Abstract
Under high light conditions or UV radiation, tea plant leaves produce more flavonols, which contribute to the bitter taste of tea; however, neither the flavonol biosynthesis pathways nor the regulation of their production are well understood. Intriguingly, tea leaf flavonols are enhanced by UV-B but reduced by shading treatment. CsFLS, CsUGT78A14, CsMYB12, and CsbZIP1 were upregulated by UV-B radiation and downregulated by shading. CsMYB12 and CsbZIP1 bound to the promoters of CsFLS and CsUGT78A14, respectively, and activated their expression individually. CsbZIP1 positively regulated CsMYB12 and interacted with CsMYB12, which specifically activated flavonol biosynthesis. Meanwhile, CsPIF3 and two MYB repressor genes, CsMYB4 and CsMYB7, displayed expression patterns opposite to that of CsMYB12. CsMYB4 and CsMYB7 bound to CsFLS and CsUGT78A14 and repressed their CsMYB12-activated expression. While CsbZIP1 and CsMYB12 regulated neither CsMYB4 nor CsMYB7, CsMYB12 interacted with CsbZIP1, CsMYB4, and CsMYB7, but CsbZIP1 did not physically interact with CsMYB4 or CsMYB7. Finally, CsPIF3 bound to and activated CsMYB7 under shading to repress flavonol biosynthesis. These combined results suggest that UV activation and shading repression of flavonol biosynthesis in tea leaves are coordinated through a complex network involving CsbZIP1 and CsPIF3 as positive MYB activators and negative MYB repressors, respectively. The study thus provides insight into the regulatory mechanism underlying the production of bitter-tasting flavonols in tea plants.
Similar content being viewed by others
Introduction
Tea plants (Camellia sinensis) synthesize diverse flavonoids, such as catechins, flavonols, and anthocyanins and their derivatives, at significant levels in tender tissues, such as apical buds and young leaves. These flavonoids, together with caffeine and theanine, constitute the major bioactive secondary metabolites in teas, contributing to their pleasant flavors, rich tastes, and multiple health benefits, features that are of vast importance given that tea is the most consumed nonalcohol beverage in the world1,2,3. Both catechins, primarily epigallocatechin-3-gallate (EGCG), and flavonols, mainly kaempferol glycosides, are the major contributing factors to the bitter and astringent tastes, with very low sensory doses being recognized by the human tongue4,5. The tender shoot tips of tea plants are usually picked during early spring, when the weather is still cool and misty with less sunlight radiation, to ensure the highest quality of teas. It has been well documented that these tender shoot tips contain higher levels of amino acids, mainly theanine, and fewer bitter-tasting catechins and flavonols in spring6. Indeed, tea plant leaves often accumulate higher levels of flavonols and catechins, which may result from high-intensity light irradiation during the summer-autumn seasons7. Light intensity and light quality significantly affect the accumulation of characteristic secondary metabolites in tea plant leaves8,9. Both red and blue light promote the production of catechins and caffeine9, while UV-A and UV-B promote the production of anthocyanins10. Thus, shading of tea plants has been frequently applied in tea gardens to reduce the contents of these bitter-tasting and astringent flavonoids in tea plant leaves8,11,12. Transcriptome and metabolite profiling revealed that transcription factors (TFs) involved in light perception and signaling may be connected with TFs regulating flavonoid biosynthetic genes8,9. However, the genetic factors and detailed molecular mechanisms underlying how light exposure promotes and shading reduces the accumulation of tea flavonoid contents in tea plant leaves are not yet understood8,13,14. Since the levels of flavonols significantly affect tea flavor and health function, it is highly desirable to understand how environmental factors regulate their biosynthesis.
Flavonols are a particular class of flavonoids that are present in most green leaves. The biosynthesis and regulation of flavonol glycosides in tea plant leaves attracted our attention, as they are the major bitter-tasting substances in tea leaves grown under strong light conditions. The branched flavonol pathway has been studied extensively, including work on common shared enzymes such as F3H, F3’H, F3′5′H, as well as specific enzymes such as flavonol synthase (FLS) and UDP-glucose: flavonol glycosyltransferases (UGTs)6,15. Flavonol-specific FLS competes for the precursor dihydroflavonols with dihydroflavonol 4-reductase (DFR), leading to varying amounts of anthocyanin and proanthocyanidin synthesis16,17. Flavonol synthesis is also highly regulated at the transcriptional level by several tissue-specifically expressed R2R3-MYB transcriptional activators, such as Arabidopsis AtMYB11, 12, and 11118, apple MYB12 and MYB2219, and grapevine VvMYBF120. Meanwhile, R2R3-MYB repressors, such as Arabidopsis AtMYB7 and AtMYB4, have been demonstrated to be regulators of flavonoid biosynthesis in plants21. These activators and repressors, as well as other TFs, are specifically responsive to certain environmental cues and together form a regulatory network to fine tune flavonol biosynthesis in plants22,23.
Light is a crucial signal that affects plant growth and development and involves light receptor phytohormones, signaling proteins, and many downstream effectors, including metabolic enzymes and developmental regulators24,25. Several photoreceptors are characterized to respond to different wavelengths of light: the red/far-red light photoreceptor phytochromes, the blue/UB-A light photoreceptor cryptochromes and phototropins, and the UV-B light photoreceptor UVR826,27. These activated photoreceptors directly or indirectly modify the stability of primary TFs such as ELONGATED HYPOCOTYL 5 (HY5), PHYTOCHROME INTERACTING FACTOR 3 (PIF3), and PHYTOCHROME INTERACTING FACTOR 4 (PIF4)28. It is well known that plants accumulate increased levels of flavonols under high light conditions or UV-B irradiation than under regular light irradiation29. The E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) negatively controls photomorphogenesis by interacting with SUPPRESSOR OF PHYTOCHROME A (SPA1–SPA4) proteins to inhibit photomorphogenic growth30. HY5 is a key photomorphogenesis-promoting factor downstream of COP1 and is destabilized by COP1 in darkness31. HY5 directly regulates the promoters of thousands of genes involved in plant development and flavonoid biosynthesis32. HY5 can regulate flavonol biosynthesis by mediating UV-B or light irradiation-induced AtMYB12 activation and flavonol accumulation33. In the second branch of the pathway, a basic helix-loop-helix (bHLH) TF and PHYTOCHROME-INTERACTING FACTORs (PIFs) promote skotomorphogenesis and repress photomorphogenesis under red and far-red light conditions34. PIFs play diverse roles in plant growth and development by positively or negatively regulating a large number of downstream genes35. PIF3 plays multiple roles in light signaling as a negative factor in hypocotyl elongation and anthocyanin biosynthesis and a positive factor in the plant shading response36. In contrast to the case for HY5, light irradiation leads to PIF3 protein phosphorylation and degradation37. HY5 and PIFs are oppositely regulated by light. PIF3 and HY5 interact with cryptochromes and UVR8 to regulate light-responsive genes38. HY5 and PIF1/PIF3 interact with each other directly and antagonistically regulate reactive oxygen species-responsive genes and the greening of etiolated seedlings upon light irradiation39. However, how these factors are involved in light- or shading-regulated flavonoid biosynthesis remains unknown. An improved understanding of these mechanisms in tea plants is highly important given the common application of shading to tea plants to mediate the quality of tea production8,9,11.
This study attempts to dissect the comprehensive regulatory network mediating light- and shading-regulated biosynthesis of flavonols in tea plants. UV-B or shading treatment prominently altered bitter-tasting flavonol contents in tea plant leaves. UV-B radiation acted through CsbZIP1-CsMYB12 on the key flavonol biosynthetic genes CsFLS and CsUGT78A14, while shading repressed flavonol biosynthesis not only by inactivation of HY5-like CsbZIP1 but also via activation of CsPIF3, which further activated the MYB repressor genes CsMYB4 and CsMYB7. Transactivation assays revealed that CsMYB4 and CsMYB7 repressed CsFLS. We therefore demonstrated a complex regulatory network composed of both activators and repressors in the regulation of bitter-tasting flavonol production by UV-B exposure and shading treatment in tea plant leaves.
Materials and methods
Plant material and growth conditions
“Shu Cha Zao”, “Long Jing”, “Huang Shan Bai Cha”, “Zi Juan”, and “Huang Kui” tea plants were grown in the experimental tea garden of Anhui Agricultural University, Anhui, China (31°. 55′ North, 117°. 12′ East; Hefei City, Anhui Province, China). UV-B conditions (300 μW cm−2, photoperiod of 12 h per day) were provided using a special lamp (PHILIPS NARROWBAND TL 20 W, Poland) with a characteristic peak at 311 nm, 25/18 °C light/dark. The shading experiment consisted of two treatments: tea plants with natural growth (control) and tea plants with 90% shading treatment. The nets were placed over the plants on 27 July 2019, when a new round of bud burst started. The second leaves of tea of the same growth stage were collected throughout shading treatments (0 h, 4 h, 8 h, 12 h, 2 days, 4 days, 8 days, and 14 days after shading). All samples were stored at −80 °C until use. Methyl jasmonate (MeJA) treatment experiments were performed as described previously40. Tea plant leaves sprayed with 100 µM MeJA solution or distilled water (control) were collected at 0, 12, 24, and 48 h after the onset of treatment. The polyethylene glycol (PEG) and NaCl treatment experiments were performed as described previously41. Briefly, tea plant seedlings were treated with 25% PEG or 200 mM NaCl for 0, 24, 48, and 72 h to mimic drought and salinity stress conditions, respectively. For the cold treatment experiments, tea plant leaves were collected during the cold acclimation (CA) process. Control (CK): 25 °C; CA1-6 h: 10 °C for 6 h; CA1-7 days: from 10 °C to 4 °C for 7 days; CA2-7 days: from 4 °C to 0 °C for 7 days; DA-7d: recover under 25 °C to 20 °C for 7 days, as described previously42. Transcriptome data from experiments with tea cv. “Shu Cha Zao” were retrieved from the tea plant information archive (http://tpia.teaplant.org/index.html).
Chemical standards and other chemicals
All flavonoid standards were of analytical grade, including myricetin 3-O-galactoside (M-3-O-Gal), myricetin 3-O-glucoside (M-3-O-Glu), quercetin-3-O-glucoside (Q-3-O-Glu), quercetin-3-O-rutinoside (Q-3-O-R), quercetin-3-O-galactopyranoside (Q-3-O-Gal), kaempferol 3-O-galactosylrutinoside (K-3-O-Galact), kaempferol-3-O-glucoside (K-3-O-Glu), kaempferol-3-O-galactoside (K-3-O-Gal), and kaempferol-7-O-glucoside (K-7-O-Glu), which were purchased from Sigma Chemicals Co. (Sigma-Aldrich, USA). Methanol, acetonitrile, and acetic acid of chromatographic grade were purchased from Shanghai GuoMei Pharmaceutical Co. UPLC-grade water was prepared from distilled water using a Milli-Q system (Millipore Laboratory, Bedford, MA, USA).
Flavonols were detected by ultrahigh-performance liquid chromatography (UPLC) on an Agilent InfinityLab Poroshell HPH-C18 instrument (4.6 × 100 mm, 2.7 μm, Agilent, Santa Clara, CA, USA). The samples (5 μL injection volume) were loaded on an Inertsil ODS-3 column and eluted at a flow rate of 1.0 mL/min. Mobile phases A and B were composed of 0.1% acetic acid in distilled water and acetonitrile, respectively. The elution program was as follows: calibration with 95% A (1% acetic acid) and 5% B (100% acetonitrile), a linear gradient from 5 to 10% B for 0 − 2 min, from 10 to 20% B for 2−15 min, from 20 to 30% B for 15−30 min, and from 30 to 55% B for 30 − 55 min was performed, followed by washing and equilibration. The flavonols were detected at a wavelength of 350 nm, and the column temperature was set at 35 °C6.
Detection of flavonols from leaves
Leaves of “Shu Cha Zao”, “Long Jing”, “Huang Shan Bai Cha”, “Zi Juan”, and “Huang Kui” tea plants and different tissues of “Shu Cha Zao” were ground to a fine powder using a mortar and pestle in liquid nitrogen. The powered leaf samples (0.2 g) were extracted with 2 mL 80% methanol by sonication at room temperature for 5 min, followed by centrifugation at 4500×g for 10 min. The residues were re-extracted twice by this method. The supernatants were filtered through a 0.22-μm membrane. Flavonols were analyzed according to previously described UPLC methods6.
Quantitative real-time PCR
Total RNA was isolated from leaves with RNAiso Plus and RNAiso Mate for Plant Tissue Kits (TaKaRa, China). Double-stranded cDNA was prepared using the Super SMART PCR cDNA Synthesis Kit (Clontech, Palo Alto, USA) following the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was carried out using the SYBR Green method for the detection of double-stranded PCR products (TaKaRa, Dalian, China). An IQ5 real-time PCR detection system (Bio-Rad) was utilized in this study as previously described. The tea β-actin gene was used as an internal reference gene (HQ420251.1, https://www.ncbi.nlm.nih.gov/nuccore/). qRT-PCR data were generated using an Applied Biosystems 7900HT instrument, and analyses were performed using SDS software (Applied Biosystems). PCR efficiencies were calculated using LinReg software. The primers for representative genes in this study were designed by Primer Premier 5.0 software (PREMIER Biosoft company; Tables S1 and S2).
Sequence alignment and phylogenetic analysis
In this study, amino acid sequence alignment analysis of MYBs was conducted using DNAMAN 8.0 software (Lynnon, Quebec, Canada). A phylogenetic analysis using the amino acid sequences of MYB members was performed using MEGA 7.0 software (http://www.megasoftware.net/, Mega Software, State College, PA, U.S.A.), and a phylogenetic tree was constructed using neighbor-joining distance analysis. The tree nodes were evaluated with the bootstrap method for 1000 replicates, and the evolutionary distances were computed using the p-distance method. Sequence information used in the phylogenetic tree is shown in Table S3.
Subcellular localization
Sequence information on CsMYB4, CsMYB7, CsMYB12 and CsPIF3 was obtained from the tea plant genome (http://tpia.teaplant.org/). Sequence information of CsbZIP1 was obtained from transcriptome data8. The ORFs of CsMYB4, CsMYB7, CsMYB12, CsbZIP1, and CsPIF3 within the entry vector pDONR211 were cloned into the destination binary vector, namely, PK7WGF2.0, for subcellular localization studies. Positive vectors in which the ORF was fused at the N-terminus of GFP were obtained and named PK7WGF2.0-CsMYB4, PK7WGF2.0-CsMYB7, PK7WGF2.0-CsMYB12, PK7WGF2.0-CsbZIP1, and PK7WGF2.0-CsPIF3, respectively. As described above, the plasmids were introduced into A. tumefaciens strain GV3101 to select a positive colony for infiltration of Nicotiana benthamiana. After 48 h of infiltration, leaves were examined using an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan). GFP fluorescence signals were excited with a 488-nm laser, and the emitted light was recorded from 500 to 530 nm to display the subcellular localization of CsMYB4, CsMYB7, CsMYB12, CsbZIP1, and CsPIF3.
Overexpression of CsMYB12 in soybean hairy roots
CsMYB12 was constructed in pB2WG7 for overexpression and GUS as a control. These confirmed constructs were transformed into Agrobacterium rhizogenes strain K599 by electroporation. Positive colonies were selected on LB-agar medium containing selective antibiotics at 28 °C. Positive K599 colonies were used to generate hairy roots from germinated soybean (Glycine max) seeds. Soybean cultivar “Tianlong #1” seeds were surface sterilized and germinated in Petri dishes containing sterilized filter paper. The surfaces of 7-day-old green cotyledons were wounded and infected with K599 harboring the vectors for overexpression. The transgenic hairy roots were subjected to semiquantitative or qRT-PCR analyses to validate their identity. The transgenic hairy roots were maintained on half-strength Murashige and Skoog medium (MS medium) containing 7.5 mg L−1 phosphinothricin (ppt) for selection in a growth chamber at 23 °C with a 16 h/8 h light/dark photoperiod and subculture every 3–4 weeks.
Yeast one-hybrid and two-hybrid assays
Yeast one-hybrid (Y1H) and two-hybrid assays were conducted as previously described43. Yeast one-hybridization assays were performed using the Matchmaker Gold Yeast One-Hybrid System (Clontech). To construct transcription factor-expressing cassettes, the ORFs of CsMYB4, CsMYB7, CsMYB12, CsPIF3, and CsbZIP1 were recombined into the pGADT7 vector (Clontech, Palo Alto, USA). The cloned promoter fragments of CsMYB12, CsMYB7, CsFLS, and CsUGT78A14 were inserted into the pHIS2.1 vector. The yeast strain Y187 containing the recombinant pHIS2.1 vector was grown on –Trp–Leu (–T–L) screening medium for 3 days at 30 °C. Then, the interactions between the MYB TFs and promoter fragments were detected on medium lacking Trp, Leu and His (–T–L–H) for 3–5 days at 30 °C. Empty pGADT7 vectors were used as controls.
For yeast two-hybrid assays (Y2H), the ORFs of the CsMYB4, CsMYB7, CsbZIP1, and CsMYB12 genes were recombined into the pGBKT7 and pGADT7 vectors, respectively (Clontech, Palo Alto, USA). The recombinant plasmids were cotransformed into the yeast strain AH109 and cultured on medium lacking Trp and Leu (–T–L) for 3 days at 30 °C. For interaction screening, the yeast cells were transferred to medium lacking Trp, Leu, His and adenine (–T –L–H–A) with X-gal for 3–5 days at 30 °C. Empty pGADT7 and pGBKT7 vectors were used as controls.
Luciferase reporter assay
The ORFs of CsMYB4, CsMYB7, CsMYB12, CsbZIP1, and CsPIF3 were recombined into the P2GW7 effector expression system, as described previously43. The cloned promoter fragments of CsMYB12, CsMYB7, and CsFLS were inserted into the pGreen-0800-LUC reporter. Protoplasts derived from Arabidopsis thaliana were used as the materials for transient transfection. Each transfection contained the GUS plasmid for normalization. For transient transfection, 1 μL of GUS plasmid, 5 μL of LUC reporter, and 10 μL of effector were mixed together and transformed into Arabidopsis thaliana protoplasts using 40% polyethylene glycol. After reaction at 24 °C for 12 h, the LUC and GUS activities were tested using a Multimode Plate Reader (Victor X4, PerkinElmer, http://www.perkinelmer.com/). The promoter activity was calculated by the ratio of LUC to GUS activity.
Suppression of CsMYB12 and CsbZIP1 in tea shoot tips by using candidate antisense oligonucleotides
Since tea plant transformation technology has not yet been developed, knockdown of the target gene with antisense oligonucleotides (asODN) containing the segment complementary to the target gene was used to examine how CsbZIP1 and CsMYB12 affect flavonol synthesis in tea shoot tips44,45. The antisense oligonucleotides (asODN) were selected by using Soligo software (http://sfold.wadsworth.org/cgi-bin/soligo.pl) with CsMYB12 and CsbZIP1 as input sequences (Table S2). To silence the genes, fresh shoot tips (with the apical bud and 1st leaf) of the tea plant variety “Shu Cha Zao” were incubated in 2 ml Eppendorf tubes containing 40 μM asODN-CsMYB12 or asODN-CsbZIP1 solution for various times. Shoot tips incubated with the same concentrations of sense oligonucleotides (sODN) were used as the control. Shoot tips were sampled at different time intervals for RNA and flavonol analysis.
Bioinformatic analysis
The GenBank accession numbers for genes characterized in the study were as follows: CsMYB12 (MT498592), CsMYB4 (MT498593), CsMYB7 (MT498594), CsbZIP1 (MT498595), and CsPIF3 (MT498596). A multiple sequence alignment of the amino acid sequences of the CsMYB TF proteins of tea plant, rice and Arabidopsis was generated with ClustalW. An unrooted phylogenetic tree based on the sequence alignments was constructed using MEGA 7.0 software (http://www.megasoftware.net/) and the neighbor-joining method with the following parameters: pairwise alignment and 1000 bootstrap replicates. All resulting heatmaps of expression were structured by the pheatmap R package.
Statistical analysis
All experimental data are taken from at least three independent experiments. For C. sinensis shoot tip antisense inhibition experiments, at least 10 independent plants were analyzed with three repeats each. For Y2H assays, subcellular localization, and transgenic hairy root experiments, representative pictures are shown. Differences at the 95% confidence level in two-tailed Student’s t test were considered significant.
Results
UV-B and shading treatments regulated flavonol biosynthesis in tea plant leaves
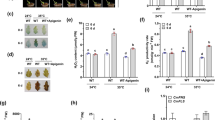
To understand the molecular regulatory mechanisms underlying the regulation of flavonol synthesis by light exposure, we conducted both UV-B radiation and shading treatment experiments on tea plant seedlings hydroponically grown in SK nutrient solution and on tea plants grown in tea gardens. In tea plant seedlings grown hydroponically under UV-B conditions (300 μW cm−2, photoperiod of 12 h per day, provided with a special lamp (PHILIPS NARROWBAND TL 20 W, Poland; Fig. 1a and S1), M-3-O-Gal, M-3-O-Glu, Q-3-O-Glu, Q-3-O-Gal, K-3-O-Glu, and K-7-O-Glu were increased by 1.5- and 2-fold compared to the control (Fig. 1b). In addition, when we shaded the tea plants grown in tea gardens under sunlight with a thick cloth (Figs. 2a and S2), which allowed only 10% of sunlight to pass through, the flavonol contents in young leaves of the tea plants decreased. Metabolite profiling revealed that light and shading treatments significantly affected flavonoid accumulation in tea leaves. Levels of kaempferol, quercetin, and myricetin glycosides, such as M-3-O-Gal, M-3-O-Glu, Q-3-O-Glu, Q-3-O-R, Q-3-O-Gal, K-3-O-Gal, K-3-O-Glu, K-3-O-Gal, and K-7-O-Glu, were reduced in plants that had received shading compared with controls (Figs. 2b and S3). Several key enzymes are involved in the biosynthesis of flavonols in tea plants, including flavanone 3-hydroxylase (CsF3H, TEA023790.1), flavonoid 3′-hydroxylase (CsF3′H, TEA006847.1), flavonoid 3′,5′-hydroxylase (CsF3'5′H, TEA026294.1), chalcone synthase (CsCHS, TEA023331.1), flavonol synthase (CsFLS, TEA006643.1), and UDP-glycose: flavonoid 3-O-glycosyltransferase (CsUGT, TEA007509.1; Fig. 2c). The expression patterns of genes associated with flavonol biosynthesis were next analyzed to understand how light regulates flavonol accumulation in tea plant leaves. Two structural genes and a MYB TF (Fig. 1d) of the flavonol biosynthetic pathway, namely, CsFLS, CsUGT78A14, and CsMYB12, were revealed by qRT-PCR analysis to be significantly activated by UV-B radiation (Fig. 1c, e). In addition, flavonol biosynthetic pathway genes displayed significantly lower expression levels in shaded leaves than in tea plant leaves fully exposed to sunlight (Fig. 2d). Moreover, CsCOP1 transcript levels were upregulated by shading treatment, but CsUVR8 transcript levels were repressed (Fig. S4a, b).
a UV-B radiation of tea plant seedlings. UV-B conditions (300 μW cm−2, photoperiod of 12 h per day, provided with a special lamp (PHILIPS NARROWBAND TL 20 W, Poland) with a characteristic peak at 311 nm. at 25/18 °C (light/dark) with those under regular white light as controls. b Content changes in M-3-O-Gal, M-3-O-Glu, Q-3-O-Glu, Q-3-O-Gal, K-7-O-G, and K-3-O-G in tea plant leaves under UV-B radiation in comparison with control (under regular white light). c Expression levels of CsFLS and CsUGT78A14 as a function of UV-B radiation compared with the control. d Phylogenetic identification of CsMYB12 and R2R3 MYB homologs from other plants. Vitis vinifera MYBF1: VvMYBF1; Arabidopsis thaliana MYB111, 12, 11: AtMYB111, AtMYB12, AtMYB11. e Expression patterns of CsMYB12 in tea plant leaves under UV-B treatment compared with controls. Differences between UV-B irradiated and untreated (control) samples were analyzed. Data were from three independent experiments and expressed as the means ± SD. (n = 3). The differences were analyzed in two-tailed comparisons with the control, and *p < 0.05; **p < 0.01 in Student’s t test
a Tea plants grown in tea gardens treated with shading (SD, 90% blocked light) in comparison with plants grown under regular light radiation (LT). b Changes in major flavonols in tea plant leaves under shading treatment. Data are expressed as the ratio of SD:LT (shading:light). c Key genes involved in the flavonoid pathway for the biosynthesis of flavonols in tea plants. d Effects of shading on metabolic gene expression in tea plant leaves. Differences between SD and LT (control) were analyzed. Data were from three independent experiments and expressed as the means ± S.D. (n = 3). Differences were analyzed in two-tailed comparisons of treatment and control, *p < 0.05; **p < 0.01 in Student’s t test
CsMYB12 mediated light-induced flavonol biosynthesis
We next identified the TFs that may regulate the light-induced or shading treatment-repressed biosynthesis of flavonol. When analyzing transcriptome data from previous experiments, we identified an AtMYB12 homolog MYB TF TEA009412 (tentatively named CsMYB12), which is more highly expressed in tea plant varieties Longjing (LJ) and Shu Cha Zao (SCZ) than in Huang Shan Bai Cha (HSBC), Huang Kui (HK), and Zi Juan (ZJ), corresponding to the higher flavonol contents in LJ and SCZ than in HSBC, HK, and ZJ varieties (Fig. S5a, b). Another AtMYB12 homolog, TEA016401, was expressed at very low levels in most tissues and did not respond to light radiation or shading treatment (Figs. S6, S7). Light and shading treatment experiments with SCZ and qRT-PCR verification of CsMYB12 transcripts also showed that CsMYB12 was repressed by shading treatment, coincident with the reduced total flavonols (Fig. 3a), and that CsMYB12 transcript levels in various tissues of tea plants were tightly associated with the total flavonol contents in these tissues (Fig. 3b). When CsMYB12 was overexpressed in soybean hairy roots (Fig. S8), it also triggered significant increases in flavonol and flavonone biosynthesis (Figs. 3c, S9). Metabolite profiling revealed that K-3-O-Glu, K-7-O-Glu, A-7-O-Glu, and A-8-C-O-Glu levels were significantly higher in the CsMYB12-overexpression (OE) hairy root lines than in the GUS control lines (Fig. 3d). Naringenin, kaempferol, and eriodictyol contents were markedly increased in CsMYB12-OE hairy root lines compared with the GUS lines (Fig. 3e). Thus, CsMYB12 is a flavonol biosynthesis regulator in tea plants. Consistent with this, GFP-CsMYB12 fusion protein signals in tobacco epidermal cells were observed in the nucleus, suggesting its function as a TF (Fig. 3f). We further investigated the regulatory function of CsMYB12. Yeast one hybrid (Y1H) studies showed that as a nuclear R2R3-MYB TF, CsMYB12 could bind to the promoters of the critical flavonol synthetic genes CsFLS and CsUGT78A14, whose promoter regions contain several putative MYB-binding cis-elements (Fig. 3g). Transactivation assays using a dual luciferase reporter system showed that CsMYB12 resulted in 3-fold activation of CsFLS (Fig. 3h).
a Effects of shading (SD) on CsMYB12 expression in comparison with light (LT). b Expression patterns of CsMYB12 in different stages and total flavonoid contents (sums of the contents at different stages). c UPLC chromatograms of flavonoid profiles in CsMYB12-overexpressing hairy root lines. d, e Flavonoid compounds in CsMYB12-overexpressing hairy root lines and GUS (control). f Nuclear localization of the CsMYB12-GFP fusion protein in leaf epidermal cells of Nicotiana benthamiana. Bar = 50 µm. g Interaction of CsMYB12 with the promoters of CsFLS and CsUGT78A14. h Effects of CsMYB12 on the promoter activity of CsFLS with the luciferase reporter assay. Differences between SD and LT (control) were analyzed. Data were from three independent experiments and expressed as the mean ± S.D. (n = 3). Differences were analyzed in two tailed with the control, *p < 0.05; **p < 0.01 in Student’s t test
The light signaling bZIP TF CsbZIP1 regulates CsMYB12 and CsFLS
We next analyzed the transcriptome data of tea plant leaves under shading treatment and found that several bZIP TFs were downregulated, including three HY5 homologs, TEA012075, TEA014348, and TEA032623 (Fig. S10). However, the bZIP gene most significantly downregulated following shading is a nonannotated transcript in the tea plant genome46. We cloned it and found that it shared 72.62% similarity with Arabidopsis HY5 at the amino acid sequence level; therefore, we named it HY5-like TF CsbZIP1 (Fig. S11). Phylogenetic analysis revealed that CsbZIP1 clustered together with VvHY5 but apart from three other HY5 orthologs, AtHY5, HaHY5, and AaHY5 (Fig. 4a). Furthermore, CsbZIP1 transcript levels were repressed by shading treatment (Fig. 4b). qRT-PCR analysis results showed that CsbZIP1 was expressed at higher levels in the first, second and third leaves than in the buds, flowers, stems, fruits and roots (Fig. 4c). GFP-CsbZIP1 fusion protein signals in tobacco epidermal cells were observed in the nucleus, suggesting its nuclear localization as a TF (Fig. 4d). Moreover, Y1H assays revealed that CsbZIP1 could bind to the promoters of CsMYB12 and two flavonol biosynthetic genes, CsFLS and CsUGT78A14 (Fig. 4e). Furthermore, CsMYB12 and CsbZIP1 physically interacted with one another in a yeast two-hybrid assay (Fig. 4f), indicating a possible synergistic activation effect on CsFLS, CsUGT78A14, and other genes associated with flavonol biosynthesis. Furthermore, a transactivation assay revealed that CsbZIP1 bound to the promoters of CsMYB12 and CsFLS and markedly activated proCsMYB12 and CsFLS expression in a transactivation assay (Fig. 4g, h). These results suggested that CsbZIP1 bound directly to the CsMYB12 promoter via the C region that contained the G-box to indirectly regulate flavonol biosynthesis in tea plants.
a Phylogenetic analysis of CsbZIP1 and other bZIP TFs from different plants. Vitis vinifera HY5: VvHY5; Populus trichocarpa HY5: PtHY5; Arabidopsis thaliana HY5: AtHY5; Artemisia annua HY5: AaHY5; Helianthus annuus HY5: HaHY5. b Effects of shading (SD) compared with regular light (LT) on CsbZIP1 expression in tea plant leaves. c Expression profiles of CsbZIP1 in different tea plant tissues. d Nuclear localization of GFP-CsbZIP1 fusion protein in tobacco leaf epidermal cells. Bar = 50 µm. e Binding of CsbZIP1 to the CsMYB12, CsFLS and CsUGT78A14 promoters in a Y1H assay. f CsbZIP1 interaction with CsMYB12 in a Y2H assay. g CsbZIP1 transactivated the promoter of CsMYB12 in a luciferase reporter assay. h CsbZIP1 transactivated the promoter of CsFLS in a luciferase reporter assay. Differences between SD and LT (control) were analyzed. Data are from three independent experiments and are expressed as the means ± S.D. (n = 3). The differences in comparison with the control were analyzed by two-tailed Student’s t test, *p < 0.05; **p < 0.01
R2R3-MYB repressors mediated the shading treatment-repression of flavonol synthesis
During the analysis of the transcriptome data from several light-treatment experiments8,9,10,11 we observed that two other R2R3-MYB TFs, CsMYB4 and CsMYB7, could be markedly activated by shading treatment (Fig. 5a, c). Both CsMYB4 and CsMYB7 clustered together with VvMYBC2-L1, PtoMYB156, and other R2R3-MYB repressors in our sequence phylogeny (Fig. S12). Furthermore, both CsMYB4 and CsMYB7 repressors contain a conserved LxLxL sequence within the C-terminal region (Fig. S13). We next tested whether these proteins acted as negative regulators of flavonol synthesis during shading- or light treatment-modified flavonol biosynthesis. Both CsMYB4 and CsMYB7 were expressed in green tissues in tea plants (Fig. 5b, d), and both CsMYB4 and CsMYB7 were localized to the nuclei, as shown by GFP-CsMYB4 and GFP-CsMYB7 fusion expression in tobacco leaf epidermal cells (Fig. 5e, f). Furthermore, they also bound to the promoters of the CsFLS and CsUGT78A14 genes, suggesting that they could regulate flavonol synthesis (Fig. 5g). The Y2H experiment results revealed that CsMYB12 interacted with MYB4 and MYB7 and that CsMYB4 and CsMYB7 interacted with each other (Figs. 5h and S14). Using the dual luciferase reporter gene system, 0800-LUC vectors of proCsFLS, as well as p2GW7 vectors of CsMYB4, CsMYB7, CsbZIP1, and CsMYB12, were constructed and transferred into Arabidopsis thaliana for promoter activation experiments (Fig. 5i). Transactivation assays with proCsFLS-driven LUC reporters showed that while CsMYB12 and CsbZIP1-activated proCsFLS, CsMYB7, or CsMYB4 individually or together synergistically repressed the CsMYB12- or CsMYB12 + CsbZIP1-activated proCsFLS. From these analyses, CsMYB7 appeared to have stronger repression activity than CsMYB4 (Figs. 5j and S15). To further understand how these MYB TF genes respond to UV-B radiation, we also examined CsMYB7 and CsMYB4 expression in UV-B radiation experiments (Fig. 5k). Indeed, CsbZIP1 was significantly upregulated by UV-B radiation, CsMYB4 and CsMYB7 were less changed, and only CsMYB7 was upregulated at 48 h after radiation (Fig. 5k).
a, c Effect of shading (SD) on CsMYB4 and CsMYB7 expression in tea leaves compared with regular light (LT). b, d qRT-PCR data show the expression profiles of CsMYB4 and CsMYB7 in different stages. e, f Nuclear localization of GFP-CsMYB4 and GFP-CsMYB7 fusion in tobacco leaf epidermal cells. Bar = 50 µm. g Binding of CsMYB4 and CsMYB7 to the promoters of CsFLS and CsUGT78A14 in Y1H assays. h Interactions among CsMYB12, CsMYB4, and CsMYB7 in Y2H assays. i Constructions of reporter and effector expression vectors for dual luciferase assays. j Transactivation of CsMYB12, CsMYB4, and CsMYB7 individually or in combination on the promoter activity of CsFLS in the luciferase reporter assay. k Effect of UV-B radiation on the expression of CsMYB4, CsMYB7, and CsbZIP1. Differences between shading (SD) and light (LT) (control) were analyzed. Data were from three independent experiments and expressed as the means ± S.D. (n = 3). Differences comparison with the control were analyzed by two-tailed Student’s t test, *p < 0.05; **p < 0.01
CsPIF3 activated CsMYB7 and thereby repressed flavonol synthesis
We further asked how CsMYB7 and CsMYB4 in tea plant shoot tips were activated by shading treatment. Since CsPIF3 genes have been shown to be upregulated by shading, we examined whether this essential light signaling gene can activate CsMYB7 and CsMYB4. Of the two Arabidopsis AtPIF3 homologs, TEA006216 and TEA007077 (Fig. 6a), only the latter was dramatically upregulated by shading (Fig. S16). We thus named it CsPIF3. CsPIF3, AtPIF1, AtPIF3, and AtPIF8 all had conserved APB and APA elements (Fig. S17), and qRT-PCR results showed that CsPIF3 could be upregulated in tea plant leaves by shading treatment compared with the control (Fig. 6b). CsPIF3 displayed higher expression levels in stems, roots, and leaves (Figs. 6c and S16). Because the CsMYB4 promoter was not assembled in the reference tea plant genome46, we cloned only the CsMYB7 promoter, which contained a G-box cis-element that is reported as a binding site by AtPIF3 (Fig. S18). CsPIF3 was localized to nuclei, as shown by GFP-CsPIF3 fusions expressed in tobacco leaf epidermal cells (Fig. 6d). Y1H experiments showed that CsPIF3 can bind to and activate the promoter of the CsMYB7 gene (Fig. 6e). Additionally, as a nucleus-localized TF, CsPIF3 activated the CsMYB7 promoter in a transactivation assay (Fig. 6f). These results showed that under shading, CsPIF3 could activate CsMYB7, through which CsPIF3 repressed flavonol synthesis. Under UV-B radiation, CsUVR8 expression was slightly upregulated, and CsCOP1 was significantly upregulated by UV-B radiation. However, in contrast to CsbZIP1, CsPIF3 expression was almost unchanged by UV-B radiation (Fig. 6g).
a A phylogenetic tree generated by MEGA analysis using amino acid sequences of CsPIF3 and other bHLH members. Arabidopsis thaliana PIFs: AtPIF2, AtPIF6, AtPIF3, AtPIF4, AtPIF5, AtPIF8, and AtPIF7. b The shading effects on CsPIF3 expression in tea leaves analyzed by qRT-PCR. c qRT-PCR data showing the expression profiles of CsPIF3 in different stages. d Nuclear localization of GFP-CsPIF3 fusion protein in the leaf epidermal cells of Nicotiana benthamiana. Bar = 50 µm. e Binding of CsPIF3 to promoters of CsMYB7 in Y1H assays. f Transactivation of CsPIF3 activity on CsMYB7 with the luciferase reporter assay. g Effect of UV-B radiation on the expression of CsUVR8, CsCOP1, and CsPIF3. Differences between shading treatment (SD) and light (LT) control were analyzed. Data were from three independent experiments and expressed as the means ± S.D. (n = 3). Differences in comparison with the control were analyzed in two-tailed Student’s t test, *p < 0.05; **p < 0.01, *p < 0.05; **p < 0.01
Suppression of CsMYB12 and CsbZIP1 in shoot tips affected flavonol biosynthesis
To further analyze the physiological role of CsMYB12 as a regulator of flavonol biosynthesis in tea plants, the expression level of CsMYB12 was suppressed in the C. sinensis bud and 1st leaves by using an antisense oligodeoxynucleotide (asOND)-interfering gene-specific suppression strategy (Fig. 7a)45. asODN knockdown resulted in CsMYB12 and two structural genes of the flavonol biosynthetic pathway, CsFLS and CsUGT78A14, which were markedly repressed over the treatment period, as verified by qRT-PCR (Fig. 7b). The contents of K-3-O-Glu and Q-3-O-Glu in the apical bud and 1st leaf were reduced by up to 1.5-fold compared with those in the control (Fig. 7c). Furthermore, CsbZIP1 was knocked down by using a similar asODN approach to understand its regulation of CsMYB12 following UV-B treatment (Fig. 7d). Obvious asODN suppression of CsbZIP1 was observed only under UV radiation (Fig. 7e). In general, CsMYB12 transcripts did not fluctuate when CsbZIP1 was repressed under normal light intensity. However, UV-B treatment resulted in a significant upregulation of CsMYB12 in the untreated asODN control, while CsMYB12 was unchanged in asODN-CsbZIP1-treated shoot tips (Fig. 7f). Correspondingly, the flavonol contents did not change in asODN-bZIP1 compared with the control, in which flavonoid contents increased upon UV-B radiation (Fig. 7g). Thus, CsMYB12 appears to play a key role in the regulation of flavonol biosynthesis in tea plants. Moreover, CsbZIP1 regulates UV-B-stimulated CsMYB12 expression and flavonol biosynthesis.
a Tea plant shoot tips fed with asODN-CsMYB12. b Expression of CsFLS, CsUGT78A14, and CsMYB12 in the shoot tips of tea plants. c Contents of flavonols after asODN-CsMYB12 treatment. d Tea plant shoot tips fed with asODN-CsbZIP1. e Repression of CsbZIP1 transcript levels in tea plant shoot tips by asODN-CsbZIP1 treatment. f Changes in CsMYB12 transcripts with asODN-CsbZIP1 treatment upon UV-B radiation. g Changes in flavonol contents in asODN-CsbZIP1 in comparison with sODN treatment. Differences between asODN treatments and sODN treatments (control) were analyzed. Data are expressed as means ± s.d. from at least three replicates. Differences in comparison with the control were analyzed in two-tailed Student’s t tests, *p < 0.05; **p < 0.01
Responsive expression of CsMYB4, CsMYB7, CsMYB12, CsbZIP1, CsUVR8, CsCOP1, and CsPIF3
To better understand the functions of CsMYB4, CsMYB7, CsMYB12, CsbZIP1, CsUVR8, CsCOP1, and CsPIF3 in tea plants, we analyzed their expression patterns in different tissues or in response to UV-B, cold and salt stresses or PEG and MeJA treatments. Under UV-B treatments, CsMYB12, CsbZIP1, CsUVR8, and CsCOP1 were upregulated (Figs. 1d, 5k, and 6g). However, CsMYB4 was repressed under UV-B treatment (Fig. 5k). CsMYB7 and CsPIF3 were initially slightly induced and then repressed by UV-B, when CsMYB12, CsbZIP1, and CsUVR8 transcripts reached the highest levels (Figs. 1e, 5k and 6g). These results are similar to those of previous studies10.
Other studies showed that flavonol contents in tea plant tissues also increased under MeJA treatment and salinity and PEG stresses, coupled with significant changes in the structural genes involved in flavonol biosynthesis40,41 (Fig. S19). CsMYB12 and CsMYB7 were generally more highly expressed in leaves than in roots (Figs. 3b and 5d); however, CsMYB7 was expressed at low levels following cold and MeJA treatments (Fig. S20a, c). CsMYB12 expression was induced by cold and MeJA treatment of leaves (Fig. S20a, c) but repressed by salt and PEG treatment (osmotic stress; Fig. S20b, d)40,41. Meanwhile, CsPIF3 and CsMYB4 showed the opposite behavior; CsPIF3 was repressed by MeJA treatment, while CsMYB4 was initially slightly induced and then repressed by MeJA when CsMYB12 transcripts reached their highest levels (Fig. S20c)41. These results indicate that light and abiotic stress regulation of flavonol synthesis and accumulation occurs at the level of transcription.
Discussion
Characteristics of tea, such as color, taste, smell, and levels of health-conferring metabolites, are regarded as the major tea quality parameters that guide tea plant cultivation, breeding, and tea processing. These qualities depend primarily on the types and contents of tea plant-specific secondary metabolites present in the fresh tea plant leaves and the ways these starting materials are processed into teas5. Flavonol glycosides, such as myricetin 3-O-galactoside and quercetin-3-O-rutinoside, although present at relatively low levels in tea plant leaves compared with catechins and caffeine, have recently been recognized as among the major contributors to the bitter and astringent tastes of tea47,48. Therefore, their biosynthesis and regulation in tea plant leaves have been the focus of considerable attention. FLS and UGT are two critical and specific genes involved in flavonol glycoside biosynthesis6,49. While flavonol glycosides present in tea plant leaves grown under strong light in the spring-summer season are the major contributors to teas with stronger bitter tastes7, a reduction in light radiation by various measures, such as shading, has been shown to effectively improve tea quality8,11. Studies have revealed the biosynthetic pathways and enzymes involved in the production of a wide array of flavonol glycosides that are present at significant levels in strong light-radiated plant leaves or fruits50,51. Despite the fact that the regulatory mechanism underlying the high light or UV-B-radiation induction of flavonol glycoside biosynthesis is well known51, the repression of this activity by shading treatment is poorly understood. Significant progress has been made in understanding plant responses to high light or UV-B and shading treatments, including the characterization of several photoreceptors, COPIs, HY5, and PIFs, and other downstream effectors52. Our current study attempted to elucidate the transcriptional regulatory mechanisms underlying light-regulated flavonol accumulation in tea plants, providing new insights into the complex regulatory network controlling light- and shading-regulated flavonol biosynthesis.
High light or UV-B radiation regulated flavonol biosynthesis in tea plant leaves
Many reports have investigated tea metabolites under shade or altered light conditions. Previous studies have focused predominantly on the effect of shading treatment on catechin biosynthesis in tea cultivars8,11. In the present study, we found that flavonols decreased even more significantly in tea leaves than catechins did upon shading treatment (Fig. 2b). Consistent with these transcriptome data, our qRT-PCR data showed that the expression of key flavonol pathway genes, including CsF3H, CsF3′H, CsF3′5′H, CsFLS, and CsUGT78A14, was markedly reduced by shading treatment and conversely was upregulated by UV-B. We further revealed that the MYB TF CsMYB12 directly regulated these flavonol synthesis genes and that two light signaling TFs, CsbZIP1 and CsPIF3, worked upstream of CsMYB12, thereby acting in concert to translate UV-B and high-light radiation or shading treatment into effects on flavonol biosynthesis. Furthermore, we uncovered an even more complex regulatory network by naturally or deliberately regulating lighting or shading treatments. These studies likely reveal the two sides of the same coin. That is, UV-B or high-light radiation induces CsbZIP1 and CsMYB12 and dominantly upregulates flavonol synthetic genes, thereby promoting the accumulation of bitter-tasting flavonols in tea plant leaves in the spring-summer season, whereas under shading treatment, both CsbZIP1 and CsMYB12 are repressed as the result of both COP1-mediated CsbZIP1 degradation and the regulation of two R2R3 repressors, CsMYB7 and CsMYB4, by another bHLH light signaling protein (CsPIF3) to further effectively repress CsMYB12 activity and thus repress flavonol biosynthesis (Fig. 8). Furthermore, upregulated CsPIF3, CsMYB7, and CsMYB4 mediate red- or far-red light signaling in darkness. Thus, it is suggested that shading treatment, similar to red light radiation, can effectively reduce the biosynthesis of flavonols and that two mechanisms explain the reduced flavonol content in tea plant leaves under shading treatment.
Under high light conditions, UVR8 activates CsbZIP1, which enhances CsMYB12 transcription and activity. Both CsbZIP1 and CsYMB12 synergistically activate CsFLS and CsUFGT expression and promote bitter-tasting flavonol biosynthesis and accumulation, thereby decreasing the processing suitability of tea plant leaves. Meanwhile, CsPIF3, CsMYB7, and CsMYB4 are expressed at low levels and inactivated. However, under shading conditions, CsbZIP1 is repressed, while CsPIF3, CsMYB7, and CsMYB4 are activated. The repression of CsPIF3 by PhyA and PhyB is relieved, allowing CsPIF3 to interact with and repress CsbZIP1 activity. Shading upregulates the expression of the repressor genes CsMYB7 and CsMYB4. CsMYB7 and CsMYB4 can interact with CsMYB12 and thus interfere with CsMYB12 activation of flavonol synthesis. CsMYB7 and 4 also directly repressed CsFLS expression and, as a consequence, flavonol biosynthesis under shading conditions
MYB activators or repressors as regulators of phenylpropanoid metabolism in plants
Many R2R3-MYB activators of flavonol biosynthesis have been characterized. AtMYB11, 12, and 111 control flavonol accumulation in different parts of the Arabidopsis seedling18. VvMYBF1 was confirmed to complement the flavonol-deficient phenotype of the AtMYB12 mutant20. In addition, under abiotic stress conditions, MYB repressor TFs are particularly important, and R2R3-MYB repressors contain a conserved LxLxL sequence within the C-terminal region53. In grapevine, three flavonoid repressor MYBs, namely, MYBC2-L1, MYBC2-L2, and MYBC2-L3, were identified22. CsMYB4a, as a lignin synthesis repressor, was identified in tea plants54. Therefore, activator–repressor systems coordinate the fine tuning of critical metabolite biosynthesis and accumulation22. Subgroup 4 of R2R3-MYB transcription factors in Arabidopsis consists of repressors MYB3, MYB4, MYB7, and MYB32, possessing the conserved EAR repression motif54. In this study, we isolated three potential genes, CsMYB12, CsMYB4, and CsMYB7, from tea plants that were hypothesized to positively and negatively regulate flavonol biosynthesis. Then, overexpression of CsMYB12 promoted the accumulation of flavonol in soybean roots (Fig. S9). The luciferase reporter assay results showed that CsMYB4 and CsMYB7 had a significant effect on the negative regulator CsFLS (Fig. 5j). Thus, CsMYB4 and CsMYB7 can affect flavonol biosynthesis, rendering them repressors with potentially broad impacts on tea plant secondary metabolism. It seems likely that the coordinated action of repressor and activator MYBs could be important for the fine tuning of flavonoid biosynthesis during development or following stress.
CsbZIP1 and CsPIFs differentially regulate flavonol synthesis under normal light and shading conditions
Light is one of the most important environmental factors regulating flavonoid biosynthesis55. HY5 and activator MYB TFs regulate flavonoid biosynthesis and are required for UV-B tolerance. Given the importance of HY5 in a broad panel of photomorphogenic responses downstream of multiple photoreceptors26,52, it is of interest to characterize the CsbZIP1 gene and its signaling pathway in the context of light-induced and developmental regulation of tea plant secondary metabolism. In contrast, PIF3 is a bHLH TF with a light regulation mechanism on the other darkness side that binds to the palindromic G box motif CACGTG, which is common to many plant genes56,57. In this study, we functionally characterized two light signaling genes, CsbZIP1 and CsPIF3, in the regulation of flavonol biosynthesis in light- and shading-treated tea plants through their binding and activating and repressing CsMYB12, as well as two negative MYB regulator genes, CsMYB4 and CsMYB7. The complex regulatory network composed of both activators and repressors of various kinds of TFs related to the light and shading responses in tea plants can explain the increased levels of bitter-tasting flavonols under high light (including stronger UV-B radiation during the early summer and late autumn), as well as the drastic reduction in flavonol levels in tea plant leaves under shading treatment.
The interaction between CsbZIP1 and CsMYB12 and the direct binding of CsbZIP1 to the promoter of the CsMYB12 gene for its activation play a dominant role in connecting light signal perception to flavonol biosynthesis in tea plants. Even under shading treatment, reduced CsbZIP1 transcript levels remain a critical factor maintaining certain levels of flavonols in tea plant leaves. CsPIF3 is expressed at significantly higher levels in tea plant leaves under shading treatment, and CsPIF3 binds to the G-box in the promoter of CsMYB7 to upregulate the CsMYB7 repressor and, more likely, CsMYB4. Both CsMYB7 and CsMYB4 repressed CsFLS transcription and thus interfered with CsMYB12 function as activators of CsFLS and other flavonol biosynthetic genes under shading treatment. The regulatory function of CsMYB12 seems highly specific to flavonoid biosynthetic genes, such as CsFLS and CsUGT78A14. However, the regulatory targets of CsMYB4 and CsMYB7 may be nonspecific, since their upregulation under shading treatment or changes in lighting could also be negatively correlated with catechin levels. Under shading treatment, catechin contents also generally decreased when tea plant shoot tips became less bitter. Further study will be needed to demonstrate how these negative MYB regulators work in the regulation of flavonoid biosynthesis in tea plants.
It is possible that AtPIF3 and HY5 interact directly; alternatively, their antagonistic effects may be mediated through another factor, such as cryptochromes and UVR839. HY5 binding to the promoters of UV-B-responsive genes is enhanced by UV-B in a UVR8-dependent manner in Arabidopsis thaliana. In agreement with this observation, overexpression of REPRESSOR OF UV-B PHOTOMORPHOGENESIS2, a negative regulator of UVR8 function, blocks UV-B-responsive HY5 enrichment at target promoters58. A T/G-box in the HY5 promoter is required for its UV-B responsiveness. HY5 and its homolog HYH bind to the T/G(HY5)-box cis-acting element to activate its own expression redundantly upon UV-B exposure. HY5 and HYH interact directly with a T/G-box cis-acting element of the HY5 promoter, mediating the transcriptional activation of HY5 in response to UV-B59.
In summary, UV-B radiation promoted and shading repressed flavonol biosynthetic genes and consequently flavonol production in tea plant leaves. We demonstrated here that the different effects of light and shading involved CsbZIP1 and CsPIF3, a flavonol biosynthesis activator CsMYB12, and two MYB repressors CsMYB7 and CsMYB4. UV-B radiation of tea plants upregulated CsbZIP1 and CsMYB12 (Figs. 1d and 5k), whereas 90% shading treatment clearly upregulated CsCOP1 and repressed CsbZIP1 and CsMYB12 (Figs. 1d, 4b, and S4a). CsbZIP1 acted as an activator of CsMYB12 and CsFLS and CsUGT78A14 genes with CsMYB12 to promote UV-B-induced flavonol production. However, after shading treatment, CsbZIP1 and CsMYB12 were repressed to lower expression levels. Meanwhile, shading treatment activated CsPIF3, CsMYB7, and CsMYB4 to antagonize the effect of CsbZIP1 and repress the CsMYB12, CsFLS, and CsUGT78A14 genes. Both CsMYB7 and CsMYB4 repressed CsFLS and CsUGT78A14 by directly binding to their promoters. CsMYB7 and CsMYB4 also directly interact with CsMYB12 and may interfere with CsMYB12 activation activity. Furthermore, CsPIF3 activated CsMYB7 through binding to its promoter. This study provides new insights into the mechanism of how light regulates the production of bitter-tasting flavonols in tea plants, which may provide molecular tools for the genetic improvement of tea quality and flavor.
Data availability
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is Jian Zhao (jianzhao@ahau.edu.cn).
References
Lakenbrink, C., Lapczynski, S., Maiwald, B. & Engelhardt, U. H. Flavonoids and other polyphenols in consumer brews of tea and other caffeinated beverages. J. Agric. Food Chem. 48, 2848–2852 (2000).
Yang, C. S. & Hong, J. G. Prevention of chronic diseases by tea: possible mechanisms and human relevance. Annu. Rev. Nutr. 33, 161–181 (2013).
Gilbert, N. The science of tea’s mood-altering magic. Nature 566, S8–S9 (2019).
Scharbert, S., Holzmann, N. & Hofmann, T. Identification of the astringent taste compounds in black tea infusions by combining instrumental analysis and human bioresponse. J. Agric. Food Chem. 52, 3498–508 (2004).
Zhuang, J. et al. Evaluation of astringent taste of green tea through mass spectrometry-based targeted metabolic profiling of polyphenols. Food Chem. 305, 125507 (2020).
Cui, L. et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 67, 2285–2297 (2016).
Zhu, J. et al. Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis). Plant Sci. 290, 110306 (2019).
Liu, L. et al. Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 18, 233 (2018).
Zheng, C. et al. Regulation of growth and flavonoid formation of tea plants (Camellia sinensis) by blue and green light. J. Agric. Food Chem. 67, 2408–2419 (2019).
Li, W. et al. The effects of ultraviolet A/B treatments on anthocyanin accumulation and gene expression in dark-purple tea cultivar ‘ziyan’ (Camellia sinensis). Molecules 25, 354 (2020).
Wang, Y. S. et al. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 141, 7–16 (2012).
Ji, H. G. et al. Diverse metabolite variations in tea (Camellia sinensis L.) leaves grown under various shade conditions revisited: a metabolomics study. J. Agric. Food Chem. 66, 1889–1897 (2018).
Zhao, J., Li, P., Xia, T. & Wan, X. Exploring plant metabolic genomics: chemical diversity, metabolic complexity in the biosynthesis and transport of specialized metabolites with the tea plant as a model. Crit. Rev. Biotechnol. 40, 667–688 (2020).
Xia, En-Hua et al. Tea plant genomics: achievements, challenges and perspectives. Hortic. Res. 7, 7 (2020).
Irmisch, S. et al. Flavonol biosynthesis genes and their use in engineering the plant antidiabetic metabolite montbretin A. Plant Physiol. 180, 1277–1290 (2019).
Luo, P. et al. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 13, 1257 (2016).
Owens, D. K. et al. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147, 1046–1061 (2008).
Stracke, R. et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677 (2007).
Wang, N. et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292 (2017).
Czemmel, S. et al. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530 (2009).
Wang, X. et al. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 101, 637–652 (2020).
Cavallini, E. et al. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470 (2015).
Ma, D., Reichelt, M., Yoshida, K., Gershenzon, J., Constabel, C. & Two, P. R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 96, 949–965 (2018).
Lau, O. S. & Deng, X. W. Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577 (2010).
Sun, J., Qi, L., Li, Y., Chu, J. & Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Gene 8, e1002594 (2012).
Chen, M., Chory, J. & Fankhauser, C. Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87–117 (2004).
Legris, M., Nieto, C., Sellaro, R., Prat, S. & Casal, J. J. Perception and signaling of light and temperature cues in plants. Plant J. 90, 683–697 (2017).
Nguyen, N. H. et al. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 84, 1192–1205 (2015).
Solovchenko, A. & Schmitz-Eiberger, M. Significance of skin flavonoids for UV-B-protection in apple fruits. J. Exp. Bot. 54, 1977–1984 (2003).
Zhu, D. et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20, 2307–23 (2008).
Osterlund, M. T., Hardtke, C. S., Wei, N. & Deng, X. W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000).
Zhang, H. et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358 (2011).
Stracke, R. et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103 (2010).
Zhang, X. et al. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174, 2487–2500 (2017).
Leivar, P. & Monte, E. PIFs: systems integrators in plant development. Plant Cell. 26, 56–78 (2014).
Kim, J. et al. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 15, 2399–2407 (2003).
Al-Sady, B., Ni, W., Kircher, S., Schäfer, E. & Quail, P. H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006).
Pedmale, U. V. et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245 (2016).
Chen, D. et al. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25, 1657–1673 (2013).
Shi, J. et al. Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol. 15, 233 (2015).
Zhang, Q. et al. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet. Genomes 13, 78 (2017).
Ahmad, M. Z. et al. Genome-wide analysis of serine carboxypeptidase-like acyltransferase gene family for evolution and characterization of enzymes involved in the biosynthesis of galloylated catechins in the tea plant (Camellia sinensis). Front. Plant Sci. 11, 848 (2020).
Li, P. et al. Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 14, 1604–1618 (2016).
Dinç, E. et al. Synthetic antisense oligodeoxynucleotides to transiently suppress different nucleus-and chloroplast-encoded proteins of higher plant chloroplasts. Plant Physiol. 157, 1628–1641 (2011).
Xie, Z. et al. A selection strategy in plant transformation based on antisense oligodeoxynucleotide inhibition. Plant J. 77, 954–961 (2014).
Wei, C. et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl Acad. Sci. USA 115, E4151–E4158 (2018).
Xu, Y. et al. Quantitative analyses of the bitterness and astringency of catechins from green tea. Food Chem. 258, 16–24 (2018).
Narukawa, M., Kimata, H., Noga, C., & Watanabe, T. Taste characterization of green tea catechins. Int. J. Food Sci. Technol. 45, 1579–1585 (2010).
Vu, T. T. et al. Characterization of Brassica napus flavonol synthase involved in flavonol biosynthesis in Brassica napus L. J. Agric. Food Chem. 35, 7819–7829 (2015).
Neugart, S., Krumbein, A., & Zrenner, R. Influence of light and temperature on gene expression leading to accumulation of specific flavonol glycosides and hydroxycinnamic acid derivatives in kale (Brassica oleracea var. sabellica). Front. Plant Sci. 7, 326 (2016).
Matus, J. T. et al. Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60, 853–867 (2009).
Jiao, Y., Lau, O. S. & Deng, X. W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 (2007).
Kranz, H. D. et al. Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276 (1998).
Li, M. et al. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 8, 943 (2017).
Zoratti, L., Karppinen, K., Escobar, A. L., Häggman, H. & Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 5, 534 (2014).
Menkens, A. E., Schindler, U. & Cashmore, A. R. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510 (1995).
Ni, M., Tepperman, J. M. & Quail, P. H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667 (1998).
Binkert, M. et al. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200–4213 (2014).
Loyola, R. et al. The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signaling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 18, 5429–5445 (2016).
Acknowledgements
The authors acknowledge support from the National Key Research and Development Program of China (2018YFD1000601), the Key Research and Development (R&D) Program of Anhui Province (18030701155), Funding from Anhui Agricultural University, and Funding from the State Key Laboratory of Tea Plant Biology and Utilization. The Postgraduate Foundation of Anhui Agricultural University, Anhui Province, China (2020ysj-33).
Author information
Authors and Affiliations
Contributions
J.Z. planned and designed the research. X.C.Z., X.S.Z., N.L., and S.Y. performed the experiments; X.C.Z., N.L., and X.S.Z. analyzed the data. J.Z., X.C.Z., and A.R.F. wrote and revised the article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, X., Zeng, X., Lin, N. et al. CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator–repressor network. Hortic Res 8, 110 (2021). https://doi.org/10.1038/s41438-021-00545-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00545-8
This article is cited by
-
The CmMYB3 transcription factors isolated from the Chrysanthemum morifolium regulate flavonol biosynthesis in Arabidopsis thaliana
Plant Cell Reports (2023)
-
α-Tocopherol application as a countermeasure to UV-B stress in bread wheat (Triticum aestivum L.)
Environmental Science and Pollution Research (2023)