Abstract
Evolution of xeno nucleic acid (XNA) world essentially requires template-directed synthesis of XNA polymers. In this study, we demonstrate template-directed synthesis of an acyclic XNA, acyclic l-threoninol nucleic acid (l-aTNA), via chemical ligation mediated by N-cyanoimidazole. The ligation of an l-aTNA fragment on an l-aTNA template is significantly faster and occurs in considerably higher yield than DNA ligation. Both l-aTNA ligation on a DNA template and DNA ligation on an l-aTNA template are also observed. High efficiency ligation of trimer l-aTNA fragments to a template-bound primer is achieved. Furthermore, a pseudo primer extension reaction is demonstrated using a pool of random l-aTNA trimers as substrates. To the best of our knowledge, this is the first example of polymerase-like primer extension of XNA with all four nucleobases, generating phosphodiester bonding without any special modification. This technique paves the way for a genetic system of the l-aTNA world.
Similar content being viewed by others
Introduction
The RNA world is widely thought to be a stage in the origin of life1,2,3,4. At this stage, RNA self-replication must have occurred from simple inorganic and organic molecules that existed on ancient Earth. To provide support for the RNA world hypothesis, nonenzymatic self-replication of oligoribonucleotide via efficient chemical ligation of monomer units or short fragments under aqueous conditions in the presence of metal ions and simple activators has been investigated. Orgel and co-workers have primarily reported that activated nucleotide monomers bearing a leaving group on phosphate can be polymerized on a template oligonucleotide to produce a complementary strand5,6. Richert’s group Hagenbuch et al. and Deck et al. utilized azabenzotriazolide as an effective leaving group and a helper strand to achieve effective primer extension7,8. Orgel and Szostak introduced an amino group into 2′- and 3′-positions9, which enabled effective RNA copying due to acceleration of the ligation reaction10,11. These investigations indicated that prebiotic conditions may have allowed ribo-oligonucleotide elongation on a template strand in the absence of a protein enzyme.
Why nature chose ribofuranosyl nucleic acids rather than some other family of molecular structure as the carrier of genetic material12 has prompted chemists to synthesize various xeno nucleic acids (XNAs) composed of unnatural scaffolds13,14. Cyclic XNAs such as hexitol nucleic acid15, cyclohexenyl nucleic acid16, tricyclo-DNA17, arabinonucleic acid18, and (3′,2′)-α-L-threose nucleic acid (TNA)12,19 form homoduplexes and cross-pair with DNA and/or RNA. Due to the structural similarities to DNA and RNA, cyclic XNAs can be polymerized on a template using native and engineered polymerases, and functional XNAs that act as ribozymes and aptamers have been identified using in vitro selection20,21,22,23,24,25,26. TNA has particularly attracted attention because it is chemically simpler than RNA and may have existed prior to RNA19,27,28,29,30. Interestingly, a ring structure is not necessary for stable duplex formation. Several XNAs composed of acyclic structures have been synthesized31,32,33,34,35,36,37,38. An example is glycerol nucleic acid (GNA), designed by the Zhang et al.34. Although GNA does not have a ring structure, it forms very stable homoduplex and can cross-pair with RNA, depending on the sequence. As other examples, we have reported serinol nucleic acid39 and acyclic l-threoninol nucleic acid (l-aTNA; Fig. 1a)40,41, which can cross-hybridize with DNA and RNA and has high nuclease durability, promising application to wide variety of situations such as living cell system42,43,44. Not only our group but Kumar, Gothelf, et al. Kumar et al., Kumar and Gothelf, and Kumar et al. have also reported unique properties of l-aTNA45,46,47.
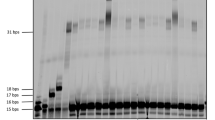
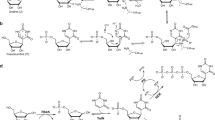
a Chemical structures of DNA and l-aTNA. b Sequences of 8-mer fragments and 16-mer templates. c Schematic illustration of the chemical ligation reaction. Denaturing PAGE analysis of d l-aTNA ligation reaction and e DNA ligation reaction. Lane 1, X8a as a marker; lane 2, reaction products after 1 h; lane 3, reaction products after 4 h; lane 4, reaction products after 24 h; lane 5, products after reaction in the absence of template; lane 6, products after reaction in the absence of CNIm; and lane 7, products after reaction in the absence of Mn2+. Conditions: [X8a] = 0.9 μM, [X8b-p] = 1.1 μM, [X16t] = 0 or 1.0 μM, [NaCl] = 100 mM, [MnCl2] = 0 or 20 mM, [CNIm] = 0 or 20 mM, 4 °C. Source data are provided as a Source Data file. f Chemical ligation yield of l-aTNA (red circles) and DNA (black circles) after indicated times of reaction. g Chemical ligation yields of l-aTNA at indicated temperatures. Conditions: [oligomers] = 1.0 μM each, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM. Source data are provided as a Source Data file. h Ligation yields of l-aTNA at indicated concentrations of CNIm and Mn2+. Conditions: [T8a] = 0.9 μM, [T8b-p] = 1.1 μM, [T16t] = 1.0 μM, [NaCl] = 100 mM 25 °C, 30 min. Source data are provided as a Source Data file.
If template-directed synthesis of an XNA could occur in the absence of enzymes, the XNA world would become a more likely candidate for a pre-RNA world. Szostak’s group has used chemical ligation to polymerize 2′-amino-modified GNA48. Liu and co-workers reported that peptide nucleic acid (PNA) aldehydes could be chemically ligated on a DNA template, generating a complementary PNA strand49. Inspired by these works, we focused on template-mediated chemical ligation of l-aTNA as the first step of achieving the acyclic XNA world, because (1) l-aTNA is synthetically and structurally simple, (2) l-aTNA can form very stable homoduplex in an antiparallel manner, and (3) l-aTNA with clockwise helicity can also form stable duplex with RNA as well as DNA. A condensing reagent N-cyanoimidazole (CNIm) can connect a phosphate with a neighboring hydroxyl group to generate a phosphodiester bond, the same type of bond formed with natural enzymatic ligation50,51. For DNA, ligation of various types of structures were achieved using CNIm, due to relatively high reaction yields52,53,54,55,56. We therefore expected that CNIm could be used to ligate l-aTNA. If short fragments can be sequentially ligated on a template, selective and effective synthesis of complementary strands is possible as shown for other types of nucleic acids11,49,57,58.
Herein, we demonstrate template-directed chemical ligation of l-aTNA fragments, which is of interest from the viewpoint of the prebiotic world and from the standpoint of establishment of XNA-based artificial life. The chemical ligation of l-aTNA gives considerably higher yield and rate than DNA ligation. This effective ligation enables a pseudo-primer extension reaction using a pool of random l-aTNA trimers as substrates.
Results and discussion
Template-directed chemical ligation of l-aTNA in the presence of N-cyanoimidazole and divalent metal ion
We synthesized an 8-mer l-aTNA fragment with the fluorophore Cy3 conjugated at the 3′ terminus (T8a) and a fragment carrying a phosphate at the 3′ terminus (T8b-p). Both are complementary to a 16-mer l-aTNA template (T16t) (Fig. 1b). These three strands can form a duplex with single nick at the center. Upon addition of CNIm, we expected that the 1′-OH of T8a and the 3′-phosphate of T8b-p would be connected to generate a phosphodiester linkage (Fig. 1c). DNA oligonucleotides of almost the same sequences (D8a, D8b-p, and D16t) were prepared as a comparison. First, we evaluated ligation efficiency of the l-aTNA fragments upon addition of CNIm in the presence of Mn2+, conditions used in previous reports50,54. The ligation reaction was performed at 4 °C, a temperature at which both l-aTNA and DNA strands form duplexes. Products were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). Over time, the intensity of the band corresponding to T8a decreased and that of a band with slower mobility increased (Fig. 1d). The analysis of the reaction mixture by MALDI-TOF MS revealed that this new band contained the desired 16-mer ligation product of T8a and T8b-p (Calcd: 5879.6, Obs: 5877.3, Supplementary Fig. 1). This ligated product was not observed in the absence of T16t, Mn2+, or CNIm. The ligation reaction efficiency was high with 95% yield after 4 h. In contrast, the DNA ligation occurred in only 10% under the same conditions, and only 47% yield of DNA product was detected after 24 h (Fig. 1e, f). The observed rate constants (kobs) were estimated as 1.4 h−1 for l-aTNA and 0.027 h−1 for DNA. Thus, the l-aTNA ligation rate was about 50-fold higher than the DNA ligation rate.
Examination of ligation reaction yields over a range of temperatures revealed that the reaction rate was accelerated when the temperature was elevated to 50 °C (Fig. 1g and Supplementary Fig. 2): kobs was 6.4, 16, and 21 h−1 at 25, 40, and 50 °C, respectively, resulting in almost 90% yield within 20 min at temperatures from 25 to 50 °C. Note that the reaction rate of l-aTNA octamer ligation was comparable to that of 3′-amino oligonucleotides with 2-methylimidazole-activated phosphate (about 7 h−1 at optimized conditions for decamer fragment)11, both are much faster than the rate of the chemical ligation to form a phosphodiester bond. In contrast, the l-aTNA octamer ligation reaction was suppressed at temperatures higher than the melting temperatures (Tm) of the T8b-p/T16t duplex (59.5 °C), and thus no reaction was observed at 70 °C. This suggests that duplex formation between fragments and template is necessary for ligation, although CNIm may degrade at high temperatures. Examinations of the concentration dependences on CNIm and Mn2+ showed that 20 mM CNIm and 20 mM Mn2+ were sufficient for effective ligation (Fig. 1h and Supplementary Fig. 3). We also determined yields in Cd2+, Co2+, Ni2+, and Zn2+ and showed that each of these metal ions could substitute for Mn2+ (Supplementary Fig. 4). Thus, a wide variety of divalent metals induce ligation of l-aTNA.
Position of phosphate and thermal stability of fragment duplex influence ligation rate
In order to examine the reasons for the remarkably high efficiency of l-aTNA ligation, we prepared and analyzed ligation reactions of l-aTNA and DNA of different sequences (Fig. 2a). T8a-p and T8b were designed to examine the effect of the position of the phosphate position on the ligation. Several 16-mer DNA fragments (D16a, D16b-p, D16a-p, and D16b) and a 32-mer template (D32t) were synthesized to prepare DNA duplexes with Tm values similar to that of 8-mer l-aTNA fragments and the 16-mer l-aTNA template (Fig. 2b and Supplementary Fig. 5). Interestingly, the position of terminal phosphate influenced the reaction rate. In the case of l-aTNA, the ligation was faster for the 1′-OH/3′-phosphate pair (T8a/T8b-p/T16t, kobs = 6.4 h−1) than for the 1′-phosphate/3′-OH pair (T8a-p/T8b/T16t, kobs = 0.61 h−1; Fig. 2c and Supplementary Fig. 6). For DNA, the ligation was faster for the 5′-OH/3′-phosphate pair (D16a-p/D16b/D32t, kobs = 0.71 h−1) than for the 5′-phosphate/3′-OH pair (D16a/D16b-p/D32t, kobs = 0.22 h−1, Fig. 2c and Supplementary Fig. 6). These differences could be partially explained by steric hindrance around the OH group, which acts as a nucleophile to attack the phosphorous atom of the phosphate activated by CNIm50. Both the 1′-OH on l-aTNA and the 5′-OH on DNA are less bulky primary hydroxyl groups than are the 3′-OH groups, which are bulky secondary hydroxyl groups (Fig. 2d). The same trend was reported for the chemical ligation of DNA using BrCN59 and EDC60 with nick ligation between 3′-phosphate/5′-OH resulting in higher yields than ligation between the 5′-phosphate/3′-OH pair. Thus, the primary hydroxyl group is an important factor in reactivity of chemical ligation as Ashley et al. predicted60.
a Sequences of 8-mer fragments and 16-mer template of l-aTNA and 16-mer fragments and 32-mer template of DNA. b Melting temperatures of fragment/template duplexes. c Yield as a function of time for the chemical ligation reactions of l-aTNA (red circles) and of DNA (black circles) with either primary (closed circles) or secondary OH termini (open circles). Conditions: [oligomers] = 1.0 μM each, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM, 25 °C. Source data are provided as a Source Data file. d Schematic illustration of chemical ligation focusing on condensation between OH and phosphate.
For the ligation of substrates with primary OH termini, T8a/T8b-p was much faster than that of D16a-p/D16b (Fig. 2c). Those containing a secondary OH group had the same tendency with l-aTNA more reactive than DNA. These results indicate that a mechanism in addition to the reactivity of the OH enhances l-aTNA ligation. The energy minimized structure of the l-aTNA duplex obtained from a computer calculation suggested that there is formation of a hydrogen bond between the carbonyl oxygen and the terminal OH group (Supplementary Fig. 7a–e), this would enhance nucleophilicity of the oxygen of the OH group. Another possibility is that Mn2+ complexes with the carbonyl of the amide, the phosphate, and the OH group to optimally position the phosphate and OH for ligation (Supplementary Fig. 7f).
Homo- and heteroligations of DNA and l-aTNA are mediated by templates
In order to determine whether interconversion of sequence information between l-aTNA and DNA or RNA is possible, ligation efficiencies of combinations of l-aTNA, DNA, and RNA fragments and templates were evaluated (Fig. 3a). Ligation yields after 24 h reactions indicated that T8a was more reactive than D8a (Fig. 3b and Supplementary Fig. 8), expected as the 1′-OH of l-aTNA has higher nucleophilicity than does the 3′-OH of DNA. Almost complete ligation of T8a/T8b-p was observed irrespective of the templates (Fig. 3b), indicating the possibility of transfer of sequence information encoded in DNA and RNA into l-aTNA. D8a/D8b-p ligation occurred on the l-aTNA template (T16t) although the yield was not high (Fig. 3b). Thus, this chemical ligation technique possibly enables interconversion of genetic information between DNA and l-aTNA. Ligation of fragments of different scaffolds was also observed. T8a/D8b-p was ligated in high yield on D16t and R16t templated and with slightly lower yield on T16t, whereas ligation of D8a/T8b-p was observed only on T16t (Fig. 3b). The ligation on D16t and on R16t gave comparable efficiency, presumably indicating that the helical structure of the duplexes did not so much affect the efficacy of ligation.
a Sequences of 8-mer fragments and 16-mer template. b Ligation yields of indicated pairs of fragments on different templates. T8a/T8b-p (red bars), T8a/D8b-p (yellow bars), D8a/T8b-p (green bars), and D8a/D8b-p (blue bars) were ligated. Conditions: [X8a] = 0.9 μM, [X8b-p] = 1.1 μM, [X16t] = 1.0 μM, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM, 4 °C, 24 h. Source data are provided as a Source Data file. c Sequences of 8-mer primer and fragments and 16-mer template. d Ligation yields for fragments of different lengths as a function of time. 5-mer fragment (red circles), 4-mer fragment (blue circles), 3-mer fragment (yellow circles), and 2-mer fragment (green circles) were used. Conditions: [T8primer] = 0.9 μM, [T16t] = 1.0 μM, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM, concentration of Tnb-p strands is indicated in legend, 4 °C. Source data are provided as a Source Data file.
l-aTNA ligation with short fragments
We reasoned that the high reaction rate and yield of l-aTNA ligation should enable the elongation of shorter fragments in a template-directed fashion. Ligation of l-aTNA fragments of 2-, 3-, 4-, and 5-mer (T2b-p, T3b-p, T4b-p, and T5b-p, respectively) to the 8-mer primer strand (T8primer) on T16t template was evaluated (Fig. 3c). T5b-p and T4b-p were almost completely ligated to the primer within 6 h at 4 °C with almost equivalent reaction rates of 0.82 h−1 for T5b-p and 0.78 h−1 for T4b-p (Fig. 3d and Supplementary Fig. 9). This indicates that rate determining step is the ligation and not hybridization when sufficiently long fragments are used. In contrast, about 60% of T3b-p was ligated after 24 h and the observed reaction rate was 0.056 h−1, and only about 10% of T2b-p was ligated under these conditions (kobs = 0.0034 h−1; Fig. 3d and Supplementary Fig. 9). By increasing the concentration of fragments to 50 μM, the reaction efficacy was considerably higher; about 80% ligation of T3b-p and about 20% ligation of T2b-p was observed after 24 h (kobs = 0.22 h−1 for T3b-p and kobs = 0.018 h−1 for T2b-p; Fig. 3d and Supplementary Fig. 9). This suggests that the rate determining step for trimer and dimer fragments is binding to the template. Undesired intrastrand cyclization of fragments between phosphate and OH would be chemically possible58,61, however l-aTNA fragments did not show such cyclization under the present ligation conditions (Supplementary Fig. 10). These data indicated that enzyme-free primer extension of trimer fragments should be possible.
l-aTNA trimers can be sequentially ligated
We first sought to demonstrate sequential ligation using the 8-mer primer (T8primer) and three trimer fragments (T3b-p, T3c-p, and T3d-p), all of which are complementary to a 17-mer template (T17t) (Fig. 4a, b). In the presence of all three trimers, the signal due to T8primer as resolved by PAGE gradually decreased and three new bands appeared that migrated more slowly (Fig. 4c), indicating elongation of T8primer. From the comparison to products of the reactions in the absence of both T3c-p and T3d-p and in the absence of T3d-p, the product bands were assigned to 11-mer and 14-mer intermediates and full-length 17-mer product. The yield of full-length product increased with time (Fig. 4d). Initially, 11- and 14-mer intermediates accumulated, then levels of the intermediates were reduced, and concurrently full-length product signal increased. After 8 h, almost all the intermediates and T8primer were converted into full-length 17-mer product. Ligation did not proceed at all without T3b-p which hybridizes to the template immediately adjacent to the primer, this demonstrated the template-directed specificity of the reaction.
a Sequences used for sequential ligation of trimers. b Schematic illustration of the sequential, template-directed ligation of trimers. c Denaturing PAGE analysis of sequential ligation. Lane 1, T8primer as a marker; lane 2, reaction products after 1 h; lane 3, reaction products after 2 h; lane 4, reaction products after 4 h; lane 5, reaction products after 8 h; lane 6, reaction products after 24 h; lane 7, reaction products in the absence of T3c-p and T3d-p; lane 8, reaction products in the absence of T3d-p; lane 9, reaction products in the absence of T3b-p and T3d-p; and lane 10, reaction products in the absence of T3b-p and T3c-p. Conditions: [T8primer] = 0.9 μM, [T3b-p] = [T3c-p] = [T3d-p] = 50 μM, [T17t] = 1.0 μM, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM, 4 °C. Source data are provided as a Source Data file. d Ligation yield determined from PAGE. Blue triangle, green square, and red circle represent 11-mer intermediate, 14-mer intermediate, and 17-mer full-length product, respectively. Source data are provided as a Source Data file. e MALDI-TOF MS spectra and observed m/z of reaction mixture containing all components analyzed after 24 h (red), of reaction products in the absence of T3c-p and T3d-p, which yielded only 11-mer (blue line), and of reaction products in the absence of T3d-p, which yielded 14-mer (green line). Peaks corresponding to the template (T17t) and T8primer were also observed. Source data are provided as a Source Data file.
Interestingly, the reaction rate in the presence of all three fragments was much faster than that of the single ligation of T3b-p under the same conditions; the single ligation of T3b-p gave about 40% conversion of the primer after 2 h, and the reaction was not completed even after 24 h. In contrast, the conversion of the primer in the presence of T3b-p, T3c-p, and T3d-p was more than 90% after 2 h and nearly quantitative production of the full-length product was observed after 24 h (Supplementary Fig. 11). The kobs clearly reflected the difference in the efficacy: kobs was 1.2 h−1 in the presence of all three fragments, whereas kobs for ligation of the single trimer was 0.27 h−1. The trimers that hybridized downstream sequences presumably acted as helper strands as previously described8,62,63. In our case, trimer fragments would mutually assist hybridization with the template via terminal stacking interactions resulting in acceleration of the ligation through preorganized complex formation (Supplementary Fig. 12). MALDI-TOF mass spectrometry confirmed that the desired 17-mer product was successfully generated in the presence of all components; 14-mer and 11-mer intermediates were also detected (Fig. 4e). Tetramer fragments were also ligated to the primer in a sequential, template-directed manner with higher reaction rate and yield than observed with trimers (Supplementary Fig. 13). It is worth noting that almost no impurities derived from nontemplated reactions were detected by either PAGE or mass spectroscopy, indicating that complementary strand production occurs with high fidelity.
Template-mediated synthesis of complementary l-aTNA strand from random trimer pool
Finally, we demonstrate template-mediated synthesis of l-aTNA from random trimer pool (T3mix-p) that was chemically synthesized from l-aTNA phosphoramidites of four nucleobases. The extension reaction of the T8primer on the T17t template was performed in the presence of the T3mix-p (Fig. 5a). As expected, elongation of random trimer fragments into 17-mer full-length product was observed on denaturing PAGE: intermediates were produced at early stage of the reaction, whereas full-length product was observed after the consumption of intermediates (Fig. 5b). More than 75% of T8primer was converted to 17-mer full-length product after 24 h. Although a low intensity band (less than 10%) derived from unspecific ligation between template and trimer fragment was observed at the upper position, sufficiently selective elongation reaction was confirmed. MALDI-TOF mass spectrometry of the reaction mixture indicated that elongation resulted in an oligomer of sequence complementary to the template (Fig. 5c). Neither intermediates nor full-length product were observed in the absence of the template T17t. These results clearly demonstrated that template-directed polymerization of l-aTNA was achieved nonenzymatically utilizing a simple chemical catalyst without any special modification on the polymerized trimers. To the best of our knowledge, this is the first example of template-directed polymerase-like, chemical primer extension of acyclic XNA from a random fragment pool. Importantly, the natural phosphodiester linkage was generated without any special modification of the l-aTNA fragments through a fast and effective chemical ligation.
a Illustration of primer extension in the presence of T3mix-p pool. b Denaturing PAGE analysis of reaction with randomized trimers. Lane 1, T8primer as a marker; lane 2, reaction with only T3b-p as substrate to yield 11-mer intermediate; lane 3, reaction with T3b-p and T3c-p as substrates to yield 14-mer intermediate; lane 4, reaction with T3b-p, T3c-p, and T3d-p as substrates to yield full-length 17-mer; lane 5, reaction with T3mix-p as substrates after 1 h; lane 6, reaction with T3mix-p as substrates after 3 h; lane 7, reaction with T3mix-p as substrates after 6 h; lane 8, reaction with T3mix-p as substrates after 24 h; and lane 9, control without template. Conditions: [T8primer] = 0.9 μM, [T3mix-p] = 100 μM (total), [T17t] = 1.0 μM, [NaCl] = 100 mM, [MnCl2] = 20 mM, [CNIm] = 20 mM, 4 °C. Source data are provided as a Source Data file. c MALDI-TOF MS of reaction with T3mix-p as substrates after 24 h. Source data are provided as a Source Data file.
The template-directed, chemical ligation of l-aTNA using CNIm was considerably faster than ligation of DNA under the same conditions. Homo- and heteroligations of l-aTNA and DNA were also observed. This indicates that genetic information encoded in DNA and RNA sequence could possibly be transcribed into l-aTNA, and also reverse transcribed into DNA. The l-aTNA ligation was also observed when dimers were used as substrates although the reaction was less efficient than was ligation of longer fragments. Sequential ligation of l-aTNA with three trimer fragments was accomplished, and almost complete conversion into full-length product was attained. No products noncomplementary to the template were detected. The selective, template-directed elongation of l-aTNA was demonstrated in the presence of a random l-aTNA trimer pool, establishing that synthesis of a complementary strand was possible. To the best of our knowledge, this is the first report of template-directed synthesis of an acyclic XNA containing all four nucleobases by generating phosphodiester linkages without enzymes.
High reaction rate and yield was observed in the presence of condensation reagent but without any special modification on the fragments. As XNAs have acyclic scaffolds very different structurally from natural nucleic acid, it might be difficult to engineer a polymerase for primer extension of XNA. Our strategy for enzyme-free copying of sequence information of acyclic XNA is an attractive system for replication for construction of artificial life64,65,66 and highly functional biological tools composed of acyclic XNA via in vitro selection67,68.
Our data indicate that l-aTNA could have been a precursor of RNA, because the ligation of l-aTNA on DNA and RNA templates and of DNA on an l-aTNA template was possible using CNIm, a possible prebiotic condensation agent69. l-threoninol, a scaffold of l-aTNA, could be generated by reduction of l-threonine or its precursor that was possibly generated by prebiotic chemistry70. Possibility of prebiotic synthesis of nucleobase-acetic acid has already been shown as potential component of PNA71. Therefore, l-aTNA has a potential to be synthesized at prebiotic conditions. Our results show that exchange of sequence information between l-aTNA and the nucleic acids of the modern world is possible. The nonenzymatic, template-directed l-aTNA elongation is the foundation for a new genetic system of the XNA world.
Methods
All experimental procedures and characterization data are provided in Supplementary Information.
General procedure for chemical ligation by CNIm
A typical reaction in 30 μL volume was performed as follows: an aliquot of solution containing the required amount of oligomer was added to 6.0 μL of 500 mM NaCl aq and 3.0 μL of 200 mM MnCl2 aq. The mixture was diluted with autoclaved Milli-Q water to 27 μL before starting the reaction by the addition of 3.0 μL 200 mM N-cyanoimidazole aq. The desired temperature was maintained using a thermal cycler. For the time-course evaluation, 4.0 μL of the reaction mixture was taken into another microtube, and 4.0 μL of quenching mix (aqueous solution of 12.5 mM NaOH and 125 mM EDTA) was added. After the addition of 2.0 μL loading buffer (glycerol:formamide:0.5 M EDTA, 6:6:1v/v/v, containing 0.06 % bromophenol blue), 2.5 μL of the quenched sample was applied to a denaturing PAGE (20% acrylamide, 8 M urea). Electrophoresis was performed at 4 W for 2–3 h. We found that denaturing PAGE does not completely dissociate an l-aTNA/l-aTNA duplex when the sequence is long, due to high duplex stability. We therefore kept the temperature of electrophoresis apparatus below room temperature so that only unreacted 8-mer primer dissociated from the template. Under these PAGE conditions, elongation products of longer than 11-mer formed duplexes with the template during PAGE, resulting in a large difference in migration distance between the primer and the products. After the run, gel was analyzed using a Typhoon FLA 9500, and intensities of gel bands were quantified using an ImageQuant TL. kobs was estimated by data fitting assuming first-order reactions, using yield in the first three time points at time-course experiment.
MALDI-TOF MS analysis of reaction mixtures
A 50-μL aliquot of the reaction mixture was added to 7.0 μL of 3.0 M sodium acetate aq and 120 μL of absolute ethanol, and the solution was mixed well. After centrifuging at 16,000 × g for 5 min at 4.0 °C, 120 μL aqueous solution of 70% (v/v) ethanol was added and mixed before centrifuging at 16,000 × g for 5 min at 4.0 °C. The supernatant was decanted, and the pellet was air-dried. The residue was dissolved in 5.0 μL of water and was analyzed by MALDI-MS using 3-hydroxypicolinic acid as a matrix.
Statistics and reproducibility
Experiments in Figs. 1d, e, 4c, and 5b and Supplementary Fig. 6 were repeated three times and experiments in Supplementary Figs. 2–4, 8, 9, 11a, and 13c were repeated twice, which gave independently similar results.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data presented in the manuscript are available upon request from the corresponding authors. Raw data are provided as a Source Data file. Source data are provided with this paper.
References
Gilbert, W. The RNA world. Nature 319, 618 (1986).
Orgel, L. Prebiotic chemistry and the origin of life. Crit. Rev. Biochem. Mol. Biol. 39, 99–123 (2004).
Robertson, M. P. & Joyce, G. F. The origins of the RNA World. Cold Spring Harb. Perspect. Biol. 4, 1 (2012).
Adamala, K. & Szostak, J. W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342, 1098–1100 (2013).
Weimann, B. J., Lohrmann, R., Orgel, L. E., Schneider-Bernloehr, H. & Sulston, J. E. Template-directed synthesis with adenosine-5′-phosphorimidazolide. Science 161, 387 (1968).
Inoue, T. & Orgel, L. A nonenzymatic RNA polymerase model. Science 219, 859–862 (1983).
Hagenbuch, P., Kervio, E., Hochgesand, A., Plutowski, U. & Richert, C. Chemical primer extension: efficiently determining single nucleotides in DNA. Angew. Chem. Int. Ed. 44, 6588–6592 (2005).
Deck, C., Jauker, M. & Richert, C. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat. Chem. 3, 603–608 (2011).
Lohrmann, R. & Orgel, L. E. Template-directed synthesis of high molecular weight polynucleotide analogues. Nature 261, 342–344 (1976).
Mansy, S. S. et al. Template-directed synthesis of a genetic polymer in a model protocell. Nature 454, 122–125 (2008).
Zhou, L., O’Flaherty, D. K. & Szostak, J. W. Template-directed copying of RNA by non-enzymatic ligation. Angew. Chem. Int. Ed. Engl. https://doi.org/10.1002/anie.202004934 (2020).
Scöning, K.-U. et al. Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3′→2′) oligonucleotide system. Science 290, 1347–1351 (2000).
Leumann, C. J. DNA analogues: from supramolecular principles to biological properties. Bioorg. Med. Chem. 10, 841–854 (2002).
Chaput, J. C. & Herdewijn, P. What is XNA? Angew. Chem. Int. Ed. 58, 11570–11572 (2019).
Hendrix, C. et al. 1′, 5′ -Anhydrohexitol oligonucleotides: synthesis, base pairing and recognition by regular oligodeoxyribonucleotides and oligoribonucleotides. Chem. Eur. J. 3, 110–120 (1997).
Wang, J. et al. Cyclohexene nucleic acids (CeNA): Serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. J. Am. Chem. Soc. 122, 8595–8602 (2000).
Steffens, R. & Leumann, C. J. Tricycio-DNA: a phosphodiester-backbone based DNA analog exhibiting strong complementary base-pairing properties. J. Am. Chem. Soc. 119, 11548–11549 (1997).
Noronha, A. M. et al. Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA·RNA hybrid duplexes. Biochemistry 39, 7050–7062 (2000).
Orgel, L. A simpler nucleic acid. Science 290, 1306–1307 (2000).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Taylor, A. I. et al. Catalysts from synthetic genetic polymers. Nature 518, 427–430 (2015).
Houlihan, G., Arangundy-Franklin, S. & Holliger, P. Exploring the chemistry of genetic information storage and propagation through polymerase engineering. Acc. Chem. Res. 50, 1079–1087 (2017).
Mei, H. et al. Synthesis and evolution of a threose nucleic acid aptamer bearing 7-deaza-7-substituted guanosine residues. J. Am. Chem. Soc. 140, 5706–5713 (2018).
Eremeeva, E. et al. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res. 47, 4927–4939 (2019).
Levi-Acobas, F. et al. Compatibility of 5-ethynyl-2′F-ANA UTP with: In vitro selection for the generation of base-modified, nuclease resistant aptamers. Org. Biomol. Chem. 17, 8083–8087 (2019).
Pinheiro, V. B. & Holliger, P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 16, 245–252 (2012).
Joyce, C. F. & Orgel, L. E. Progress toward understanding the origin of the RNA world. in The RNA World 3rd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F.) 23–56 (Cold Spring Harbor Laboratory, 2006).
Herdewijn, P. TNA as a potential alternative to natural nucleic acids. Angew. Chem. Int. Ed. 40, 2249–2251 (2001).
Wu, X., Delgado, G., Krishnamurthy, R. & Eschenmoser, A. 2,6-Diaminopurine in TNA: effect on duplex stabilities and on the efficiency of template-controlled ligations. Org. Lett. 4, 1283–1286 (2002).
Blain, J. C., Ricardo, A. & Szostak, J. W. Synthesis and nonenzymatic template-directed polymerization of 2′-amino-2′-deoxythreose nucleotides. J. Am. Chem. Soc. 136, 2033–2039 (2014).
Schneider, K. C. & Benner, S. A. Oligonucleotides containing flexible nucleoside analogues. J. Am. Chem. Soc. 112, 453–455 (1990).
Nielsen, P. E., Egholm, M., Berg, R. H. & Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–1500 (1991).
Nielsen, P., Dreiøe, L. H. & Wengel, J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Med. Chem. 3, 19–28 (1995).
Zhang, L., Peritz, A. & Meggers, E. A simple glycol nucleic acid. J. Am. Chem. Soc. 127, 4174–4175 (2005).
Asanuma, H., Toda, T., Murayama, K., Liang, X. & Kashida, H. Unexpectedly stable artificial duplex from flexible acyclic threoninol. J. Am. Chem. Soc. 132, 14702–14703 (2010).
Karri, P., Punna, V., Kim, K. & Krishnamurthy, R. Base-pairing properties of a structural isomer of glycerol nucleic acid. Angew. Chem. Int. Ed. 52, 5840–5844 (2013).
Kumar, V., Gore, K. R., Pradeepkumar, P. I. & Kesavan, V. Design, synthesis, biophysical and primer extension studies of novel acyclic butyl nucleic acid (BuNA). Org. Biomol. Chem. 11, 5853–5865 (2013).
Luo, M. et al. Invading Escherichia coli genetics with a xenobiotic nucleic acid carrying an acyclic phosphonate backbone (ZNA). J. Am. Chem. Soc. 141, 10844–10851 (2019).
Kashida, H., Murayama, K., Toda, T. & Asanuma, H. Control of the chirality and helicity of oligomers of serinol nucleic acid (SNA) by sequence design. Angew. Chem. Int. Ed. 50, 1285–1288 (2011).
Murayama, K., Kashida, H. & Asanuma, H. Acyclicl-threoninol nucleic acid (l-aTNA) with suitable structural rigidity cross-pairs with DNA and RNA. Chem. Commun. 51, 6500–6503 (2015).
Kamiya, Y. et al. Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of hetero duplexes of L-aTNA/RNA and SNA/RNA. Commun. Chem. https://doi.org/10.1038/s42004-020-00400-2 (2020).
Le, B. T. et al. Antisense oligonucleotide modified with serinol nucleic acid (SNA) induces exon skipping in: Mdx myotubes. RSC Adv. 7, 34049–34052 (2017).
Kamiya, Y. et al. Introduction of 2,6-diaminopurines into serinol nucleic acid improves anti-miRNA performance. ChemBioChem 18, 1917–1922 (2017).
Murayama, K., Kamiya, Y., Kashida, H. & Asanuma, H. Ultrasensitive molecular beacon designed with totally serinol nucleic acid (SNA) for monitoring mRNA in cells. ChemBioChem 16, 1298–1301 (2015).
Kumar, V., Kesavan, V. & Gothelf, K. V. Highly stable triple helix formation by homopyrimidine (L)-acyclic threoninol nucleic acids with single stranded DNA and RNA. Org. Biomol. Chem. 13, 2366–2374 (2015).
Kumar, V. & Gothelf, K. V. Synthesis and biophysical properties of (L)-aTNA based G-quadruplexes. Org. Biomol. Chem. 14, 1540–1544 (2016).
Kumar, V., Nguyen, T. J. D., Palmfeldt, J. & Gothelf, K. V. Formation of i-motifs from acyclic (L)-threoninol nucleic acids. Org. Biomol. Chem. 17, 7655–7659 (2019).
Chen, J. J., Cai, X. & Szostak, J. W. N2′→ P3′ phosphoramidate glycerol nucleic acid as a potential alternative genetic system. J. Am. Chem. Soc. 131, 2119–2121 (2009).
Brudno, Y., Birnbaum, M. E., Kleiner, R. E. & Liu, D. R. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat. Chem. Biol. 6, 148–155 (2010).
Kanaya, E. & Yanagawa, H. Template-directed polymerization of oligoadenylates using cyanogen bromide. Biochemistry 25, 7423–7430 (1986).
Ferris, J. P., Huang, C. H. & Hagan, W. J. N-cyanoimidazole and diimidazole imine: water-soluble condensing agents for the formation of the phosphodiester bond. Nucleosides Nucleotides 8, 407–414 (1989).
Luebke, K. J. & Dervan, P. B. Nonenzymatic ligation of oligodeoxyribonucleotides on a duplex DNA template by triple-helix formation. J. Am. Chem. Soc. 111, 8733–8735 (1989).
Li, T. & Nicolaou, K. C. Chemical self-replication of palindromic duplex DNA. Nature 369, 218–221 (1994).
Zuber, G. & Behr, J. P. Nonenzymatic plasmid ligation mediated by minor groove-binding molecules. Biochemistry 33, 8122–8127 (1994).
Li, T. et al. Construction of circular oligodeoxyribonucleotides on the new structural basis of i-motif. J. Am. Chem. Soc. 123, 12901–12902 (2001).
Chen, H. et al. Template-directed chemical ligation to obtain 3’-3’ and 5’-5’ phosphodiester DNA linkages. Sci. Rep. 4, 5–10 (2014).
Chen, Z., Lichtor, P. A., Berliner, A. P., Chen, J. C. & Liu, D. R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem. 10, 420–427 (2018).
Sosson, M., Pfeffer, D. & Richert, C. Enzyme-free ligation of dimers and trimers to RNA primers. Nucleic Acids Res. 47, 3836–3845 (2019).
Sokolova, N. I., Ashirbekova, D. T., Dolinnaya, N. G. & Shabarova, Z. A. Chemical reactions within DNA duplexes cyanogen bromide as an effective oligodeoxyribonucleotide coupling agent. FEBS Lett. 232, 153–155 (1988).
Ashley, G. W. & Kushlan, D. M. Chemical synthesis of oligodeoxynucleotide dumbbells. Biochemistry 30, 2927–2933 (1991).
Ferris, J. P., Ertem, G. & Agarval, V. Mineral catalysis of the formation of dimers of 5’-AMP in aqueous solution: the possible role of Montmorillonite clays in the prebiotic synthesis of RNA. Origins Life. Evol. Biosph. 19, 165–178 (1989).
Vogel, S. R., Deck, C. & Richert, C. Accelerating chemical replication steps of RNA involving activated ribonucleotides and downstream-binding elements. Chem. Commun. 4922–4924 (2005).
Prywes, N., Blain, J. C., Del Frate, F. & Szostak, J. W. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife 5, 1–14 (2016).
Balaram, P. Synthesizing life. Curr. Sci. 85, 1509–1510 (2003).
Rasmussen, S., Chen, L., Nilsson, M. & Abe, S. Bridging nonliving and living matter. Artif. Life 9, 269–316 (2003).
Blain, J. C. & Szostak, J. W. Progress toward synthetic cells. Annu. Rev. Biochem. 83, 615–640 (2014).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Fahrenbach, A. C. et al. Common and potentially prebiotic origin for precursors of nucleotide synthesis and activation. J. Am. Chem. Soc. 139, 8780–8783 (2017).
Ritson, D. J. & Sutherland, J. D. Synthesis of aldehydic ribonucleotide and amino acid precursors by photoredox. Chem. Angew. Chem. Int. Ed. 52, 5845–5847 (2013).
Nelson, K. E., Levy, M. & Miller, S. L. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc. Natl Acad. Sci. USA 97, 3868–3871 (2000).
Acknowledgements
This work was supported by AMED under Grant number 20am0401007. The authors acknowledge the support from JSPS KAKENHI (grant JP18H03933 to H.A. and grant JP20K15399 to K.M.), from the Asahi Glass Foundation (H.A.), and from the Public Interest of Tatematsu (K.M.).
Author information
Authors and Affiliations
Contributions
K.M. and H.A. conceived and designed experiments. K.M., H.O., and T.K. performed all experiments. All authors analyzed data, discussed results, and cowrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Murayama, K., Okita, H., Kuriki, T. et al. Nonenzymatic polymerase-like template-directed synthesis of acyclic l-threoninol nucleic acid. Nat Commun 12, 804 (2021). https://doi.org/10.1038/s41467-021-21128-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-21128-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.