Abstract
Flavonols and other phenylpropanoids protect plants from biotic and abiotic stress and are dietarily desirable because of their health-promoting properties. The ability to develop new potatoes (Solanum tuberosum) with optimal types and amounts of phenylpropanoids is limited by lack of knowledge about the regulatory mechanisms. Exogenous sucrose increased flavonols, whereas overexpression of the MYB StAN1 induced sucrolytic gene expression. Heterologous StAN1 protein bound promoter fragments from sucrolytic genes (SUSY1 and INV1). Two additional MYBs and one microRNA were identified that regulated potato flavonols. Overexpression analysis showed MYB12A and C increased amounts of flavonols and other phenylpropanoids. Endogenous flavonol amounts in light-exposed organs were much higher those in the dark. Expression levels of StMYB12A and C were high in flowers but low in tubers. Transient overexpression of miR858 altered potato flavonol metabolism. Endogenous StmiR858 expression was much lower in flowers than leaves and correlated with flavonol amounts in these organs. Collectively, these findings support the hypothesis that sucrose, MYBs, and miRNA control potato phenylpropanoid metabolism in a finely tuned manner that includes a feedback loop between sucrose and StAN1. These findings will aid in the development of potatoes with phenylpropanoid profiles optimized for crop performance and human health.
Similar content being viewed by others
Introduction
Potato (Solanum tuberosum) is one of the world’s staple foods, along with rice, wheat and corn, and will be instrumental in achieving global food security, given projections that crop yields need to double by 20501. One aspect of food security is the ability to meet energetic and nutritional needs. In addition to providing energy from complex carbohydrates, potatoes contain various vitamins, minerals and phytonutrients, including Vitamins B6, B9, and C, potassium and carotenoids2,3,4,5,6,7,8. Potatoes also contain phenylpropanoids, which are dietarily desirable because of their beneficial effects on health thought to include cardiovascular health, longevity, gut and eye health, along with anti-inflammatory and chemoprotective properties9,10,11,12.
Phenylpropanoids accumulate constitutively and may also be induced by environmental conditions. They have diverse roles in plants such as growth and development, and in modulating plant interactions with environment, including microbes, insects, and abiotic stress13,14,15,16. Potatoes, especially those with purple, blue, or red-flesh can contain high amounts of phenylpropanoids such as chlorogenic acids and anthocyanins, but low amounts of flavonols, which can be induced by wounding17,18,19,20,21,22.
With over 8000 metabolites, flavonoids are the largest class of polyphenols and about 20% of the total carbon flux in a plant cell goes through the flavonoid pathway23. Flavonoids such as anthocyanins and flavonols are synthesized from p-coumaroyl-CoA and three malonyl-CoA molecules by chalcone synthase, a type III polyketide synthase, in the first committed step (Fig. 1a). Chalcone isomerase (CHI) catalyzes the intramolecular cyclization of chalcones to (2S)-flavanones, which are a common substrate for multiple branches of the flavonoid pathway. Ectopic expression of CHI in tomato increased flavonol amounts over 70-fold24,25. Flavanone 3-hydroxylase (F3H) converts flavanones into dihydroflavonols, which are precursors for both anthocyanins and flavonols26. Flavonols have a C3 hydroxyl group and a C2–C3 double bond, with the committed step in the pathway catalyzed by flavonol synthase (FLS), a member of the 2-oxoglutarate-dependent dioxygenase (2-ODD) superfamily27. The first committed step in the anthocyanin pathway is catalyzed by dihydroflavonol reductase (DFR), which competes with FLS for dihydroflavonol precursors. FLS expression is regulated by transcription factors, including MYB1228.
PAL phenylalanine ammonia lyase, C4H cinnamate 4-hydroxylase, 4CL 4-coumaroyl:CoA-ligase, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, F3′H flavonoid 3′ hydroxylase, F3′5′H flavonoid 3′,5-hydroxylase, FLS flavonol synthase, DFR dihydroflavonol 4-reductase, ANS anthocyanin synthase
MYBs regulate numerous processes in plants, including reproduction, cell division, responses to biotic and abiotic stresses, and secondary metabolism29,30,31,32. In silico analysis were identified over 230 candidate MYB genes in potato, several of which were induced by hormone treatments, salt or drought stress33,34. Overexpression of MYB12 in tomato increased the flavonol content35. Expression of CHS, CHI, and FLS was upregulated in Arabidopsis by overexpressing MYB12, but drastically decreased in a myb12 mutant28. Both chlorogenic acid and anthocyanin synthesis in potato are regulated by the R2R3 MYB transcription factor stAN1, but little is known about the endogenous MYB12 in potato.
In addition to MYBs, small RNAs can regulate phenylpropanoid metabolism15. MicroRNAs control gene expression at the transcriptional and post-transcriptional levels, and regulate many biological processes including secondary metabolism36,37,38,39,40,41. Increasing miR857 levels in Arabidopsis decreased the lignin content42, while overexpression of miR408 specifically downregulated phenylalanine ammonia lyase (PAL) genes in transgenic Populus43. MiR828 regulates plant phenylpropanoid accumulation by targeting R2R3 MYBs, and is associated with anthocyanin expression in potato15,44. Expression of miR858 is associated with flavonol metabolism in various plants, and also with susceptibility to cyst nematodes45,46,47,48,49.
Potatoes with higher amounts of flavonols are dietarily desirable. However, while potatoes high in phenylpropanoids such as anthocyanins and chlorogenic acid are known, no high-flavonol potato has been identified. The reason for this is not clear, and the flavonol branch of the potato phenylpropanoid pathway has been less studied than the anthocyanin or hydroxycinnamic acid branches. A lack of knowledge about how tuber phenylpropanoid metabolism is regulated, impedes the ability to develop new cultivars with the optimized types and amounts. Previously, a functional analysis of the MYB transcription factor StAN1 (Anthocyanin 1) showed its effect were not specific to anthocyanins, but that hydroxycinnamic acids and flavonols were also affected. We hypothesized a self-regulatory loop may exist between sucrose and StAN150. In the current study, we present functional evidence that StAN1 interacts with the promoters of sucrolytic genes, and identified three additional regulators of potato flavonol metabolism, StMYB12A, C, and miR858.
Results
StAN1 and sucrose in potato phenylpropanoid metabolism
Previous work showed StAN1 overexpression increased not only anthocyanins, but other phenylpropanoids, including flavonols. The StAN1 promoter was found to have multiple sucrose responsive elements (SURE) and sucrose feeding induced phenylpropanoids50. We hypothesized that StAN1 overexpression affected multiple phenylpropanoid branches due, in part, to its effect on sucrose. Growing potato plantlets in tissue culture in the presence of 120 mM sucrose increased the amount of flavonols present, and infiltration of potato leaves with StAN1 increased expression of genes involved in sucrose breakdown (Fig. 2). The promoter regions of potato SUSY1, SUSY4, INV1, and INV2 genes have MYB binding domains (Fig. S1), suggesting that StAN1 may interact directly with the promoters of sucrolytic genes. To examine whether StAN1 directly interacts with sucrolytic genes, electrophoretic mobility shift assays were conducted using heterologous StAN1 protein purified from E. coli (Fig. S2) and promoter fragments from SUSY1 and INV1. StAN1 interacted with the promoters of both genes (Fig. 2c, d). Similar results were seen for SUSY4 and INV2 (data not shown).
a Flavonol content in potato plantlets grown in tissue culture with or without 120 mM sucrose. b Effect of AN1 infiltration on expression of genes involved in sucrose metabolism. c EMSA gels. All lanes have biotin labeled SUSY1 promoter fragment. Lane 1, minus StAN1 (control); Lane 2, StAn1 purified protein; Lane 3, StAN1 protein plus a 5-fold excess of cold competitor; Lane 4, StAN1 with a 50× excess of cold competitor. d EMSA gels. All lanes have biotin labeled INV1 promoter fragment. Lane 1, minus StAN1 (control); Lane2, StAn1 purified protein; Lane 3, StAN1 protein plus a 5-fold excess of cold competitor; Lane 4, StAN1 with a 50× excess of cold competitor
MYB12 A, B, and C in potato tubers
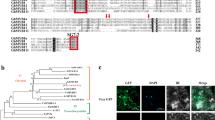
While StAN1 overexpression could be used to increase flavonols, anthocyanins are its main target and would also increase, which is not desirable in many types of commercial potatoes. Nor is it obvious how sucrose levels could be used commercially to increase flavonols. To identify other factors that regulate potato flavonol biosynthesis, a BLAST search was conducted for potato homologs of the MYB12 transcription factors known to be involved in flavonol regulation in plants like tomato and Arabidopsis. The three most homologous StMYB12 candidates were named StMYB12A, B, and C and were located on chromosomes 6, 12, and 1, respectively. Relative to the Arabidopsis flavonol transcription factors, AtMYB12, AtMYB111, and AtMYB11, StMYB12C and StMYB12A were most similar to AtMYB12 and AtMYB111, respectively (Fig. S3 and Table S1). StMYB12B had more similarity with AtMYB12 (47.6.6%) than it did AtMYB11 or AtMYB111, but its similarity with AtMYB12 was less than that shared by StMYB12A (50.6%) or C (51.9%). A phylogenetic tree was generated by aligning the potato MYB12s with MYBs involved in flavonol biosynthesis in other plant species (Fig. 3). This showed that StMYB12A and B were more closely related to each other than to StMYB12C, and that StMYB12B was most closely related to SlMYB11. StMYB12C was most related to SlMYB12. StMYB12A, B, and C were all more related to each other than to any of the AN1 MYBs. All the flavonol-related MYB transcription factors were highly conserved in the N-terminus, which contains the R2 and R3 domains (Fig. S3). The DW motif adjacent to the C-terminus was also conserved. Similar to what was observed with StAN1, analysis of the MYB12 promoters detected multiple sucrose responsive elements (SURE) in StMYB12 A and C, but not B (Table S2).
At Arabidopsis thaliana, Gt Gentiana trifloral, Md Malus domestica, Sl Solanum lycopersicum, St Solanum tuberosum, Vv Vitis vinifera, Zm Zea mays, Og Oncidium Gower, Hv Hordeum vulgare, Fa Fragaria ananassa, Lh Lilium hybrid, Am Antirrhinum majus, Ph Petunia hybrida, Nt Nicotiana tabacum, Bo Brassica oleracea, Mr Myrica rubr, Es Epimedium sagittatum, Pp Prunus persica, Pc Pyrus calleryana, Le Lycopersicum esculentum, In Ipomoea nil, GMYB10 is MYB10 in Gerbera. Bootstrap value from 0 to 1 are shown in different colors
Phenylpropanoids concentrations and MYB12 expression in different cultivars
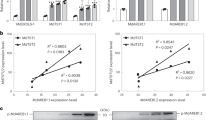
As the first step to determine if these candidate MYB12s were involved in potato flavonol regulation, we examined whether correlations existed between endogenous flavonol amounts and StMYB12 expression in different genotypes. StMYB12C was the most homologous to AtMYB12 and SlMYB12, so flavonol amounts and MYB12C expression were measured in ten potato genotypes with white flesh. MYB12C expression varied over 8-fold among these genotypes, and was highest in La Belle Russet, which also had the highest amounts of rutin and kaempferol (Fig. S4a, b, c). Relatively low flavonol amounts were present in all of these white-flesh potatoes. A positive correlation was observed between MYB12C expression and flavonol amounts (Fig. S4d, e).
Potatoes with blue-flesh, red-flesh, or purple-flesh are known to contain much higher amounts of phenylpropanoids, so we analyzed whether flavonol content and MYB expression was higher in color-flesh potatoes with more active phenylpropanoid metabolism. Flavonol content was analyzed in tubers from 11 additional cultivars representing potatoes with yellow, red, and purple flesh color in addition to white. Flavonol, chlorogenic acid, and total phenolic amounts varied markedly among these genotypes (Fig. 4), but flavonol amounts were unrelated to the amount of total phenylpropanoids in a genotype. AmaRosa and Magic Molly are cultivars with red-flesh or purple-flesh and had much higher amounts of total phenolics (Fig. 4a) and chlorogenic acid (Fig. 4b) than the white-flesh cultivars, yet Magic Molly had among the lowest amounts of flavonols (Fig. 4b).
Total phenolics (a) and total flavonols (b) in different genotypes. Chlorogenic acid amounts are shown in the inset. Relative expression of c MYB12A, d 12B, and e 12C in potatoes of varying flesh-color. f PAL and early flavonoid pathway gene expression. g Flavonol amounts in flowers, leaves or tubers; h FLS expression level in flowers, leaves and tubers; i Expression analysis of Myb12 A, B, and C in different organs. The data represents the means ± standard deviation of three biological replicates
Although StMYB12C had the highest similarity to AtMYB12 and its expression correlated with flavonol amounts in white potatoes (Fig. S4d, e), StMYB12A and B also had similarity to AtMYB12, so expression of all three transcription factors was examined. MYB12A and C were more highly expressed than MYB12B in all cultivars (Fig. 4c–e). Generally, potatoes with higher level of MYB12A and C expression tended to have higher amounts of flavonols, although there were exceptions such as in AmaRosa.
Expression of PAL and four genes involved in early flavonoid biosynthesis before the anthocyanin and flavonol branch point, were measured in seven of these cultivars (Fig. 4f). PAL expression was highest in AmaRosa and Magic Molly, the two cultivars with the highest amounts of total phenolics and lowest in Russet Burbank, the cultivar with the least total phenolics (Fig. 4a). Expression of the early flavonoid pathway genes, including CHS that encodes the first committed enzyme in flavonoid biosynthesis, were generally highest in La Ratte (Fig. 4f), which had the highest amounts of flavonols (Fig. 4b). Other than F3′H, expression of these genes was lowest in the cultivar with the lowest flavonol amounts, Russet Burbank and F3′H expression seemed quite variable.
MYB12 expression and flavonol amounts in different organs
Relative to flavonol amounts in some other crops including tomatoes, onions and broccoli51,52, tubers had low amounts, which might suggest potato lacks the necessary genes for efficient flavonol synthesis (Figs. 4b and S4). However, flavonol amounts were much higher in leaves and flowers, which are organs exposed to light during growth, in contrast to tubers that develop in the dark (Fig. 4g). This showed that potato has the necessary genes and biosynthetic capacity to synthesize large amounts of flavonols. Flowers had the highest concentration of flavonols, over 700-fold higher than in tubers, and leaves had about 75-fold higher amounts than tubers. FLS catalyzes the committed step in flavonol biosynthesis, and flavonol amounts were consistent with the amount of FLS expression in flowers, leaves, and tubers (Fig. 4h). Given these dramatic differences in flavonols, the expression of StMYB12A, B, and C was compared among flowers, leaves and tubers. StMYB12C was the most highly expressed of the three MYBs, whereas MYB12B had very low expression in all organs (Fig. 4i). MYB12A was more highly expressed than MYB12B, with expression levels in leaves and tubers similar to that of MYB12C, but it had much lower expression in flowers than MYB12C.
Agroinfiltration of MYB12 in potato leaves
The above results provide correlative evidence for a role for StMYB12A and C in potato flavonol regulation. To functionally characterize StMYB12 transcription factors, plant expression constructs containing MYB12A, B, and C were infiltrated into potato leaves to examine the effect of transient expression. Flavonols increased markedly in leaves infiltrated with MYB12A and C (Fig. 5d) as did chlorogenic acid (Fig. S5). No increase in anthocyanins was observed. Expression of early pathway genes such as PAL and flavonoid biosynthesis genes such as CHS, F3H, F3′H, and FLS were increased in leaves infiltrated with MYB12A or C (Fig. 5a–c). FLS expression was highest in leaves infiltrated with MYB12C. In contrast, infiltration with MYB12B had little effect on phenylpropanoid gene expression or metabolites.
StmiR858 negatively regulated flavonol biosynthesis through repressing MYB12
An additional potential regulatory mechanism for flavonol biosynthesis is via microRNAs. The microRNA, miR858, is involved in flavonol biosynthesis in Arabidopsis, apple (Malus domestica), muskmelon (Cucumis melo), and peach (Prunus persica)45,46,47,48. However, this has not been reported in potato. By searching the microRNA database of potato (mpss.danforthcenter.org/dbs), a putative miR858 homolog was located on chromosome 5 of potato (Fig. S6a). The mature sequence of StmiR858 contained 21 nucleotides with only a one nucleotide difference from the peach miR858 (Fig. S6b). The miR858 precursor was predicted to contain 85 nucleotides (Fig. S6c) using the software miRNAfold tanuki.ibisc.univ-evry.fr53.
An analysis was then conducted for potential targets of StmiR858 in the potato genome by using psRNATarget (plantgrn.noble.org/psRNATarget/home). The results showed that most of the potential targets are MYB transcription factors and included MYB12A and C (Table 1). To examine whether StmiR858 affected MYB12 expression and inhibits flavonol biosynthesis, a StmiR858 transient overexpression construct was agroinfiltrated into potato leaves. Infiltration increased miR858 levels ~2.5-fold (Fig. 6a). Expression of MYB12A in miR858 infiltrated leaves was 72% lower than the empty vector control (Fig. 6b). MYB12C was repressed more than 50% (Fig. 6d). However, a significant difference in MYB12B expression was not detected in leaves infiltrated with miR858 (Fig. 6c). Additional evidence MYB12A and C regulate potato flavonol metabolism and are affected by miR858, was that flavonol concentrations in the miR858 overexpression leaves decreased by ~57% (Fig. 6e). These results suggested that StmiR858 directly repressed MYB12 to regulate flavonol biosynthesis in potato.
a Expression level of mature miR858 after infiltration with the overexpressing construct or empty vector. b–d represent the expression level of MYB12A, B, and C in miR858 and empty pORE-O2 vector infiltrated leaves respectively. e Flavonol content in potato leaves after infiltration with and without overexpression of miR858. The data represents the means ± SD (n = 3). Asterisks indicates significant difference (p < 0.05)
The concentration of flavonols in flowers and leaves was determined, along with levels of MYB12C and StmiR858. Unlike white flowers, purple potato flowers have high amounts of anthocyanins, which compete with the flavonol branch of the pathway for common precursors. To assess potential differences between white and purple flowers on flavonol metabolism, both were examined. Expression of miR858 was higher in leaves than flowers (Fig. 7a). There was not a notable difference in miR858 expression in leaves from plants with purple flowers versus white flowers. Conversely, MYB12C was expressed 2.7 to 4.3-fold higher in flowers than in leaves (Fig. 7b). In particular, white flowers had higher expression of MYB12C relative to leaves from the same plants (Fig. 7b). Purple flowers had the lowest expression of miR858, but highest expression of MYB12C. Rutin and kaempferol were present in much higher concentrations in flowers than leaves (Fig. 7c, d), which is consistent with the higher expression of miR858 in leaves (Fig. 7a). StmiR858 expression was barely detectable in tubers (data not shown).
a Endogenous expression level of miR858 in potato leaves and flowers from three genotypes. b MYB12C endogenous expression levels in leaves and flowers from three potato genotypes. c Rutin content. d kaempferol content. -L, leaf; -F, flower. PAO7NCK10-57 and PAO7NCK17-66 are genotypes with white flowers, and Jester has purple flowers. The data represents the means ± SD of three biological replicates
Discussion
One potential explanation for the low amount of flavonols in tubers (Figs. 4b and S4) is that potato lacked the genes necessary for efficient flavonol metabolism. However, the high levels of flavonols in leaves and flowers do not support this explanation (Fig. 4g). Consequently, we sought to better characterize flavonol regulation in potato. While StAN1 affects multiple phenylpropanoid branches including flavonols, its primary target is anthocyanins. Additional factors must regulate potato flavonol metabolism. MYB12 regulates flavonol biosynthesis in Arabidopsis and tomato28,54. Transient overexpression of StMYB12A or C increased expression of flavonoid pathway genes including CHS, which encodes the first committed enzyme in flavonoid biosynthesis, CHI, which converts chalcone into naringenin, the precursor for all flavonoids, and F3H, which synthesizes dihydroflavonols, the direct substrate for FLS14,55.
The large increase in FLS expression and flavonol amounts in leaves infiltrated with StMYB12 A and C (Fig. 5) supports these MYBs being key regulators of potato flavonol metabolism, but do not preclude a role for additional, as yet unidentified, MYBs to regulate potato flavonols. In addition to flavonols, MYB12 A and C increased PAL expression and general phenylpropanoid metabolism, similar to what we observed previously with StAN1, the overexpression of which was not specific to the anthocyanin pathway20. Whereas overexpression of MYB12 had some overlapping effects to what we previously reported for StAN1, there were also differences. StAN1 infiltration in leaves increased anthocyanin synthesis, but StMYB12 infiltration did not.
Different phenylpropanoid regulatory mechanisms appear to be involved in different potato organs. In potato leaves, the anthocyanin pathway may be more tightly regulated than the flavonol branch. StAN1 and StMYB12 overexpression both increased activity of the overall general pathway and amounts of various leaf phenylpropanoids including flavonols, but only AN1 increased anthocyanins50. Thus, in potato leaves a consequence of increased general phenylpropanoid activity was increased flavonols but not anthocyanins. This is consistent with what was seen in tobacco leaves, where overexpressing AtPAL2 caused a two-fold increase in chlorogenic acid, a 5-fold increase in flavonols but no increase in anthocyanins56. In contrast, a general increase in tuber phenylpropanoid metabolism does not lead to increased flavonols. In potatoes with red or purple flesh, extremely high amounts of anthocyanins and various non-anthocyanin phenylpropanoids are present showing the pathway is very active, yet this is not enough to lead to elevated amounts of flavonols (Fig. 4). This is consistent with previous work in which potato phenylpropanoid metabolism was rerouted by suppressing hydroxycinnamic acid synthesis, and anthocyanins but not flavonols increased in rerouted purple tubers, whereas both increased in flowers20. Collectively, such data suggests high activity of the general phenylpropanoid pathway leads to elevated flavonols in leaves but not tubers, and flavonol synthesis is more tightly regulated in tubers than in leaves or flowers.
The effect of these R2R3 MYBs on phenylpropanoids outside of the specific anthocyanin or flavonol branches they target may be explained in part by coordinate regulation of the pathway57 and the need to increase PAL expression, which is rate limiting for the pathway, so that increased carbon flow is available to supply precursors for downstream anthocyanin and flavonol biosynthesis14. An additional explanation for why StMYB12 and StAN1 have a broad effect on phenylpropanoid metabolism, may be their interaction with sucrose.
Besides its role in primary metabolism, sucrose acts as a signal molecule that is responsive to environmental factors including light and temperature, including for phenylpropanoid metabolism58,59,60. The regulatory role of sucrose, which involves a mitogen-activated protein kinase (MAPK) signaling cascade, has been especially studied in plant anthocyanin metabolism, including grapes and Arabidopsis61,62,63. Previous work with potato showed correlations between sucrose and phenylpropanoid amounts during development and environmental responses19.
Previous work showed StAN1 overexpression decreased amounts of sucrose and hypothesized that this may be a consequence of putative MYB binding sites in the promoters of sucrolytic genes50. Here, we showed that purified StAN1 directly interacts with promoter fragments from sucrolytic genes (Fig. 2), and that the promoters of MYB12A and C each had four sucrose responsive elements (Table S2). Why MYB12B had little impact on flavonol metabolism is not clear, but it is interesting to note its lack of sucrose responsive elements. These data further support a model (Fig. 8) in which sucrose and MYBs are involved in a self-feedback loop to mediate phenylpropanoid expression, and may in part explain why both MYB12 and AN1 overexpression tend to increase general phenylpropanoid metabolism.
Sucrose has both a regulatory role and a metabolic role by supplying hexoses that can be channeled to phenylpropanoid biosynthesis. In the left green panel, sucrose increases MYB expression, which in turn increases phenylpropanoid metabolism. StAN1 concurrently increases expression of sucrolytic genes that generate hexoses to feed increased phenylpropanoid metabolism. In the right red panel, phenylpropanoid synthesis decreases either due to reduced StAN1 expression resulting from falling sucrose levels caused by StAN1 induced sucrolysis, or from increased expression of MiR858, a negative regulator. Sucrose concentrations and MYB expression form a self-regulatory cycle, where sucrose promotes MYB expression but MYB expression promotes sucrolysis. The circular arrows connecting sucrose in the two panels, indicate that the concentration of sucrose cycles and influences phenylpropanoid metabolism. Green indicates conditions that increase phenylpropanoid metabolism and red conditions that do not. Solid arrows indicate direct effects on phenylpropanoid regulation and dashed arrows indirect
In this proposed model, upregulated MYB expression increases sucrose catabolism. This may allow a finely tuned regulatory cycle in which sucrose amounts are balanced between promoting MYB expression and channeling hexoses into phenylpropanoid metabolism via the glycolytic and shikimate pathways. By promoting sucrose catabolism, MYB expression would be predicted to downregulate its own expression by decreasing the supply of sucrose if carbohydrate metabolism does not maintain sufficient levels of sucrose. This interaction between MYBs, sucrolytic genes, and sucrose suggests a mechanistic link between phenylpropanoid metabolism and starch-sucrose metabolism. Carbohydrate metabolism in potato, including starch-sucrose interconversion, is finely regulated64, and this linkage may allow an additional fine mechanism of control for tuber phenylpropanoid regulation.
This work also showed miR858 affects potato flavonol metabolism, further indicating the complexity of flavonol regulation. MiRNAs are involved in a variety of processes, such as plant growth and development, host–pathogen interactions, apoptosis and phenylpropanoid metabolism65,66,67. MiR828 is involved in phenylpropanoid metabolism and interacts with MYB transcription factors such as MYB113, PAP1, and PAP2 to coordinate anthocyanin biosynthesis in Pinus resinosa68. Overexpression of miR828 reduced anthocyanin levels in Arabidopsis and led to early flowering and downregulation of MYB transcription factors related to flavonoid biosynthesis69. However, others suggest miR828 is a positive regulator of anthocyanin synthesis in potato and grapes44,70. Similar contradictory findings have been reported about miR858, where it has been reported as a negative regulator of anthocyanins in Kiwi fruit and tomato71,72 but a positive regulator in Arabidopsis73.
Based on evidence linking it to flavonol regulation in other plants, miR858 seemed a logical candidate in potato45,46,47,48. Unlike its effect on MYB12A and C, StmiR858 overexpression did not affect MYB12B, which is consistent with the other data that did not support a role for MYB12B in potato flavonol metabolism. Moreover, flavonol content was much higher in flowers than in leaves, and endogenous StmiR858 levels in leaves were significantly higher than that in flowers, which would be expected if miR858 was a negative regulator. However, unlike in leaves or flowers, barely detectable levels of miR858 were present in tubers (data not shown). So, while StmiR858 was involved in flavonol regulation, it did not appear to be the explanation for why flavonols are present in high amounts in potato flowers and leaves, but not in tubers. Additional research is needed to determine if MiR858 affects additional genes, such as other candidates in Table 1.
Additional, unknown regulatory mechanisms must be involved in the regulation of tuber flavonol biosynthesis. Given the high amount of flavonols in flowers, the lack of necessary genes in potato is not an explanation, nor is an inability of tubers to synthesize flavonol precursors, given that tubers can have very high amounts of anthocyanins that share immediate precursors with flavonols. One possible mechanism might involve light, given that potato organs that develop in the light (leaves and flowers) had up to 700-fold higher amounts of flavonols than organs that develop in the dark, (tubers) and numerous light-responsive elements were present in the MYB12 promoters (Fig. 4g and Table S3). Light is widely known to promote flavonol synthesis in plants74,75 and has also been shown to induce expression of miR858 in plants69,76, suggesting microRNAs could be another mechanism that transduce environmental signals to mediate potato phenylpropanoid metabolism.
In summary, the direct interaction of StAN1 with promoter sequences of multiple sucrolytic genes and the presence of sucrose responsive elements in the promoters of MYB12A and C support an important role for sucrose in regulating potato phenylpropanoids. MYB12A and C were positive regulators of FLS, the expression of which appears to be a key determinant of flavonol concentrations in potato. StmiR858 functioned as a negative regulator of flavonol metabolism and adds another layer of complexity to potato phenylpropanoid regulation. The presence of various environmentally responsive cis-acting elements in the StAN1 and StMYB12 promoters, along with the proposed MYB-sucrose regulatory loop suggest mechanisms for how potato phenylpropanoid amounts are modulated. Additional studies are needed to delineate why tubers have much lower flavonol amounts than leaves, and to devise ways to selectively increase individual phenylpropanoid branches without increasing others. Understanding the regulatory mechanisms that control phenylpropanoid metabolism in potato will facilitate developing plants with optimized phenylpropanoid profiles for human health and crop performance.
Materials and methods
Isolation of MYB12 homologs from potato and phylogenetic analysis
MYB12 sequences from Arabidopsis and tomato were blasted in potato genome sequence databases to identify putative potato homologs77. Protein blast searches showed three putative MYB12 homologs in potato. StMYB12A, B, and C were amplified from potato cDNA using gene specific primers designed to isolate the full-length cDNA. The amplified gene fragments were cloned into TOPO T/A cloning vector and transformed in E. coli. Constructs were verified by sequencing. Sequences were aligned using Clustal omega alignment software and a phylogenetic tree was constructed with neighbor joining method using TOL78.
Gene expression analysis
Gene expression was analyzed by qRT-PCR analysis. PCR was carried out using cDNA prepared from 2 µg of total RNA and further diluting the cDNA ten times. Specific primers were used for expression of different genes (Table S4). PCR reactions were setup in 10 µl reactions with 3 µl diluted cDNA, 1 µl forward primer, 1 µl reverse primer, and 5 µl SYBR Green mix. Samples had a 2 min preincubation at 95 °C followed by 40 cycles of 10 s denaturation at 95 °C, 15 s annealing at 60 °C, and 25 s extension at 72 °C, in a LightCycler 480 (Roche). Relative expression of the genes was calculated by the ΔΔCT method by normalizing the expression levels of target genes to the expression mean of the housekeeping gene actin. Delta CT = CT value of target gene—CT value of housekeeping gene (Actin). Expression = 2(-delta delta CT). The expression level of miR858 was detected with stem-loop RT-PCR method79.
Agrobacterium infiltration studies
Full-length coding sequences of the transcription factors StMYB12 A, B, C, and AN1 were cloned into the modified binary vector pOREO2 with a 35S promoter80. The artificial miR858 expression plasmid was constructed following the protocol on website wmd3.weigelworld.org81. The pRS300 plasmid was used to generate artificial miR858. Positive clones were confirmed by PCR and sequencing. An infiltration approach was adopted based on published studies82,83. A single positive colony was cultured in 5 ml of LB medium overnight and used to inoculate 50 ml of medium. Cells were harvested and adjusted to O.D. 0.5 in 10 mM MgCl2 containing 100 μM acetosyringone. Cultures were then diluted (1:1) with the gene silencing suppression vector p19TBSV of tomato bushy stunt virus37. Samples were infiltrated into leaves of 4-week-old potato plants (Solanum tuberosum cv. La Ratte). Leaves were harvested at 2 days of post-infiltration. Empty vector was used as control along with non-infiltrated samples.
Phenylpropanoid quantification
Phenylpropanoids were measured in light and dark grown organs (leaves, flowers, tubers). Total phenolics were extracted from 50 mg freeze-dried samples with a total of 1.5 ml of 50% methanol, 1 mM EDTA, and 2.5% metaphosphoric acid. Total phenolics were quantitated as gallic acid equivalents (mg gallic acid/g extract) using the FC method84. Reactions were measured at 755 nm using a Synergy 2 Multi-Mode microplate reader (BioTek Instruments). Flavonols were analyzed by using HPLC-MS, with 20 μl of sample injected onto an Agilent 1200 HPLC equipped with an Onyx 100 × 4.6 mm column (Phenomenex) at 35 °C with a flow rate of 1 ml min−1 and a gradient elution of 0–1 min 100% A, 1–9 min 0–30% B, 9–10.5 min 30% B, 10.5–14 min 35–65% B, 14–16 min at 65–100% B, 16–16.5 min 100% B (Buffer A: 10 mM formic acid pH 3.5 with NH4OH; Buffer B: 100% methanol with 5 mM ammonium formate as described50). Flavonols were quantified as rutin equivalents.
Heterologous expression of StAN1 protein in E. coli
The potato AN1 cDNA comprising of start codon and stop codon was amplified with forward and reverse primers having BamHI and SacI sites, respectively. Amplified product was cloned first in T/A cloning vector. After sequence confirmation, the full-length cDNA fragment was ligated into the pET28a vector (Novagen). These constructs were transformed into the host strain E. coli (Shuffle cells) for IPTG-induced expression. Recombinant his-tagged StAN1 was obtained from cells grown in 200 ml Luria Broth with shaking at 37 °C until an O.D. of 0.4. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.4 mM, the incubation continued for 3 h, after which cells were harvested by centrifugation at 5000×g for 5 min at 4 °C. The pellet was resuspended in 5 ml cold protein extraction buffer (20 mM Tris-Cl (pH-8.0), 200 µg/ml of lysozyme, 150 mM NaCl and 20 µg/ml DNase1, 5 mM imidazole and 2 M urea). Protein was isolated using a repetitive freeze thaw method and purified on a NTA column (Qiagen) at 4 °C according to manufacturer instructions, with stepwise imidazole elution at 25, 50, 100, and 200 mM. The eluted fractions were analyzed on SDS page85.
Isolation of promoter fragments for SUSY1, SUSY4, INV1, and INV2
Cultivar Purple Majesty genomic DNA was used to isolate promoter fragments (~1.5 kb) of SUSY1, SUSY4, INV1, and INV2. Specific primers were designed from promoter sequences obtained from the potato genome database77. Isolated promoter fragments were sequenced and MYB binding consensus sequences identified within them (Fig. S1). Cloned fragments of SUSY1, SUSY4, INV1, and INV2 were used as templates, and nested PCR was carried out using specific primers designed to amplify 100–150 bp regions containing multiple MYB binding sequences. The amplified regions were cloned and sequenced to confirm their identity.
Electrophoretic mobility shift assay for StAN1 and sucrolytic gene promoters
Fifty nanogram of amplified DNA fragments (SUSY1, SUSY4, INV1, and INV2) were end-labeled with biotin. A LightShift chemiluminescent EMSA kit was used according to directions (Thermo Scientific) and blots were photographed with a CCD camera. Two microgram of purified protein was used for binding. For competition assays a 5-fold or 50-fold excess of unlabeled transcript was used.
References
Ray, D. K., Mueller, N. D., West, P. C. & Foley, J. A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8, e66428 (2013).
Morris, W. L. et al. Carotenogenesis during tuber development and storage in potato. J. Exp. Bot. 55, 975–982 (2004).
Love, S. L. et al. Stability of expression and concentration of ascorbic acid in North American potato germplasm. HortScience 39, 156–160 (2004).
Mooney, S. et al. Genotype-specific changes in vitamin B6 content and the PDX family in potato. Biomed. Res. Int. 2013, 389723 (2013).
Burrowes, J. D. & Ramer, N. J. Changes in the potassium content of different potato varieties after cooking. J. Ren. Nutr. 18, 249 (2008).
Robinson, B., Sathuvalli, V., Bamberg, J. & Goyer, A. Exploring folate diversity in wild and primitive potatoes for modern crop improvement. Genes 6, 1300–1314 (2015).
Navarre, D. A., Brown, C. R. & Sathuvalli, V. R. Potato vitamins, minerals and phytonutrients from a plant biology perspective. Am. J. Potato Res. 96, 111–126 (2019).
Bagri, D. S., Upadhyaya, D. C., Kumar, A. & Upadhyaya, C. P. Overexpression of PDX-II gene in potato (Solanum tuberosum L.) leads to the enhanced accumulation of vitamin B6 in tuber tissues and tolerance to abiotic stresses. Plant Sci. 272, 267–275 (2018).
Cardona, F., Andrés-Lacueva, C., Tulipani, S., Tinahones, F. J. & Queipo-Ortuño, M. I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 24, 1415–1422 (2013).
Scalbert, A., Manach, C., Morand, C., Remesy, C. & Jimenez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 45, 287–306 (2005).
Tohge, T. & Fernie, A. R. An overview of compounds derived from the Shikimate and phenylpropanoid pathways and their medicinal importance. Mini Rev. Med. Chem. 17, 1013–1027 (2017).
Bischoff, S. C. Quercetin: potentials in the prevention and therapy of disease. Curr. Opin. Clin. Nutr. Metab. Care 11, 733–740 (2008).
Ferrer, J.-L., Austin, M., Stewart, C. Jr. & Noel, J. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370 (2008).
Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20 (2010).
Deng, Y. & Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 36, 257–290 (2017).
Sheehan, H. et al. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nat. Genet. 48, 159 (2016).
Andre, C. M. et al. Antioxidant profiling of native Andean potato tubers (Solanum tuberosum L.) reveals cultivars with high levels of beta-carotene, alpha-tocopherol, chlorogenic acid, and petanin. J. Agric. Food Chem. 55, 10839–10849 (2007).
Navarre, D. A., Pillai, S., Shakya, R. & Holden, M. J. HPLC profiling of phenolics in diverse potato genotypes. Food Chem. 127, 34–41 (2011).
Navarre, D. A., Payyavula, R. S., Shakya, R., Knowles, N. R. & Pillai, S. Changes in potato phenylpropanoid metabolism during tuber development. Plant Physiol. Biochem. 65, 89–101 (2013).
Payyavula, R. S. et al. Synthesis and regulation of chlorogenic acid in potato: rerouting phenylpropanoid flux in HQT-silenced lines. Plant Biotechnol. J. 13, 551–564 (2015).
Valiñas, M. A., Lanteri, M. L., ten Have, A. & Andreu, A. B. Chlorogenic acid, anthocyanin and flavan-3-ol biosynthesis in flesh and skin of Andean potato tubers (Solanum tuberosum subsp. andigena). Food Chem. 229, 837–846 (2017).
Tudela, J. A., Cantos, E., Espín, J. C., Tomás-Barberán, F. A. & Gil, M. I. Induction of antioxidant flavonol biosynthesis in fresh-cut potatoes. Effect of domestic cooking. J. Agric. Food Chem. 50, 5925–5931 (2002).
Tohge, T., de Souza, L. P. & Fernie, A. R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028 (2017).
Colliver, S. et al. Improving the nutritional content of tomatoes through reprogramming their flavonoid biosynthetic pathway. Phytochem. Rev. 1, 113–123 (2002).
Muir, S. R. et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 19, 470 (2001).
Holton, T. A. & Cornish, E. C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083 (1995).
Holton, T. A., Brugliera, F. & Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4, 1003–1010 (1993).
Mehrtens, F., Kranz, H., Bednarek, P. & Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096 (2005).
Dubos, C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010).
Rotman, N. et al. A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15, 244–248 (2005).
Liu, J., Osbourn, A. & Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708 (2015).
Liu, M. et al. CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J. Exp. Bot. 70, 69–84 (2018).
Sun, W., Ma, Z., Chen, H. & Liu, M. MYB gene family in potato (Solanum tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 20, 4847 (2019).
Li, X. et al. Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 9, 317 (2019).
Luo, J. et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326 (2008).
Vaucheret, H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771 (2006).
Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 (2009).
Zhang, Y. et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6, 8635 (2015).
Yates, L. A., Norbury, C. J. & Gilbert, R. J. The long and short of microRNA. Cell 153, 516–519 (2013).
Zeng, Y. & Cullen, B. R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32, 4776–4785 (2004).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004).
Zhao, Y. et al. MicroRNA857 is involved in the regulation of secondary growth of vascular tissues in arabidopsis. Plant Physiol. 169, 2539–2552 (2015).
Shi, R., Yang, C., Lu, S., Sederoff, R. & Chiang, V. L. Specific down-regulation of PAL genes by artificial microRNAs in Populus trichocarpa. Planta 232, 1281–1288 (2010).
Bonar, N. et al. Potato miR828 is associated with purple tuber skin and flesh color. Front. Plant Sci. 9, 1742 (2018).
Fahlgren, N. et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2, e219 (2007).
Garcia-Mas, J. et al. The genome of melon (Cucumis melo L.). Proc. Natl Acad. Sci. USA 109, 11872–11877 (2012).
Xia, R., Zhu, H., An, Y. Q., Beers, E. P. & Liu, Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13, R47 (2012).
Zhu, H. et al. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12, 149 (2012).
Piya, S., Kihm, C., Rice, J. H., Baum, T. J. & Hewezi, T. Cooperative regulatory functions of miR858 and MYB83 during Cyst nematode parasitism. Plant Physiol. 174, 1897 (2017).
Payyavula, R. S., Singh, R. K. & Navarre, D. A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 64, 5115–5131 (2013).
Justesen, U., Knuthsen, P. & Leth, T. Quantitative analysis of flavonols, flavones, and flavanones in fruits, vegetables and beverages by high-performance liquid chromatography with photo-diode array and mass spectrometric detection. J. Chromatogr. 799, 101–110 (1998).
Crozier, A., Lean, M. E. J., McDonald, M. S. & Black, C. Quantitative analysis of the flavonoid content of commercial tomatoes, onions, lettuce, and celery. J. Agric. Food Chem. 45, 590–595 (1997).
Tav, C., Tempel, S., Poligny, L. & Tahi, F. miRNAFold: a web server for fast miRNA precursor prediction in genomes. Nucleic Acids Res. 44, W181–W184 (2016).
Adato, A. et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 5, e1000777 (2009).
Nakayama, T., Takahashi, S. & Waki, T. Formation of flavonoid metabolons: functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant. Sci. 10, 1–12 (2019).
Chang, J., Luo, J. & He, G. Regulation of polyphenols accumulation by combined overexpression/silencing key enzymes of phenylpropanoid pathway. Acta Biochim. Biophys. Sin. 41, 123–130 (2009).
Dixon, R. A. & Paiva, N. L. Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097 (1995).
Gollop, R., Even, S., Colova-Tsolova, V. & Perl, A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J. Exp. Bot. 53, 1397–1409 (2002).
Koch, K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246 (2004).
Tognetti, J. A., Pontis, H. G. & Martínez-Noël, G. M. Sucrose signaling in plants: a world yet to be explored. Plant Signal. Behav. 8, e23316 (2013).
Boss, P. K., Davies, C. & Robinson, S. P. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111, 1059–1066 (1996).
Meng, L. S. et al. Sucrose signaling regulates anthocyanin biosynthesis through a MAPK cascade in Arabidopsis thaliana. Genetics 210, 607–619 (2018).
Teng, S., Keurentjes, J., Bentsink, L. N., Koornneef, M. & Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852 (2005).
Geigenberger, P., Stitt, M. & Fernie, A. R. Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ. 27, 655–673 (2004).
Hong, Y. & Jackson, S. Floral induction and flower formation–the role and potential applications of miRNAs. Plant Biotechnol. J. 13, 282–292 (2015).
Leung, A. K. & Sharp, P. A. MicroRNA functions in stress responses. Mol. Cell 40, 205–215 (2010).
Rajagopalan, R., Vaucheret, H., Trejo, J. & Bartel, D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425 (2006).
Luo, Q.-J., Mittal, A., Jia, F. & Rock, C. D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80, 117–129 (2012).
Sharma, D. et al. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 171, 944–959 (2016).
Tirumalai, V., Swetha, C., Nair, A., Pandit, A. & Shivaprasad, P. V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 70, 4775–4792 (2019).
Jia, X. et al. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293 (2015).
Li, Y. et al. MicroRNA858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta) based on small RNA sequencing. PLoS ONE 14, e0217480 (2019).
Wang, Y., Wang, Y., Song, Z. & Zhang, H. Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405 (2016).
Fgaier, S., de Almeida Lopes, M. M., de Oliveira Silva, E., Aarrouf, J. & Urban, L. Xenon lamps used for fruit surface sterilization can increase the content of total flavonols in leaves of Lactuca sativa L. without any negative effect on net photosynthesis. PLoS ONE 14, e0223787 (2019).
Zoratti, L., Karppinen, K., Luengo Escobar, A., Häggman, H. & Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 5, 534 (2014).
Sánchez-Retuerta, C., Suaréz-López, P. & Henriques, R. Under a new light: regulation of light-dependent pathways by non-coding RNAs. Front. Plant Sci. 9, 962 (2018).
PGSC. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Kramer, M. F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 15, 10 (2011).
Coutu, C. et al. pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 16, 771–781 (2007).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006).
Aguilar, E., del Toro, F. J., Chung, B.-N., Canto, T. & Tenllado, F. Infection of Nicotiana benthamiana Plants with potato virus X (PVX). Bio-Protoc. 6, e2063 (2016).
Du, J., Rietman, H. & Vleeshouwers, V. G. A. A. Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. JoVE 83, e50971–e50971 (2014).
Singleton, V. L. & Rossi, J. A. Jr. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 16, 144–158 (1965).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Acknowledgements
We thank Rich Quick and Launa Hamlin (USDA-ARS) for help with harvesting and providing the plant material. We thank Joanne M. Holden for help with measurement of phenylpropanoids. We thank the Northwest Potato Research Consortium and Andy Jensen for support. This study was supported by grants from the Northwest Potato Research Consortium, and funding from the USDA-Agricultural Research Service 2092-21220-001-00D.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, S., Singh, R.K., Moehninsi et al. R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Hortic Res 8, 25 (2021). https://doi.org/10.1038/s41438-021-00463-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00463-9
This article is cited by
-
Combined analysis of mRNA and miRNA transcriptomes reveals the regulatory mechanism of Xanthomonas arboricola pv pruni resistance in Prunus persica
BMC Genomics (2024)
-
Tissue-Specific Expression of StMYB3 Gene Involves Regulating Potato Tuber Skin and Vascular Bundle Anthocyanin Biosynthesis
Potato Research (2023)
-
Characterization of TaSPP-5A gene associated with sucrose content in wheat (Triticum aestivum L.)
BMC Plant Biology (2022)
-
Identification of microRNAs and their target genes related to needle discoloration of evergreen tree Chinese cedar (Cryptomeria fortunei) in cold winters
Planta (2021)