecker, S., Palmer, M., Lai, D. et al. An essential role for tungsten in the ecology and evolution of a previously uncultivated lineage of anaerobic, thermophilic Archaea. Nat Commun 13, 3773 (2022). https://doi.org/10.1038/s41467-022-31452-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-31452-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.