Abstract
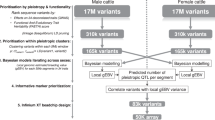
Ear size is a classical model for hot climate adaptation following the evolution, but the genetic basis of the traits associated with ear size remains to be elucidated. Here, we performed a genome-wide association study on 158 cattle to explain the genetic mechanism of ear size. One region on BTA6 between 36.79 and 38.80 Mb included 50 suggestive SNPs and 4 significant SNPs that were significantly associated with ear size. The most significant locus (P = 1.30 × 10−8) was a missense mutation (T250I) on the seventh exon of integrin-binding sialoprotein (IBSP), which had an allele substitution effect of 23.46 cm2 for ear size. Furthermore, this mutation will cause changes in the three-dimensional structure of the protein. To further identify genes underlying this typical feature, we performed a genome scan among nine cattle breeds with different ear sizes by using SweeD. Results suggested that IBSP was under positive selection among four breeds with relatively large ear sizes. The expression levels of IBSP in ear tissues of large- and small-ear cattle were significantly different. A haplotype diversity survey of this missense mutation in worldwide cattle breeds strongly implied that the origin of this missense mutation event was Bos taurus. These findings have important theoretical importance for the exploration of major genes associated with ear size and provide important molecular markers for the identification of cattle germplasm resources.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw whole-genome sequencing data were reported in our previous study (Chen et al. 2020) and are available at the NCBI Short Read Archive under the BioProject accession number PRJNA555741.
References
Alvord LS, Farmer BL (1997) Anatomy and orientation of the human external ear. J Am Acad Audiol 8:383–390
Bos EJ, Pluemeekers M, Helder M, Kuzmin N, van der Laan K, Groot M-L et al. (2018) Structural and mechanical comparison of human ear, alar, and septal cartilage. Plast Reconstr Surg Glob Open 6:e1610
Bouleftour W, Boudiffa M, Wade-Gueye NM, Bouët G, Cardelli M, Laroche N et al. (2014) Skeletal development of mice lacking bone sialoprotein (BSP)-Impairment of long bone growth and progressive establishment of high trabecular bone mass. PLoS One 9(5):e95144
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81(5):1084–1097
Chen N, Fu W, Zhao J, Shen J, Chen Q, Zheng Z et al. (2020) BGVD: Integrated Web-database for Bovine Sequencing Variations and Selective Signatures. Genom. Proteom. Bioinform 18(2):186–193
Chen Q, Zhan J, Shen J, Qu K, Hanif Q, Liu J et al. (2020) Whole-genome resequencing reveals diversity, global and local ancestry proportions in Yunling cattle. J Anim Breed Genet 137(6):641–650
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
DeGiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R (2016) SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics 32(12):1895–1897
Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC et al. (2011) Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet 7(4):e1001372
Ekdale EG (2016) Form and function of the mammalian inner ear. J Anat 228(2):324–337
Fukui N, Miyamoto Y, Nakajima M, Ikeda Y, Hikita A, Furukawa H et al. (2008) Zonal gene expression of chondrocytes in osteoarthritic cartilage. Arthritis Rheum 58(12):3843–3853
Gao L, Xu SS, Yang JQ, Shen M, Li MH (2018) Genome‐wide association study reveals novel genes for the ear size in sheep (Ovis aries). Anim Genet 49(4):345–348
Green MR, Sambrook J (2012) Molecular cloning: a laboratory manual 4th en. Cold Spring Harbor Laboratory Press: New York, NY, USA
Herath S, Dobson H, Bryant C, Sheldon I (2006) Use of the cow as a large animal model of uterine infection and immunity. J Reprod Immunol 69(1):13–22
Horwitz GC, Risner-Janiczek JR, Jones SM, Holt JR (2011) HCN channels expressed in the inner ear are necessary for normal balance function. J Neurosci 31(46):16814–16825
Kim RH, Shapiro HS, Li JJ, Wrana JL, Sodek J (1994) Characterization of the human bone sialoprotein (BSP) gene and its promoter sequence. Matrix Biol 14(1):31–40
Komori T (2010) Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res 339(1):189–195
Komori T (2017) Roles of Runx2 in Skeletal Development. Adv Exp Med Biol 962:83–93
Komori T (2018) Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem Cell Biol 149(4):313–323
Law MP, Ahier R, Field S (1979) The effect of prior heat treatment on the thermal enhancement of radiation damage in the mouse ear. Br J Radiol 52(616):315–321
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Livak KJ, Schmittgen TD (2002) Analysis of relative gene expression data using real-time quantitative PCR. Methods 25(4):402–408
Lui JC, Yue S, Lee A, Kikani B, Temnycky A, Barnes KM et al. (2019) Persistent Sox9 expression in hypertrophic chondrocytes suppresses transdifferentiation into osteoblasts. Bone 125:169–177
Ma J, Qi W, Ren D, Duan Y, Qiao R, Guo Y et al. (2009) A genome scan for quantitative trait loci affecting three ear traits in a White Duroc× Chinese Erhualian resource population. Anim Genet 40(4):463–467
Nekrutenko A, Taylor J (2012) Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet 13(9):667–672
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2(12):e190
Pavlidis P, Živković D, Stamatakis A, Alachiotis N (2013) SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol 30(9):2224–2234
Phillips PK, Heath JE (1992) Heat exchange by the pinna of the African elephant (Loxodonta africana). Comp Biochem Physiol Comp Physiol 101(4):693–699
Phoofolo P (1993) Epidemics and revolutions: the rinderpest epidemic in late nineteenth-century Southern Africa. Past Present 138:112–143
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Reefmann N, Kaszàs FB, Wechsler B, Gygax L (2009) Ear and tail postures as indicators of emotional valence in sheep. Appl Anim Behav Sci 118(3-4):199–207
Ren J, Duan Y, Qiao R, Yao F, Zhang Z, Yang B et al. (2011) A missense mutation in PPARD causes a major QTL effect on ear size in pigs. PLoS Genet 7(5):e1002043
Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E et al. (2020) De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 9(3):giaa021
Van Hemmen JL (2003) Auditory system. Biol. Cybern 89:317
Torres M, Giráldez F (1998) The development of the vertebrate inner ear. Mech Dev 71(1-2):5–21
Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Pielberg GR, Sigurdsson S et al. (2011) Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet 7(10):e1002316
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164
Webster DB (1966) Ear structure and function in modern mammals. Am Zool 6(3):451–466
Wei W, De Koning D, Penman J, Finlayson H, Archibald A, Haley C (2007) QTL modulating ear size and erectness in pigs. Anim Genet 38(3):222–226
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Wilkinson S, Lu ZH, Megens H-J, Archibald AL, Haley C, Jackson IJ et al. (2013) Signatures of diversifying selection in European pig breeds. PLoS Genet 9(4):e1003453
Wuttke M, Müller S, Nitsche DP, Paulsson M, Hanisch F-G, Maurer P (2001) Structural characterization of human recombinant and bone-derived bone sialoprotein functional implications for cell attachment and hydroxyapatite binding. J Biol Chem 276(39):36839–36848
Yang H, Zhang X, Hou L, Hong Y, Ren J, Guo Y (2013) Measurement of the ear area of pig in vivo with the pixel method. Acta Agric Univ Jiangxi 35(5):1024–1029
Zhang L, Liang J, Luo W, Liu X, Yan H, Zhao K et al. (2014) Genome-wide scan reveals LEMD3 and WIF1 on SSC5 as the candidates for porcine ear size. PLoS One 9(7):e102085
Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44(7):821–824
Acknowledgements
We would like to thank High-Performance Computing (HPC) of Northwest A&F University (NWAFU) for providing computing resources. The study was supported by the National Natural Science Foundation of China (31872317) and China Agriculture Research System of MOF and MARA (Grant No. CARS-37).
Author information
Authors and Affiliations
Contributions
YJ and CZL designed the study and supported the funding. JFS and XTX curated and analyzed the data. JFS wrote the original manuscript. QH reviewed and edited the manuscript. LYS, XHM, NBC, and HC reviewed the manuscript. BZH, KXQ, and JCZ organized sampling and conducted fieldwork. All authors commented on the manuscript and gave final approval for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor Christine Baes
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, J., Xia, X., Sun, L. et al. Genome-wide association study reveals that the IBSP locus affects ear size in cattle. Heredity 130, 394–401 (2023). https://doi.org/10.1038/s41437-023-00614-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00614-9