Abstract
Morphological scaling relationships between the sizes of individual traits and the body captures the characteristic shape of a species, and their evolution is the primary mechanism of morphological diversification. However, we have almost no knowledge of the genetic variation of scaling, which is critical if we are to understand how scaling evolves. Here we explore the genetics of population scaling relationships (scaling relationships fit to multiple genetically-distinct individuals in a population) by describing the distribution of individual scaling relationships (genotype-specific scaling relationships that are unseen or cryptic). These individual scaling relationships harbor the genetic variation in the developmental mechanisms that regulate trait growth relative to body growth, and theoretical studies suggest that their distribution dictates how the population scaling relationship will respond to selection. Using variation in nutrition to generate size variation within 197 isogenic lineages of Drosophila melanogaster, we reveal extensive variation in the slopes of the wing-body and leg-body individual scaling relationships among genotypes. This variation reflects variation in the nutritionally-induced size plasticity of the wing, leg, and body. Surprisingly, we find that variation in the slope of individual scaling relationships primarily results from variation in nutritionally-induced plasticity of body size, not leg or wing size. These data allow us to predict how different selection regimes affect scaling in Drosophila, and is the first step in identifying the genetic targets of such selection. More generally, our approach provides a framework for understanding the genetic variation of scaling, an important prerequisite to explaining how selection changes scaling and morphology.
Similar content being viewed by others
Introduction
Static morphological scaling relationships (commonly referred to as “static allometries” (Klingenberg and Zimmermann 1992) describe the size relationships among morphological traits as they co-vary with body size among individuals at the same developmental stage in a population, species, or other biological group (Shingleton 2010). In as much as the shape of an animal is determined by the relative size of its constituent body parts, differences in morphological scaling relationships capture variation in body shape within and among animal groups. Because morphological diversity is dominated by variation in animal shape, the study of morphological scaling relationships has been the focus of evolutionary biologists for well over a century (Huxley 1924, 1932; Gould 1966; Thompson and Bonner 1992; Gayon 2000). Nevertheless, until recently, almost nothing was known regarding the developmental-genetic mechanisms that regulate morphological scaling and that are the proximate targets of selection for morphological change (Tang et al. 2011; Emlen et al. 2012; Casasa et al. 2017). Even less is known of the distributions of genetic variation in these mechanisms that should determine how scaling responds to selection. This is primarily because, unlike most other morphological traits, scaling is ostensibly a characteristic of a group rather than an individual. Because groups of animals are typically genetically heterogeneous, the genetic variation of morphological scaling is therefore difficult to describe.
Historically, the literature has been concerned mostly with “population” scaling relationships (Huxley and Tessier 1936; Gould 1973; Klingenberg and Zimmermann 1992; Wilkinson 1993; Dreyer et al. 2016). Here, size variation among individuals results from genetic and environmental variation, and the line fit to the relationship between trait and body size reveals how these covary among individuals in that particular population and environment. (For clarity, here we restrict the term “trait” to morphological characteristics other than body size). When trait and body size are plotted on a log-log scale, the slope of their relationship is referred to as the allometric coefficient (Huxley and Tessier 1936) which reflects the relative sensitivity of the trait and body to the myriad environmental and genetic factors that affect their size (Shingleton et al. 2007).
More recently, attention has turned to individual scaling relationships (Fig. 1; Dreyer et al. 2016). These result from co-variation in trait and body size due to variation in a single environmental or genetic factor, with all other size regulatory factors held constant (including genotype). Variation in slopes and intercepts among individual scaling relationships reflects genetically-based differences among individuals in how trait and body size respond to the varying size-regulatory factor. When the size-regulatory factor is environmental—yielding an individual environmental scaling relationship—variation among scaling relationships is a consequence of genotype-by-environment interactions. Importantly, different environmental factors can generate different individual scaling relationships for the same genotype. For example, in Drosophila melanogaster, the effects of nutrition vs. temperature variation during development generate different morphological scaling relationships between morphological traits and the body (Shingleton et al. 2009). Similarly, when the size-regulatory factor is genetic—yielding an individual genetic scaling relationship—variation among scaling relationships is a consequence of gene-by-genotype interactions. This would be generated by allelic variation at a single locus interacting epistatically with an otherwise constant genetic background. In reality, each individual occupies only a single point on their individual scaling relationship, reflecting the particular combination of environmental and genetic effects that determine trait and body size in that individual. Fitting a line to a collection of these points from genetically heterogenous individuals in a population, each experiencing a unique combination of environmental factors, generates a population scaling relationship (Fig. 1A).
A Individual scaling relationships (thin gray lines) result from variation in body size due to variation in a single environmental or genetic factor, with all other size-regulatory factors held constant. However, because each individual has a single genotype and is exposed to a single combination of environmental factors, it occupies only a single point on its individual scaling relationship (white circles). The observed population scaling relationship (red line) is the scaling relationship among individuals in a population. B The distribution of individual scaling relationships determines how the population scaling relationship responds to selection (Frankino et al. 2019), and can be classified as speedometer, broomstick, or seesaw, depending on where the median point of intersection (green circle) lies relative to the bivariate mean of trait sizes (yellow circle).
Underlying population scaling relationships are, therefore, collections of unseen, or cryptic, individual scaling relationships. The distribution of individual scaling relationships within a population will place individuals at particular locations around the population scaling relationship, and selection on these individuals may alter the distribution of individual scaling relationships in the population, which in turn can change the slope or intercept of the population-level scaling relationship (Dreyer et al. 2016; O’Brien et al. 2017; Houle et al. 2019). Mathematical modeling suggests that the response of the population scaling relationship to selection is dependent on the distribution of the underlying individual scaling relationships (Dreyer et al. 2016). Broadly speaking, this distribution can be classified as either broomstick, seesaw, or speedometer; these names are derived from objects that move in a manner that looks like a plot of individual scaling relationships under each distribution (Fig. 1B). The distribution is “speedometer” if the slopes intercept below the average body size; the distribution is “broomstick” if they intercept above average body size; and the distribution is “seesaw” if they intercept at average body size (Fig. 1B). Importantly, two populations with statistically indistinguishable population scaling relationships may have very different underlying distributions of individual scaling relationships, and will respond very differently to the same selective pressure (Dreyer et al. 2016). Consequently, if we are to understand the evolution of population scaling relationships, we need to understand the genetic variation of the individual scaling relationships that underlie them.
While the concept of individual scaling relationships is straightforward, quantifying them is not. Individual environmental scaling relationships can be generated by fitting a line to the trait-body size combinations expressed by genetically-identical individuals reared across an environmental gradient. Individual genetic scaling relationships can be generated by fitting a line to the trait-body size combinations expressed by individuals possessing allelic variation at only a single locus in an otherwise co-isogenic background and reared in a single environment. For many animals, such environmental and genetic control is impractical or impossible to impose. The measurement of individual scaling relationships is tractable in Drosophila, however, as the long-term maintenance of (near) isogenic populations is routine and genetic variation can be generated at a single gene or locus (Frankino et al. 2019; Houle et al. 2019).
In this paper we focus on understanding the genetics of population scaling relationships by characterizing the distribution of individual scaling relationships, using isogenic lineages of D. melanogaster as a model. The individual scaling relationships for each genotype were generated by varying access to food during development; because trait and body size results from variation in developmental nutrition, we refer to these individual scaling relationships as nutritional scaling relationships (Dreyer et al. 2016). We have previously used this simple diet manipulation to generate variation in wing and body size in D. melanogaster (Stillwell et al. 2011; Frankino et al. 2019). Here we apply this approach to 197 isogenic lineages of D. melanogaster, to determine the distribution of individual nutritional wing- and leg-body scaling relationships in this population. Further, we assay the nutritionally-induced size plasticity of these traits and the body within each lineage. We use these data to explore the genetic variation of nutritional scaling within and among traits and the variation in relative trait plasticity that accounts for this variation in scaling.
Material and methods
Fly stocks
All flies used in this study came from The Drosophila Genome Resource Panel (DGRP), a library of ~200 fully sequenced inbred isogenic Drosophila lineages that originated from a single outbred population (Mackay et al. 2012) collected from Raleigh, NC, USA. Flies were maintained on standard cornmeal molasses medium (Frankino et al. 2019) and maintained on a 12:12 light cycle at 22 °C and 75% humidity.
Starvation treatment
Drosophila egg collection, rearing, and phenotyping followed our established protocols (Stillwell et al. 2011, 2016; Frankino et al. 2019). For each DGRP lineage, females oviposited for 3 days. At 24, 48 and 72 h, eggs were collected, divided into lots of 50 and placed into vials containing 10 ml of standard cornmeal molasses medium. This generated three age cohorts of flies (D0, D1 and D2, respectively). When third instar larvae from D0 began to pupariate, larvae from all cohorts were removed from the food and placed into empty food vials with a wet cotton plug to provide moisture. Pupae were removed from these vials and transferred to individual 1.5 ml Eppendorf tubes, each with a small hole in the lid, to complete development to adulthood. Larvae in the D0 cohort were starved for between 0–24 h before pupariation, larvae in the D1 cohort were starved for between 24–48 h before pupariation, and larvae in the D3 cohort were starved for between 48–72 h before pupariation. Because larvae stop feeding ~24 h before pupariation (Testa et al. 2013), D0 larvae were essentially allowed to feed ad libitum and more-or-less achieved full adult body size. In contrast, D1 and D2 larvae were starved before larval wandering, reducing adult size depending on their size at initiation of starvation. Across all cohorts, our starvation treatment therefore generated nutritionally-induced variation in body size. Flies were collected in nine temporal blocks, with five lineages repeated across all blocks as a control.
Body and trait size measurement
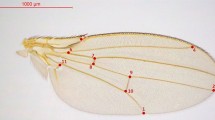
Body and trait size were measured using established protocols (Shingleton et al. 2009; Stillwell et al. 2011). Briefly, Drosophila adults were dissected, and their right wing and right first leg mounted in dimethyl hydantoin formaldehyde (DMHF). Pupal area (a proxy for body size; (Stillwell et al. 2016), wing area, and femur length (a proxy for leg length; (Shingleton et al. 2009) were measured across the full range of body size for ~50 individuals per sex per lineage; Fig. 2). All traits were measured via semi-automated custom software (Metamorph, Molecular Devices LLC) that captures images from a digital camera-equipped microscope (Leica DM6000B, Leica Microsystems Inc). Femur length was squared to put it in the same dimension as wing and pupal area, and all measurements were log transformed to ensure scale invariance across traits of different sizes.
A The wing-body population scaling relationship. B. The leg-body population scaling relationship. Points show the mean wing/leg/body size of all flies in each lineage (females: gray points; males: black points). Lines show the mean population scaling relationship, generated by sampling a single individual from each lineage, fitting the MA regression, and repeating 10,000 times by sex (females: broken lines; males: solid lines). For both wing-body and leg-body scaling relationships, there is a significant difference in intercept but not slope between females (gray) and males (black) (Table 1). The measurements taken are shown in red on the images of the wing, leg and pupa.
Statistical analysis
All data as well as the R scripts used to analyze them are provided on Dryad (doi:10.5061/dryad.98sf7m0nd). We collected data from >6 flies per sex per lineage, with a mean of 65 flies per sex per lineage, across 194 lineages. Block effects were removed by fitting the model T = K to the data, where T is body/trait size and K is block. We then used the residuals of the fit for each trait/body as a measure of trait/body size independent of block. Theoretical studies indicate that major axis (MA) model II regression best captures the developmental mechanisms that generate morphological scaling relationships (Shingleton 2019), so where possible we used this method to fit the individual scaling relationships. However, for completeness, and when testing more sophisticated models (e.g., when lineage was treated as a random factor) we fit the relationship using model I linear regression, using maximum likelihood (R package: lme4; Bates et al. 2014) and Bayesian methods (R package: MCMCglmm; Hadfield 2010). Other relationships were modeled using ordinary least square (OLS) model I linear regression. The relative importance of regressors in multiple linear regressions were calculated using the Lindeman, Merenda and Gold’s method (R package: relaimpo; Grömping 2006). Finally, as a measure of body/trait plasticity within each lineage, we first conducted a principal component analysis on the covariance matrix for wing, leg and body size within each lineage. We then used the first principal component from this analysis to select the largest and smallest 20% of individuals in each lineage, and used the difference in mean wing, leg and body size between them as a measure of trait/body-size plasticity.
Results
Population and individual morphological scaling relationships
Almost all published scaling relationships are population-level scaling relationships, where each point on a plot of body size against trait size is a genetically distinct individual. To estimate the population scaling relationship between wing or leg and body (pupal) size in our Drosophila population, we first randomly sampled measurements for one individual of each sex from each lineage (genotype), pooled these observations to create a population, and then calculated the slope and intercept of the major axis (MA) Model II regression of trait size against body size. We repeated this 10,000 times to generate 95% confidence intervals for the slope and the intercept for the female and male wing-body and leg-body population scaling relationships (Table 1 and Fig. 2). There were no differences between the sexes in the slope of either of these scaling relationships (Table 1). In contrast, the intercept for the wing-body size population-level scaling relationship was higher in females than in males, while the intercept for the leg-body population scaling relationship was higher in males than in females (Table 1). This was supported by an MA regression of mean trait size against mean body size among lineages (Fig. 2), which also detected no sex differences in the slope of either the wing-body or leg-body population scaling relationship (wing-body slope: p = 0.806; leg-body slope: p = 0.315, n = 194), but found a significant sex difference in intercept (wing-body intercept and leg-body intercept: p < 0.0001 for both, n = 194).
We next explored the individual scaling relationships across the 194 isogenic lineages. We estimated the slope and intercept of the wing-body and leg-body individual scaling relationships for each sex in each lineage using MA regression. Across lineages, the wing-body individual scaling relationships were steeper and had a higher intercept in females than in males (paired t-test, p < 0.0001 for slope and intercept, Table 1), indicating that females had larger wings than males relative to body size, and that this difference in relative wing size increased disproportionally with overall body size within lineages. In contrast, the leg-body scaling relationship had lower intercept in males than females (paired t-test, p < 0.0001, Table 1), and tended to be shallower, although the difference in slope was not significant (paired t-test, p = 0.0774, Table 1). These patterns were supported when fitting the individual scaling relationships using a linear-mixed model and treating lineage as a random factor, where slopes and intercepts were allowed to vary among lineages independently in males and females (Supplementary Tables 2 and 3).
Within sexes, there was significant variation among genotypes in slope for both the wing-body and leg-body individual scaling relationships (Fig. 3) when the relationships were fit using either an MA regression (treating lineage as a fixed factor; Supplementary Table 4) or linear mixed-model regression (treating lineage as a random factor affecting both slope and intercept; Supplementary Table 5). For females, the coefficient of variation (CV) (Houle 1992) for the wing-body and leg-body MA slopes was 17.1% and 24.2% respectively, while for males the CV for the wing-body and leg-body MA slopes was 15.2% and 18.5% respectively. An important caveat is that these estimates of genetic variation are among isogenic lineages and so may not reflect the additive genetic variation for slope in an outbred population (Houle et al. 2019). Among lineages, there was a significant correlation between male and female slopes for both the wing-body scaling relationship (ρ = 0.28, 95% CI: 0.15–0.41) and the leg-body scaling relationship (ρ = 0.40, 95% CI: 0.28–0.52). Fitting an MA regression to this correlation revealed that, for both wing-body and leg-body scaling relationships, as the slope of the individual scaling relationship increased among lineages, the female slope increased more than the male slope (Supplementary Fig. 1). Finally, within each sex, there was a significant correlation between the slope of the wing-body and leg body-scaling relationship (females: ρ = 0.52, 95% CI: 0.41–0.62; males: ρ = 0.39, 95% CI: 0.26–0.50).
The distribution of wing-body individual scaling relationships in females (A) and males (A′). Males have proportionally smaller wings and shallower slopes than females (Table 1). The distribution of leg-body individual scaling relationships in females (B) and males (B′). Males have proportionally larger legs and steeper slopes than females (Table 1). The steepness of the slope is indicated by color (blue = shallow, yellow = steep). The blue circle shows the bivariate mean of trait-body size. The red circle shows the median point of intersection (MPI) for the lines. The black line is the mean individual scaling relationship across all lineages. All the scaling relationships were fit using MA model II regression and extend two standard deviations above and below the mean body size for each lineage.
Distribution of cryptic individual scaling relationships
Theory suggests that the distribution of individual scaling relationships in a population determines the response to selection on the population scaling relationship slope (Dreyer et al. 2016). These distributions can be classified as either broomstick, seesaw, or speedometer, depending on where the morphological scaling relationships intersect on average relative to bivariate average of body/trait sizes (Fig. 1B). We therefore explored the distribution of individual scaling relationships among lineages, using MA model II regression to estimate the scaling relationships. (All subsequent analyses also use MA model II regressions to estimate the slopes and intercepts of the individual scaling relationships). Previously, we used the mean point-of-intersection among all pairs of individual scaling relationships and mean body/trait size to classify their distribution (Frankino et al. 2019). However, pairs of near-parallel individual scaling relationships can generate substantial outliers in the distribution of points-of-intersection, biasing the mean. To circumvent this problem, here we instead used the median point of intersection (MPI) relative to the bivariate medial trait/body size. For both the individual wing-body and the leg-body size scaling relationships, the MPI was close to the bivariate median trait/body size in males and in females (Fig. 3). Consequently, the distribution of individual scaling relationships appeared to be a seesaw in both sexes for both traits.
An artificial selection experiment to increase or decrease relative wing size (wing:thorax ratio) resulted in a corresponding increase or decrease in the slope of the wing-thorax scaling relationship, respectively (Robertson 1962). This finding suggests a positive correlation between relative wing size and the slope of the wing-body scaling relationship among genotypes. This would occur if the distribution of individual scaling relationships were speedometer (Fig. 1), which appears to contradict our observation that the distribution of wing-pupal individual scaling relationships is seesaw. Indeed, we found no correlation between a lineage’s relative wing size and the slope of its wing-body scaling relationship (fit using MA regression), in either males or females (OLS regression: R2 < 0.005, p > 0.08 for both).
However, unlike our study, Robertson (1962) did not use diet manipulation to increase the range of body size among flies, and so likely selected on only well-fed individuals. In our study, these are the largest flies that occupy the upper-right portion of their individual scaling relationships (black Lines, Fig. 4A). For a seesaw distribution (Fig. 1B), large size-class flies will also show a positive correlation between their wing-body slope and relative wing size among lineages. This hypothesis was supported by our data. We found there was a significant positive relationship among lineages between mean relative wing size for the largest 25% of individuals in a lineage and the slope of the lineage’s wing-pupal size scaling relationship, in both males and females (Fig. 4B’, C’). Conversely, for a seesaw distribution, the smallest flies should show a negative correlation between wing-body slope and relative wing size (gray lines, Fig. 4A), which was also supported by our data (Fig. 4B, C).
A For a seesaw distribution of individual cryptic scaling relationships (dashed lines), among the largest individuals (black lines), there should be a positive correlation between the mean relative wing size for a lineage and the slope of the lineage’s individual scaling relationship, whereas this relationship should be negative among the smallest individuals (gray lines). B, C For both females (B, B′) and males (C, C′), there was a significant positive relationship between relative wing size for the largest 25% of individuals in each lineage and slope of the lineage’s individual wing-body scaling relationship (B′, C′, black points: OLS model I regression: slope = relative wing size, F1,192 > 5.702, p < 0.0179 for both), and a significant negative relationship between relative wing size for the smallest 25% of individuals in each lineage, and slope (B, C, gray points: OLS model I regression: slope = relative wing size, F1,192 > 32.04, p < 0.001 for both). D For a speedometer distribution of individual scaling relationships (dashed lines), for the largest individuals (black lines), there should be a positive correlation between their mean relative wing size in each lineage and the slope of the lineage’s individual scaling relationship, whereas this relationship should be weaker or absent among the smallest individuals (gray lines). E, F For both females (E, E′) and males (F, F′), there was a significant positive relationship between relative leg size for the largest 25% of individuals in each lineage, and slope of the lineage’s individual leg-body scaling relationship (E′, F′, black points: OLS model I regression: slope = relative leg size, F1,192 > 5.811, p < 0.017 for both), but no relationship between relative wing size for the smallest 25% of individuals in each lineage, and slope (E, F: OLS model I regression: slope = relative leg size, F1,192 < 0.997, p > 0.319 for both).
We also examined the relationship among lineages between the leg-body slope for a lineage and relative leg size in the largest and smallest individuals from that lineage. As was the case for the wing, there was a positive correlation between mean relative leg size for the largest 25% of individuals in a lineage and the slope of the lineage’s leg-pupal scaling relationship, in both males and females (Fig. 4E’, F’). We could not detect, however, any correlation between relative leg size and slope using data from the smallest 25% of individuals in each lineage (Fig. 4E, F). This suggests that the distribution of individual scaling relationships between the leg and the body is more of a speedometer than seesaw (Fig. 1B).
Morphological scaling and size plasticity
Individual scaling relationships reflect variation in body size and covariation in trait size; that is, size variation caused by a particular environmental or genetic factor. When size variation is due to an environmental factor, the slope of an Individual scaling relationship (Δy/Δx) on a log-log scale captures the genotype-specific size plasticity of the trait (Δy) relative to that of the body (Δx) (Shingleton et al. 2007). When trait size is more plastic relative to body size (Δy > Δx), the slope of the scaling relationship is greater than one (i.e., hyperallometric); when the trait exhibits less size plasticity than the body (Δy < Δx), the slope will be less than one (i.e., hypoallometric). Variation among lineages in the slope of individual nutritional scaling relationships can therefore be due to variation in the plasticity of trait size, variation in the plasticity of body size, or some combination of both. For example, it is possible that wing size has more-or-less the same response to changes in developmental nutrition across all lineages, but that body size has a variable response. Under this scenario, variation in the slope of the wing-body individual scaling relationships among lineages would be due to variation in the plasticity of body size. Conversely, it may be variation in wing-size plasticity among lineages that generates variation in the wing-body individual scaling relationships.
To explore the relationship between trait- and body-size plasticities and the slope of individual scaling relationships, we used the difference in mean trait and body size between the largest and smallest 20% of genetically-identical individuals within a lineage as a measure of size plasticity (Fig. 5B). To identify these largest and smallest individuals, we used the first principal component from a PCA on pupal, wing and leg size as an index of overall individual size. We used the size differences between the largest and smallest individuals rather than size difference between starvation treatments as a measure of plasticity, because we are explicitly interested in whether variation in the slope of the scaling relationship among lineages is a consequence of genetic variation in the nutritionally-induced range of body (Δx) or trait (Δy) sizes expressed in each lineage (Fig. 5B).
A The slope of an individual nutritional scaling relationship reflects the nutritional plasticity of body size relative to trait size, and variation in either or both generates variation among slopes. B We used the differences in mean body/wing/leg size (red stars) between the largest (black points) and smallest (white points) 20% of flies in each lineage as a measure of plasticity. Variation in the slope of the wing-body (C) and leg-body (D) scaling relationship is primarily due to variation in body-size plasticity in females. Variation in the slope of the wing-body (E) and leg-body (F) scaling relationship is due to variation in both wing/leg-size and body-size plasticity in males. Percent variance was calculated by first fitting a multiple regression of slope against trait- and body-size plasticity among lineages, and then calculating the relative importance of each regressor using Lindeman, Merenda and Gold’s method. Error bars are 95% confidence intervals determined using 1000 bootstraps of the data.
We found significant correlations between the plasticity of trait pairs (wing vs. leg, leg vs. body, wing vs. body) both within and between sexes (Table 2). Similarly, we also found significant correlations in the plasticity of the same trait between sexes (Table 2). We may a priori expect trait plasticities to be correlated due to the systemic effects of nutrition on overall body size (Shingleton et al. 2007). We therefore regressed wing- and leg-size plasticity individually against body-size plasticity using OLS regression, and used the residual values as a measure of trait-size plasticity that was independent of body-size plasticity. This analysis revealed significant correlations in trait-size plasticity among appendages and between sexes, independent of body-size plasticity (Supplementary Table 6).
We next investigated the extent to which trait- or body-size plasticity independently explains among-genotype variation in the slope of the wing-body and leg-body individual scaling relationships. To do this, we fit the model Slope = Trait Plasticity + Pupal Plasticity using OLS model I regression and calculated the relative importance of each regressor, while controlling for the other, using Lindeman, Merenda and Gold’s method (Lindeman et al. 1980; Grömping 2006). For both sexes, variation in the plasticity of body size explained more of the variation in the slope of the individual scaling relationships than did variation in the plasticity of either the wing or the leg, although only significantly more in females (Fig. 5). Further, variation in body size plasticity explained more of the variation in the slope of individual scaling relationships in females than in males, for both wing-body and leg-body scaling. This suggests that variation in body size plasticity is greater in females than in males (Table 2). Pairwise comparisons of the wing, leg and body size plasticity variances between females and males supported this hypothesis: variance in body-size plasticity was significantly greater among females than among males (F1,193 = 1.40, p = 0.02), which was not true for variance in wing-size plasticity (F1,193 = 1.13, p = 0.10), or leg-size plasticity (F1,193 = 1.05, p = 0.31).
Discussion
Evolution of morphological scaling dominates the generation of morphological diversity among species, and yet we know little of how selection targets the developmental-genetic mechanisms that regulate trait and body size to create this diversity. Theoretical studies that distinguish between population and individual scaling relationships have hypothesized that the distribution of cryptic individual scaling relationships (seesaw vs. speedometer vs. broomstick) determines how the population-level scaling relationship will respond to selection (Dreyer et al. 2016). Our study explores variation among individual scaling relationships derived from isogenic D. melanogaster lineages, and how this variation relates to nutritionally induced size plasticity of two traits and the body. We find that, across the full range of body sizes generated by variation in access to nutrition during growth, the distribution of individual scaling relationships is approximately a seesaw for both wing-body and leg-body scaling. Further and somewhat surprisingly, we find that variation in the slope of the individual scaling relationships, which reflects the relative nutritional plasticities of trait and body size, is primarily a consequence of variation in the relative plasticity of body size. These data provide important insight into the genetic basis of variation in morphological scaling, and how this variation may respond to selection to generate morphological diversity through evolution of the population-level scaling relationship.
Evolvability and genetic variation in scaling
While the slopes of morphological scaling relationships can vary dramatically among species, particularly for exaggerated traits used to attract or compete for mates (Baker and Wilkinson 2001), this seems to be the exception rather than the rule: For most species and most traits, the slopes of morphological scaling relationships tend to be evolutionarily invariant (Gould 1966; Pelabon et al. 2014). These observations have led to the hypothesis that morphological scaling relationships are evolutionarily constrained by developmental or physiological mechanisms (Pelabon et al. 2014). Developmental studies in Drosophila, however, reveal that simple changes in the expression of a single gene are sufficient to substantially alter the slope of trait-body scaling for traits such as the wing or genitalia that otherwise maintains a near constant allometric coefficient across species (Tang et al. 2011; Shingleton and Tang 2012). Subsequent studies have changed gene expression to alter the slope of genital-body scaling in dung beetles (Casasa and Moczek 2018), mandible-body scaling in male flour beetles (Okada et al. 2019), and horn-body scaling in rhinoceros beetles (Ohde et al. 2018). Consequently, it does not appear that the evolutionary invariance of morphological scaling relationship slope is rooted in developmental or physiological constraint, at least mechanistically.
An alternative explanation for the evolutionary conservation of scaling is that there is little genetic variation in the developmental mechanisms that regulate morphological scaling relationships, and upon which selection can act. This would be evident as a lack of genetic variation in the slopes of individual morphological scaling relationships within a population. Hitherto, there have been almost no data on the genetic variation of either the slope or intercept of morphological scaling (Frankino et al. 2019). However, our study reveals considerable variation in the slope of both the wing-body and leg-body individual scaling relationships, with coefficients of variation (CV) comparable to that for overall body size (Lafuente et al. 2018). A similar study on the scaling relationship between wing-vein length and wing size also revealed considerable genetic variation in the slope of individual scaling relationships (Houle et al. 2019). Thus, any observed inertia in the evolution of trait-body scaling relationship slope does not appear to result from lack of genetic variation, at least in Drosophila. An important caveat, however, is that the variation in slope detected in both this and previous studies (Frankino et al. 2019; Houle et al. 2019) is among isogenic lineages and may not reflect the level of additive genetic variation for the slope of individual scaling relationships in natural populations.
If the slopes of morphological scaling relationships are not developmentally or physiologically constrained, and if they possess levels of genetic variation comparable to that for body size, which responds rapidly to artificial selection (Hillesheim and Stearns 1991; Partridge and Fowler 1993; Turner et al. 2011), we should expect morphological scaling to also respond rapidly to artificial selection. However, artificial selection on the slope of the wing-body scaling relationship in Drosophila revealed an erratic and weak response, with an apparent heritability of less than 0.015 (Stillwell et al. 2016). The imposed selection regime attempted to alter the slope of the population wing-body scaling relationship without changing either mean wing or mean body size (i.e., to rotate the scaling relationship approximately about the bivariate mean). To increase the slope, these investigators selected and crossed large-bodied individuals with disproportionally large wings and crossing them with small-bodied individuals with disproportionally small wings. To decrease the slope, they selected large-bodied individuals with disproportionally small wings and crossing them with small-bodied individuals with disproportionally large wings. The authors ascribed the low response to pleiotropy between the slope and mean trait and body size (Stillwell et al. 2016). However, an alternative explanation is that the selection regime failed to consider the relationship between the observed population-level scaling relationship and the underlying distribution of individual cryptic scaling relationships. That is, the individuals selected because of their disproportionally sized wings may have possessed individual wing-body scaling relationships that would not facilitate or might even impede the desired response to selection. This would occur if the slope distribution of the individual scaling relationship were of the speedometer or broomstick distribution (Dreyer et al. 2016). The same reasoning may explain why another selection experiment, which attempted to change the slope of the scaling relationship between wing-vein length and wing size, had a similarly weak and erratic response when the full range of body size was produced via diet manipulation (Bolstad et al. 2015). This latter study also ascribed the relative lack of response to pleiotropic constraints (Houle et al. 2019), rather than a failure of the selection regime to efficiently target alleles that regulate the slope of the scaling relationship.
Our data detailing the distribution of individual scaling relationships in a population—albeit among homozygous genotypes—will facilitate the design of artificial selection regimes that most efficiently target the slope of individual scaling relationships. The efficacy of such selection regimes will provide a nuanced method to test of the pace and extent to which the slopes of morphological scaling relationships can evolve. Earlier artificial selection experiments to shift the intercept of morphological scaling relationships in Drosophila (Robertson 1962) and stalk-eyed flies (Wilkinson 1993)—by selecting to change relative wing size and eye-span—rapidly and indirectly altered the slope of the relationship. A third study, that applied directional selection on body size in the tobacco hornworm Manduca sexta also indirectly altered the slope of the wing-body scaling relationship (Tobler and Nijhout 2010). Our data (Fig. 3) may reveal why such selection will be effective, at least with respect to the wing-body scaling relationship in Drosophila: Selection to increase relative wing size in large well-fed flies will indirectly select to increase the slope of the wing-body scaling relationship (Fig. 4B’, C’).
Why then, given the apparent extent of genetic variation underlying the slope of population-level morphological scaling relationships, do their slopes appear to be evolutionarily constrained? Our data, along with those of Houle et al. (2019), support the hypothesis that the evolutionary conservatism of morphological scaling relationship slopes is a consequence of natural selection favoring proportions that enable ecological performance. This may be particularly true for appendages that are involved in mobility, such as wings and legs, where changes in loading—that is total body mass divided by appendage dimensions (Gilchrist and Huey 2004; David et al. 2011)—may have substantial energetic or functional consequences. An alternative, and non-exclusive, hypothesis is that changes in the slope of scaling relationships reduce fitness due to pleiotropic effects, for example by altering the scaling relationship between other traits and the body (Houle et al. 2019). While this may be the case for the scaling relationships among traits in a highly integrated organ, for example the veins of the wing (Houle et al. 2019), this does not appear to be true for the relationship among appendages: developmental manipulation to alter the scaling relationship between wing and body size, for example, does not affect the scaling relationships between body size and other traits (Tang et al. 2011). Nevertheless, our data indicate a genetic correlation in size plasticity among traits independent of body size plasticity. Because linkage disequilibrium breaks down over short distances in the population of flies used in our study (Mackay et al. 2012), this correlation likely arises from pleiotropy, which would need to be broken for natural selection to change the slope of one trait’s morphological scaling relationship with body size independently of another. Exploring the fitness of flies that have been allometrically engineered to have atypical scaling relationships, generated using either transgenics or artificial selection, will help resolve these questions (Wilkinson and Reillo 1994; Frankino et al. 2005, 2007; Houle et al. 2019).
Size plasticity and genetic variation in scaling
The slope of nutritionally-generated individual scaling relationships reflects the relative nutritional plasticity of trait and body size (Shingleton et al. 2007). Variation among the slopes of these scaling relationships can result from genetic variation in relative body size plasticity, trait size plasticity, or both. From a developmental perspective, both trait and body size plasticity are a response to developmental nutrition, mediated through systemic growth-regulatory mechanisms, canonically the IIS and TOR signaling pathways (Vea and Shingleton 2020). Autonomous changes in a trait’s growth-sensitivity to variation in either IIS or TOR signaling is sufficient to alter the slope of the trait-body scaling relationship (Tang et al. 2011; Shingleton and Tang 2012; Luo et al. 2013; Casasa and Moczek 2018; Okada et al. 2019). If there were genetic variation in the growth-sensitivity of individual traits to changes in IIS or TOR signaling, this would generate genetic variation in the slope of the trait-body size morphological scaling relationship. Further, developmental studies suggest that the distribution of slopes (seesaw, speedometer, broomstick) would depend on the locus of genetic variation. For example, wing-autonomous changes in the expression of the Forkhead Transcription Factor (FOXO), which suppresses growth when nutrition is low but is not active when nutrition is high, generate a broomstick distribution of wing-body scaling relationships (Shingleton and Tang 2012). In contrast, wing-autonomous changes in the expression of the Insulin Receptor (InR), which promotes growth when nutrition is high but is not active when nutrition is low, generate a speedometer distribution (Shingleton and Tang 2012).
We found that in males, variation in the slope of individual scaling relationships among lineages was a consequence of variation in the plasticity of both wing/leg-size and body-size. In females, however, variation in the slope of individual scaling relationships was primarily a consequence of variation in body-size plasticity. This suggests that it is variation in the sensitivity of the body to changes in nutrition, independent of the sensitivity of individual traits, that generates variation in the slope of individual scaling relationships in females. How this is achieved seems paradoxical, since the size of the body ostensibly reflects the collective size of its constituent parts. In Drosophila, as with all fully metamorphic insects, the external appendages, such as wings, legs, genitalia, and mouthparts, develop as imaginal discs within the larval body. Pupal size, which we used a proxy for overall body size, is determined by the size of the larva when it stops feeding ~24 h before pupariation. The imaginal discs, however, continue to grow until ~24 h after pupariation (Bryant and Schmidt 1990). Consequently, the developmental mechanisms that regulate body size are potentially distinct from those that regulate the size of individual traits. Further, developmental studies demonstrate that it is possible to change the nutritional plasticity of wing size without affecting the nutritional plasticity of body size (Tang et al. 2011). Thus, genetic variation in the plasticity of body size and the plasticity of trait size can be independent of one another. Indeed, the existence of genetic variation in the slope of individual scaling relationships (Fig. 2) indicates that the developmental mechanisms that regulate body-size plasticity are at least partially independent of those that regulate trait-size plasticity. Interestingly, if variation in the slope of trait-body scaling relationships is generated by genetic variation in the plasticity of body size independent of the plasticity of trait size, then changes in body-size plasticity will affect the slope of all trait-body scaling relationships. This may, in part, account for the observed correlation in slopes between the wing-body and leg-body scaling relationships in both sexes, and explain why this correlation is stronger in females than in males.
One consideration in interpreting genetic variation in body- and trait-size plasticity is that we may have exposed different lineages to different ranges of developmental nutrition; that is, nutritional deprivation may not have been applied equally across lineages. This would result in differences in the range of body and trait sizes expressed by different lineages unrelated to genotype. This seems unlikely in our study, however. The smallest adult size for a fly is generated when a larva is starved at its minimal weight for eclosion (MVWeclosion) (Stieper et al. 2008). Larvae starved below this weight have insufficient stored nutrients to complete developmental and do not eclose as adults. Our nutritional deprivation protocol withdraws food from larvae below the MVWecolosion, all the way up to larvae that have completed development and are about to pupariate, and thus encompasses the full range of nutritional conditions a larvae could experience and still successfully eclose (Stillwell et al. 2011). Nevertheless, even if nutritional deprivation were applied unequally across lineages, this cannot account for variation in the slope of the scaling relationships, or the relative contributions of body size and trait size plasticity to this variation. This is because the wings, legs, and body all form part of the same individual, and so, across all individuals in a lineage, the range of nutritional deprivation experienced by a growing wing and leg is the same as experienced by a growing body.
While we have extensive knowledge of the developmental mechanisms that regulate nutritionally-induced size plasticity of both the body and of individual traits (Nijhout et al. 2014), it remains a hypothesis that genetic variation in this plasticity, and by extension in the slope of individual nutritional scaling relationships, lies within these mechanisms. This hypothesis appears to be supported by GWAS studies on thermotolerance and thermally induced body-size plasticity, which have identified and functionally validated genes that are involved in the response to environmental change in general and thermal change in particular (Gerken et al. 2015; Lafuente et al. 2018; Lecheta et al. 2020). Consequently, we may expect that genetic variation for nutritionally-induced size variation lies within the developmental pathways involved in the response to nutritional change. The next step is therefore to identify the genes that underlie the observed slope variation in nutritional scaling relationships, and to functionally test their role in regulating the response of body and trait size to variation in developmental nutrition.
Conclusion
Our data reveal the distribution of previously cryptic individual scaling relationships for wing and leg size against body size in Drosophila, and explore their relationship with variation in nutritionally induced plasticity of trait and body size. These data not only provide insight into the characteristics of genetic variation of the wing-body and leg-body population scaling relationships, but they also allow us to predict how the population scaling relationship will respond to selection for changes in slope and intercept. Further, future analysis promises to identify the developmental mechanisms that are responsible for the observed genetic variation in individual scaling relationships—the mechanisms that may be targeted by selection to alter population scaling.
Data availability
All data as well as the R scripts used to analyze them are provided on Dryad (https://doi.org/10.5061/dryad.98sf7m0nd).
References
Baker RH, Wilkinson GS (2001) Phylogenetic analysis of sexual dimorphism and eye-span allometry in stalk-eyed flies (Diopsidae). Evolution 55:1373–1385
Bates D, Mächler M, Bolker B, Walker S (2015). Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw 67
Bolstad GH, Cassara JA, Márquez E, Hansen TF, Linde K, van der, Houle D et al. (2015) Complex constraints on allometry revealed by artificial selection on the wing of Drosophila melanogaster. Proc Natl Acad Sci USA 112:13284–13289
Bryant PJ, Schmidt O (1990) The genetic control of cell proliferation in Drosophila imaginal discs. J Cell Sci 1990:169–189
Casasa S, Moczek AP (2018) Insulin signalling’s role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc Biol Sci 285:20181631
Casasa S, Schwab DB, Moczek AP (2017) Developmental regulation and evolution of scaling: novel insights through the study of Onthophagus beetles. Curr Opin Insect Sci 19:52–60
David JR, Yassin A, Moreteau J-C, Legout H, Moreteau B (2011) Thermal phenotypic plasticity of body size in Drosophila melanogaster: sexual dimorphism and genetic correlations. J Genet 90:295–302
Dreyer AP, Ziabari OS, Swanson EM, Chawla A, Frankino WA, Shingleton AW (2016) Cryptic individual scaling relationships and the evolution of morphological scaling. Evolution 70:1703–1716
Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC (2012) A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337:860–864
Frankino WA, Bakota E, Dworkin I, Wilkinson GS, Wolf JB, Shingleton AW (2019) Individual cryptic scaling relationships and the evolution of animal form. Integr Comp Biol 59:1411–1428
Frankino WA, Zwaan BJ, Stern DL, Brakefield PM (2005) Natural selection and developmental constraints in the evolution of allometries. Science 307:718–720
Frankino WA, Zwaan BJ, Stern DL, Brakefield PM (2007) Internal and external constraints in the evolution of morphological allometries in a butterfly. Evolution 61:2958–2970
Gayon J (2000) History of the concept of allometry. Integr Comp Biol 40:748–758
Gerken AR, Eller OC, Hahn DA, Morgan TJ (2015) Constraints, independence, and evolution of thermal plasticity: probing genetic architecture of long- and short-term thermal acclimation. Proc Natl Acad Sci USA 112:4399–4404
Gilchrist GW, Huey RB (2004) Plastic and genetic variation in wing loading as a function of temperature within and among parallel clines in Drosophila subobscura. Integr Comp Biol 44:461–70
Gould SJ (1966) Allometry and size in ontogeny and phylogeny. Biol Rev 41:587–638
Gould SJ (1973) Positive allometry of antlers in the “Irish Elk”, Megaloceros giganteus. Nature 244:375–376
Grömping U (2006) Relative Importance for Linear Regression in R: the Package relaimpo. J Stat Softw 17:1–27
Hadfield JD (2010). MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J Stat Softw 33
Hillesheim E, Stearns S (1991) The responses of Drosophila melanogaster to artificial selection on body weight and its phenotypic plasticity in two larval food environments. Evolution 45:1909–1923
Houle D (1992) Comparing evolvability and variability of quantitative traits. Genetics 130:195–204
Houle D, Jones LT, Fortune R, Sztepanacz JL (2019) Why does allometry evolve so slowly? Integr Comp Biol 59:1429–1440
Huxley JS (1924) Constant differential growth-ratios and their significance. Nature 114:895–896
Huxley JS (1932) Problems of relative growth. Methuen & Co Ltd, London p 316
Huxley JS, Tessier G (1936) Terminology of relative. Growth 137:780–781
Klingenberg C, Zimmermann M (1992) Static, ontogenic, and evolutionary allometry—a multivariate comparison in 9 species of water-striders. Am Nat 140:601–620
Lafuente E, Duneau D, Beldade P (2018) Genetic basis of thermal plasticity variation in Drosophila melanogaster body size (GP Copenhaver, Ed.). PLoS Genet 14:e1007686
Lecheta MC, Awde DN, O’Leary TS, Unfried LN, Jacobs NA, Whitlock MH et al. (2020) Integrating GWAS and transcriptomics to identify the molecular underpinnings of thermal stress responses in Drosophila melanogaster. Front Genet 11:658
Luo J, Liu Y, Nässel DR (2013) Insulin/IGF-regulated size scaling of neuroendocrine cells expressing the bHLH transcription factor dimmed in Drosophila. PLoS Genet 9:e1004052
Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D et al. (2012) The Drosophila melanogaster genetic reference panel. Nature 482:173–178
Nijhout HF, Riddiford LM, Mirth C, Shingleton AW, Suzuki Y, Callier V (2014) The developmental control of size in insects. Wiley Interdiscip Rev Dev Biol 3:113–134
O’Brien DM, Katsuki M, Emlen DJ (2017) Selection on an extreme weapon in the frog-legged leaf beetle (Sagra femorata). Evolution 71:2584–2598
Ohde T, Morita S, Shigenobu S, Morita J, Mizutani T, Gotoh H et al. (2018) Rhinoceros beetle horn development reveals deep parallels with dung beetles. PLoS Genet 14:e1007651
Okada Y, Katsuki M, Okamoto N, Fujioka H, Okada K (2019) A specific type of insulin-like peptide regulates the conditional growth of a beetle weapon. PLoS Biol 17:e3000541
Partridge L, Fowler K (1993) Responses and correlated responses to artificial selection on thorax length in Drosophila melanogaster. Evolution 47:213–226
Pelabon C, Firmat C, Bolstad GH, Voje KL, Houle D, Cassara J et al. (2014) Evolution of morphological allometry. (CW Fox and TA Mousseau, Eds.). Ann NY Acad Sci 1320:58–75
Lindeman RH, Merenda PF, Gold RZ (1980) Introduction to bivariate and multivariate analysis. Scott, Foresman, Glenview IL
Robertson FW (1962) Changing the relative size of the body parts of Drosophila by selection. Genet Res 3:169–180
Shingleton AW (2010) Allometry: the study of biological scaling. Nat Ed Knowl 3:2
Shingleton AW (2019) Symposium article: which line to follow? The utility of different line-fitting methods to capture the mechanism of morphological scaling. Integr Comp Biol 61:838
Shingleton AW, Estep CM, Driscoll MV, Dworkin I (2009) Many ways to be small: different environmental regulators of size generate distinct scaling relationships in Drosophila melanogaster. Proc Biol Sci 276:2625–2633
Shingleton AW, Frankino WA, Flatt T, Nijhout HF, Emlen DJ (2007) Size and shape: the developmental regulation of static allometry in insects. Bioessays 29:536–548
Shingleton AW, Tang HY (2012) Plastic flies: the regulation and evolution of trait variability in Drosophila. Fly 6:1–6
Stieper BC, Kupershtok M, Driscoll MV, Shingleton AW (2008) Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev Biol 321:18–26
Stillwell RC, Dworkin I, Shingleton AW, Frankino WA (2011) Experimental manipulation of body size to estimate morphological scaling relationships in Drosophila. J Vis Exp 56:e3162
Stillwell RC, Shingleton AW, Dworkin I, Frankino WA (2016) Tipping the scales: evolution of the allometric slope independent of average trait size. Evolution 70:433–444
Tang HY, Smith-Caldas MSB, Driscoll MV, Salhadar S, Shingleton AW (2011) FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet 7:e1002373
Testa ND, Ghosh SM, Shingleton AW (2013) Sex-specific weight loss mediates sexual size dimorphism in Drosophila melanogaster. PLoS ONE 8:e58936
Thompson DW, Bonner JT (1992) In: Bonner JT (ed) On growth and form. Cambridge University Press, p 345
Tobler A, Nijhout HF (2010) Developmental constraints on the evolution of wing-body allometry in Manduca sexta. Evol Dev 12:592–600
Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM (2011) Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet 7:e1001336
Vea IM, Shingleton AW (2020) Network‐regulated organ allometry: the developmental regulation of morphological scaling. Wiley Interdiscip Rev Dev Biol 10:e391
Wilkinson GS (1993) Artificial sexual selection alters allometry in the stalk-eyed fly Cyrtodiopsis dalmanni (Diptera: Diopsidae). Genet Res 62:213–222
Wilkinson G, Reillo P (1994) Female choice response to artificial selection on an exaggerated male trait in a stalk-eyed fly. Proc Biol Sci 255:1–6
Acknowledgements
This work was made possible through the assistance of undergraduate members of the Shingleton and Frankino labs, who reared and measured the flies used in the study. Additional financial support was provided by the University of Illinois at Chicago. WAF was supported by NSF IOS-1558098. AWS was supported by NSF IOS-1952385.
Author information
Authors and Affiliations
Contributions
AWS and WAF designed and oversaw execution of the study; ASW and IMV oversaw the collection of the data; ASW, IMV, WAF and AWS contributed to the data analysis and in preparing the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Rowan Barrett.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wilcox, A.S., Vea, I.M., Frankino, W.A. et al. Genetic variation of morphological scaling in Drosophila melanogaster. Heredity 130, 302–311 (2023). https://doi.org/10.1038/s41437-023-00603-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00603-y