Abstract
Molecular ecologists often rely on phylogenetic evidence for assessing the species-level systematics of newly discovered lineages. Alternatively, the extent of introgression at phylogeographic transitions can provide a more direct test to assign candidate taxa into subspecies or species categories. Here, we compared phylogenetic versus hybrid zone approaches of species delimitation in two groups of frogs from the Western Mediterranean region (Discoglossus and Pelodytes), by using genomic data (ddRAD). In both genera, coalescent analyses recovered almost all nominal taxa as “species”. However, the least-diverged pairs D. g. galganoi/jeanneae and P. punctatus/hespericus admix over hundreds of kilometers, suggesting that they have not yet developed strong reproductive isolation and should be treated as conspecifics. In contrast, the comparatively older D. scovazzi/pictus and P. atlanticus/ibericus form narrow contact zones, consistent with species distinctiveness. Due to their complementarity, we recommend taxonomists to combine phylogenomics with hybrid zone analyses to scale the gray zone of speciation, i.e., the evolutionary window separating widely admixing lineages versus nascent reproductively isolated species. The radically different transitions documented here conform to the view that genetic incompatibilities accumulating with divergence generate a weak barrier to gene flow for long periods of time, until their effects multiply and the speciation process then advances rapidly. Given the variability of the gray zone among taxonomic groups, at least from our current abilities to measure it, we recommend to customize divergence thresholds within radiations to categorize lineages for which no direct test of speciation is possible.
Similar content being viewed by others
Introduction
Whether evolutionary divergences initiated by biogeographic events regress or persist upon secondary contacts is a central issue in phylogeography and speciation biology (Avise 2000; Rosenblum et al. 2012; Garrick et al. 2019). It is well recognized that the propensity of two lineages to hybridize successfully, and thus, their fate as separate entities, depends on the amount of time spent in allopatry (Avise et al. 1998; Singhal and Moritz 2013). But how much time in isolation is needed to complete speciation, and how long can gene flow persist as nascent species diverge? The answers to these fundamental questions may greatly vary within and between taxonomic groups, raising controversy on the genetic mechanisms contributing to species formation (Gavrilets 2004; Bolnick and Fitzpatrick 2007), and causing endless debates about the status of closely related taxa (Coyne and Orr 2004; De Queiroz 2007).
Instances of cryptic divergences are particularly puzzling in this context. In the absence of obvious ecological and phenotypic differentiation, taxonomists primarily rely on phylogenetic evidence to classify novel biodiversity (Bickford et al. 2007; Adams et al. 2014). Nowadays, the common practice is to reconstruct species trees and compare candidate taxonomic scenarios under the multispecies coalescent framework (Edwards 2009; Liu et al. 2009). Thresholds of genetic distances also provide complementary support (Fouquet et al. 2007; Vieites et al. 2009), and are still widely used in the context of DNA barcoding. Yet, these “phylogenetic” approaches have been suspected to identify population structure rather than species (e.g., Sukumaran and Knowles 2017) and are contingent to the type of loci used (often mitochondrial, e.g., Zink and Barraclough 2008) and the assumption that speciation progresses at a fixed rate among taxonomic groups. Moreover, monophyly does not necessarily imply reproductive isolation, the central aspect of the biological species concept (Mayr 1942), which is increasingly overlooked in favor of the general lineage species concept (De Queiroz 2007; Fišer et al. 2018). As a consequence, the model-based species delimitation methods currently available are being questioned, as they do not provide any explicit test of speciation, and can consequently oversplit genetically compatible lineages (Sukumuran and Knowles 2017; Campillo et al. 2019).
A more direct way to delimit cryptic species is to infer reproductive isolation from the extent of genetic introgression at their phylogeographic transitions, i.e., via the study of hybrid zones. Post-zygotic isolation is expected to arise primarily from the progressive accumulation of genetic changes responsible for incompatibilities in hybrids, such as Dobzhansky–Muller incompatibilities (DMIs) (Orr 1995). While weak selection against hybrids should restrict introgression at only a few barrier loci, large parts of the genome can quickly become impermeable to gene flow due to linkage disequilibrium, as more barrier loci get recruited (the coupling hypothesis, Barton 1983). Negligible at first, reproductive isolation is thus expected to build up exponentially, as accumulating DMIs multiply their effects and cause a “snowball effect” on hybrid fitness (Orr 1995; Orr and Turelli 2001; Mendelson et al. 2004; Gourbiere and Mallet 2009; Matute et al. 2010; Moyle and Nakazato 2010). As a result, lineages widely admix up to a certain degree of genetic divergence, after which gene flow is considerably reduced, and ultimately suppressed (e.g., Singhal and Moritz 2013; Dufresnes et al. 2019a, 2019b). Nevertheless, and despite its potential, the hybrid zone approach remains largely underexploited in systematics due to the high effort required (multilocus population genetics), its inapplicability for allopatric taxa, and because the genetic structure at species transitions also depends on extrinsic factors (e.g., dispersal capabilities, time since first contact), which are usually not specifically documented.
We argue that the systematics of species complexes should ideally benefit from a combination of both approaches, namely phylogenetics and direct tests of reproductive isolation, in order to dissect the “gray zone of speciation”, i.e., the evolutionary windows where diverging lineages can still exchange genes (Roux et al. 2016). Insufficiently diverged taxa that remained conspecific should experience little genetic incompatibilities and admix freely across wide geographic areas. In contrast, valid species should exhibit greater divergence and narrower transition zones, due to their (partly) incompatible genomes, preventing their fusion and “despeciation” (Rhymer and Simberloff 1996; Wollenberg Valero et al. 2019). Phylogenetic scaling of the gray zone within radiations can, in turn, help to customize thresholds of divergence for closely related species, and predict the status of candidate allopatric lineages for which no empirical test of speciation is possible (on islands or other geographically isolated regions). Reciprocally, the direct approach of species delimitation (hybrid zone analysis) offers an ad hoc test for validating the indirect approach (phylogenetics), especially under the multispecies coalescent (e.g., Campillo et al. 2019).
Amphibians from the Western Palearctic realm offer appealing systems to implement both approaches together. First, many traditional species consist of several genetically divergent lineages that meet across multiple phylogeographic transition zones (e.g., Beddek et al. 2018). Second, episodes of vicariance may be linked to well-dated geo-climatic events, enabling to calibrate phylogenies with some comparability between taxonomic groups (Ehl et al. 2019). Third, the improvement of high-throughput sequencing techniques now allows affordable access to thousands of loci, even from the large genomes of amphibians (Brelsford et al. 2016), enabling robust phylogenetic inferences and accurate measures of introgression across hybrid zones.
Here, we compared the indirect (phylogenetic) and direct (hybrid zone) approaches of species delimitation to investigate the speciation history and systematics in two anuran groups that diversified in the Western Mediterranean basin: the painted frogs (Alytidae, Discoglossus) and the parsley frogs (Pelodytidae, Pelodytes). The genus Discoglossus includes seven taxa naturally distributed in Iberia (D. galganoi galganoi and D. galganoi jeanneae), North Africa (D. scovazzi, D. pictus auritus), Sicily (D. pictus pictus) and the Tyrrhenian archipelago (D. sardus and D. montalentii) (Fromhage et al. 2004; Zangari et al. 2006; Pabijan et al. 2012; Vences et al. 2014). Two pairs are of particular interest. First, the Iberian D. g. galganoi (west) forms multiple contact zones with its sister lineage D. g. jeanneae (east), especially in southern Spain (along the Guadalquivir River) and central Spain (García-París and Jockusch 1999; Martínez-Solano 2004). No diagnostic nuclear markers are known from this transition, and mitochondrial barcoding is possibly confounded by the presence of pseudogenes (Vences et al. 2014; see “Discussion” section). Second, the north-African D. pictus and D. scovazzi, which diverged earlier in the phylogeny (Martínez-Solano 2004; Pabijan et al. 2012), meet along the surroundings of the Moulouya valley in northeastern Morocco (Vences et al. 2014; Beddek et al. 2018), where contemporary gene flow was not detected (based on a single nuclear marker, Vences et al. 2014). These pairs of taxa thus represent distinct stages of divergence that potentially lie on the two edges of the gray zone of speciation.
Our second focal group, the genus Pelodytes, is represented by a Caucasian endemic (P. caucasicus) and four cryptic candidate species distributed across the Iberian Peninsula and France (Díaz‐Rodríguez et al. 2015, 2017). The diversification of these western taxa is supposedly of Plio-Pleistocene age, but the phylogeny remains incompletely resolved (Díaz‐Rodríguez et al. 2015, 2017). Several contact zones were identified in Portugal (P. atlanticus/P. ibericus), southern Spain (P. ibericus/P. hespericus), and north-eastern Spain (P. hespericus/P. punctatus), with local signs of introgression that suggest incomplete reproductive isolation, although comprehensive analyses are lacking (Díaz‐Rodríguez et al. 2017).
Using ddRAD sequencing, we performed phylogenomic reconstructions and applied the multispecies coalescent for phylogenetic species delimitations in each group. We then assessed the validity of these results by measuring the extent of admixture across four secondary contact zones involving candidate species of different ages. Assuming similar dispersal among the members of each genus, and no geographic barriers to gene flow at their transitions, we predict wide hybrid zones for the younger pairs if they did not yet evolve significant reproductive isolation, but narrow transitions for the older pairs, if they represent distinct species. We compared the outcomes of the two approaches, established the timeframe of speciation of Discoglossus and Pelodytes, and evaluated which divergence thresholds can be relevant to delimit incipient species when no direct evidence from hybrid zones is available.
Methods
Sampling
An extensive set of newly collected samples was complemented with data and samples available from previous studies (Martínez‐Solano 2004; Real et al. 2005; Gutiérrez-Rodríguez et al. 2014; Vences et al. 2014), chosen to cover four geographic transitions: (1) the Iberian Peninsula (D. g. galganoi/jeanneae); (2) Morocco (D. pictus/scovazzi); (3) Catalonia (P. hespericus/punctatus); and (4) southern Portugal (P. atlanticus/ibericus). For the phylogenies, we selected additional samples of D. scovazzi (Morocco), D. pictus auritus (Algeria), D. pictus pictus (Sicily, Italy), D. sardus (Corsica, France), D. montalentii (Corsica, France), and P. punctatus (France) from the EPHE-CNRS herpetological collection in the CEFE lab, Montpellier. Samples consisted of noninvasive buccal swabs, toe clips, tadpole tail tips, or muscles from voucher specimens, stored at −20 °C and/or in 70–96% ethanol. DNA was extracted by using the Qiagen BioSprint Robotic workstation. Tables S1 and S2 detail the origin of the samples analyzed in this study.
ddRAD sequencing
Four genomic libraries were prepared following the double-digest RAD (ddRAD) protocol of Brelsford et al. (2016). Library 1 included samples from Vences et al. (2014), namely 67 from D. scovazzi/pictus (17 localities) and 51 from D. g. galganoi/jeanneae (36 localities). Library 2 included 110 D. g. galganoi/jeanneae samples (17 additional localities), and 10 from other Discoglossus taxa for the phylogeny (see above). Library 3 included 124 P. hespericus/punctatus samples (28 localities). Library 4 included 80 P. atlanticus/ibericus samples (17 localities), and 3 additional P. punctatus for the phylogeny. Library preparation consisted of restriction enzyme digestion with SbfI and MseI, ligation of individual barcodes, PCR amplification and size selection of the ligated fragment (between 400 and 500 bp), and sequencing (single end) of the purified final library on an Illumina Hi-Seq 2500 (two lanes/library). For library 1, however, MspI was used instead of Mse1; it consequently did not share loci with other libraries and was analyzed separately.
Raw reads were quality-checked with FastQC v.0.10.1 and processed with STACKS 1.48 (Catchen et al. 2013), which includes demultiplexing (process_radtags), loci construction and cataloging (ustacks, cstacks, and sstacks), and SNP calling (populations). We applied the default stacking parameters, and for each system, we optimized filtering options to obtain a good balance between the number of loci and of missing data across all populations (–p and –r), while limiting paralog overmerging (–min_maf 0.05 and –max_obs_het 0.75). For the Discoglossus transitions, we kept loci present in all populations (–p as the number of populations) and in 80% of each (–r 0.8), to export (i) a matrix of 1121 SNPs for D. g. galganoi/jeanneae from library 1 (100% of completeness); (ii) a matrix of 4748 SNPs for D. g. galganoi/jeanneae from library 2 (97.6% of completeness); (iii) a matrix of 794 SNPs for D. scovazzi/pictus (99.0% of completeness). For the Pelodytes transitions, we kept loci present in all populations (–p as the number of populations) and all individuals (–r 1) to export (i) a matrix of 3788 SNPs for P. hespericus/punctatus (100% of completeness); (ii) a matrix of 3396 SNPs for P. atlanticus/ibericus (100% of completeness). For multispecies comparisons, subsets of 24 samples were selected for each genus, representing all available taxa, and sampled away from the hybrid zones (Tables S1 and S2). We generated sequence alignments of concatenated RAD tags, as well as SNP matrices, considering the loci present in all individuals: (i) 73,042 bp (2448 SNPs) for Discoglossus; (ii) 564,866 bp (9097 SNPs) for Pelodytes.
Mitochondrial barcoding
The distributions of mitotypes were examined in each system. For Discoglossus, we first harvested the cyt-b information published by Vences et al. (2014) for D. scovazzi/pictus (n = 198 from 35 localities) and D. g. galganoi/jeanneae (n = 148 from 75 localities), which included samples from our RAD library 1. Second, we sequenced partial 16 S (~600 bp) and/or cyt-b fragments (~400 bp) in 223 additional D. g. galganoi/jeanneae samples, i.e., 103 included in RAD library 2, and 120 others (77 localities). The following primers were used: 16SA (5′‐CGCCTGTTTATCAAAAACAT‐3′), 16SB (5′‐CCCGTCTGAACTCAGATCACG‐3′), Disco-cytb-F1 (5′‐CGTAATCGCAGGCGCAGCCAT-3′), and Disco-cytb-R1 (5′‐GTRATAGGTCGGAAGATAAG-3′). PCRs were carried out in 25-μl reactions containing 3 μl of template DNA, 12.5 μl of nanopure water, 7.5 μl of multiplex master mix (Qiagen, containing buffer, dNTPs, and hot‐start polymerase), and 1 μl of each primer (10 μM), and were run as follows: 95 °C for 15 min, 35 cycles of 94 °C for 30 s, 53 °C for 45 s, and 72 °C for 1 min and 72 °C for 5 min. Because of base ambiguities at D. g. galganoi/jeanneae mitochondrial amplicons, possibly due to pseudogenes (Vences et al. 2014, see “Discussion” section), we carefully inspected chromatograms to flag double peaks, and trimmed the sequences before indels (which disrupted the reading frame in heterozygotes). The resulting sequences (~230 bp for 16 S and ~330 bp for cyt-b) were manually aligned in Seaview (Gouy et al. 2010), and compared with reference haplotypes to infer their identity (Genbank JQ626663–JQ626668, KF644587–KF644736).
For Pelodytes, we used custom primers (Pd-cytb-F1: 5′-ATAAACGACTCATTYATYGACC-3′ and Pd-cytb-R2: 5′-GGTGAAAATAAGGCTAGAAGTA-3′) to partially amplify cyt-b (~600 bp) in 126 individuals (29 localities) from the P. hespericus/punctatus transition, and 97 individuals (19 localities) from the P. atlanticus/ibericus transition. PCRs were conducted as above, sequences were manually aligned in Seaview, and matched against published references (Genbank KP166402–166768).
Phylogenetic analyses
We performed phylogenetic reconstructions from the Discoglossus and Pelodytes nuclear SNP datasets by using the Bayesian framework of SNAPP (Leaché et al. 2014), implemented in the BEAST 2 platform (Bouckaert et al. 2014). We optimized model parameters and priors according to the data, following the recommendations by Leaché and Bouckaert (2018), namely coupling of mutation rates u and v, sampling the coalescent rate (with default starting value), and gamma distributions for λ (with α = 2 and β = 250) and snapprior (with α = 1 and β = 250). Test runs varying the parameter space explored by the priors yielded similar results. Chains were sampled every 1000 iterations, ran for about 3 million iterations, and stopped after confirming that they had reached long-term stationarity and large effective sample sizes of parameters (>200), using Tracer v. 1.5. Maximum-clade credibility consensus trees and cloudograms were produced by the modules TreeAnnotator and Densitree 2.0 (Bouckaert and Heled 2014), respectively, discarding the first 20% as burn-in.
SNAPP was further used to compare different coalescent models of species delimitations. Briefly, the approach, best known as Bayes factor delimitation, consists of reconstructing phylogenies with different predefined species and sample assignments, ranking them by marginal likelihood estimations (MLE), and assessing ranking support through Bayes factors (BF) (Grummer et al. 2013). To do so, we performed stepping-stone analyses with the path sampler module of SNAPP, considering twelve steps for each model. For these, chains were run 50,000 iterations, sampling every 50, and considering a conservative burn-in cutoff of 50%. Stationarity was always reached within the first thousand iterations, and increasing chain length and steps gave identical MLE values down to the decimal. Several alternative models of species hypotheses were tested against the current taxonomic arrangements (see “Results” section) by comparing MLE and BF.
We also conducted phylogenetic analyses based on concatenated RAD tag alignments in BEAST, using a GTR + G model (selected by bModelTest, Bouckaert and Drummond 2017), a relaxed lognormal clock, and a birth–death tree prior. Chains were run for 100 million iterations, sampling every 10,000, and checked with Tracer before exporting the results, with a burn-in of 20%. To obtain unconstrained phylogenies, we first ran the analyses without a molecular clock. We then dated the phylogenies by calibrating certain nodes in the light of the literature of these two radiations, as follows.
For Discoglossus, several biogeographical scenarios have been proposed (Fromhage et al. 2004). While the early split of Corsican D. montalentii was attributed to diverse plausible geotectonic events (e.g., Fromhage et al. 2004; Zangari et al. 2006), one phylogenetic pattern supported in several studies is the simultaneous divergence of the Iberian D. galganoi, the Tyrrhenian D. sardus, and the North African D. scovazzi and D. pictus (Pabijan et al. 2012, see Results). Following Busack (1986) and Zangari et al. (2006), we hypothesize that these events of vicariance were initiated by the end of the Messinian Salinity Crisis (5.33 My), when the Atlantic Ocean refilled the Mediterranean Sea (Krijgsman et al. 1999). We thus fixed the D. scovazzi/galganoi and D. pictus/sardus splits to ~5.33 My, with normally distributed priors of mean = 5.33 and σ = 0.1.
For Pelodytes, the Iberian diversification supposedly results from the Plio-Pleistocene climatic fluctuations, but no particular event can be attributed to a single split (Díaz‐Rodríguez et al. 2015, 2017). Based on the oldest fossil records, Díaz‐Rodríguez et al. (2017) assumed a divergence time no younger than ~3.5 My for the tree root, an age consistent with former phylogenetic analyses in Pelodytidae and related families, and accounting for the slow mitochondrial clock in these archaic neobatrachian frogs (Irisarri et al. 2012; Díaz‐Rodríguez et al. 2017, and references therein). We thus calibrated our Pelodytes tree root following previous inferences, with a gamma-distributed prior to α = 2, β = 0.2, and an offset of 3.4 Mya (95% prior distribution: 3.5–4.4 My, with a median of 3.7 My).
For the mitochondrial trees, we refer to previous studies that performed comprehensive multigene phylogenies of each group. This included Pabijan et al. (2012) for Discoglossus (2865 bp, four genes) and Díaz‐Rodríguez et al. (2015) for Pelodytes (1493 bp, two genes). In addition, we examined the relative genetic differentiation among taxa by computing net pairwise sequence divergence at two sets of markers in MEGA X (Kumar et al. 2018): (i) from the concatenated RAD tags; (ii) at sequences of the widely used mitochondrial gene 16 S (see previous references). Finally, for each group, we performed principal component analyses (PCA) on our phylogenetic SNP data (including all taxa) with the R package adegenet (Jombart 2008).
Analyses of contact zones
Each transition was analyzed in four different ways. First, we applied the Bayesian clustering algorithm of STRUCTURE (Pritchard et al. 2000) to assign RAD genotypes to genetic clusters, using the admixture model with correlated allele frequencies, and without pre-assignment to their geographic origins (locality). Chains were run for 100,000 iterations, after a burn-in of 10,000. Since each dataset involves two species, we primarily focused on solutions with K = 2 clusters, from which we extracted the ancestry coefficient Q to either species, as a genome-wide proxy to the hybrid index. Analyses for up to K = 5 were conducted, with five replicates for each K, in order to infer the rate of likelihood increase and the ΔK index (with STRUCTURE HARVESTER, Earl and vonHoldt 2012). Replicates were combined with CLUMPP (Jakobsson and Rosenberg 2007) and the averaged membership proportions of individuals were plotted using DISTRUCT (Rosenberg 2004). Second, to confirm the results obtained by STRUCTURE, individual genotypes were also clustered in two K groups with the model-based algorithm of ADMIXTURE 1.22 (Alexander et al. 2009). Twenty replicates were run, combined with CLUMPP, and plotted with DISTRUCT. Third, we performed PCAs on the individual SNP genotypes using the R package adegenet.
Fourth, sigmoid clines were fitted to the hybrid index (STRUCTURE’s Q) along geographic transects spanning across phylogeographic transitions with the R package hzar (Derryberry et al. 2014). These were located as follows: (1) from Andalusia (S-Spain) to Algarve (S-Portugal) for D. g. galganoi/jeanneae, along localities Ib53 to Ib21, and Ib1 to Ib3; (2) from the Moroccan-Algerian border to Ceuta (the Spanish enclave at the northernmost tip of Morocco) for D. pictus/scovazzi, along localities Af17 to Af3; (3) across Catalonia and Aragon regions in NE-Spain for P. punctatus/hespericus, along localities Cat1 to Cat29; and (4) across Beja district (S-Portugal) for P. ibericus/atlanticus, along localities Por8 to Por18. For each, we tested the different cline models available in hzar, from two (width w and center c) to eight parameters (including constrained or unconstrained exponential tails), and selected the ones with the lowest AIC values.
Results
Phylogenomics and coalescent species delimitation
The nuclear phylogenies obtained with SNAPP were fully resolved for both the painted (Fig. 1, 2448 SNPs) and parsley frogs (Fig. 2, 9097 SNPs). In Discoglossus, the topology matched those obtained with BEAST and published mitochondrial trees (Fig. S1). Our molecular clock dated the tree root (8.6 My, 95% HPD: 6.0–11.4 My), and the split for D. scovazzi/D. pictus (6.5 My, 95% HPD: 7.1–5.9 My) to the Late-Miocene, but the splits of Iberian D. g. galganoi/jeanneae (2.5 My, 95% HPD: 3.2–1.8 My) and Sicilian/African D. p. pictus/auritus (2.5 My, 95% HPD: 3.6–1.5 My) to the Plio-Pleistocene. The PCA provided a clear picture of genetic differentiation (Fig. 1), with PC1 separating the early diverged D. montalentii, and PC2 sorting Iberian, North-African, and Tyrrhenian taxa. Sequence divergences were stronger between the two African taxa than between the Iberian ones (RAD: 0.34 versus 0.06%; 16 S: 5.7 versus 2.2%, Table S3). Bayes factor delimitation rejected the current taxonomy: the best model distinguishes D. g. galganoi and D. g. jeanneae as distinct “species”, but not D. p. pictus and D. p. auritus, for a total of six “species” (Table 1).
In Iberian Pelodytes, our nuclear trees place P. atlanticus closer to P. ibericus (Fig. 2), while it is the closest to P. hespericus in the mitochondrial phylogeny (Fig. S2). The internal nodes are not fully supported in the BEAST analysis (Fig. S2), and the divergence time estimates of all species widely overlapped: 3.7 My (95% HPD: 4.1–3.4 My) for the tree root, 3.2 My (95% HPD: 3.8–2.0 My) for P. ibericus/atlanticus, and 2.8 My (95% HPD: 3.6–1.7 My) for P. hespericus/punctatus. Weaker nuclear differentiation for the latter pair is suggested by the PCA (Fig. 2) and divergence at RAD sequences (0.28 versus 0.36–0.45% in other pairwise comparisons, Table S4). As expected by the differing phylogenetic relationships, mitochondrial divergences were not congruent with the nuclear data, being actually stronger between the admixing Catalonian taxa for 16 S (Table S4). Bayes factor delimitation unambiguously favored the model retaining the current taxonomy, containing all four “species” (Table 1).
Contact zones in Discoglossus
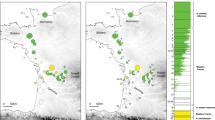
The analyses of genetic structure applied in parallel on each RAD dataset (STRUCTURE, ADMIXTURE, and PCA) were fully congruent, as shown by the highly correlated individual ancestry coefficients/scores on the first PCA component (Figs. S3–S6), suggesting robust inferences. The two Discoglossus transitions investigated revealed contrasting patterns of admixture. In Iberia, the analyses with K = 2 differentiated D. g. galganoi and D. g. jeanneae lineages in both RAD datasets (1121 SNPs and 4748 SNPs), and identified an array of introgressed individuals across Central and Southern Spain (Fig. 3). In Southern Spain, admixed populations were mainly located along the Guadalquivir River (loc 46–53), while galganoi frogs from Algarve (S-Portugal) bore faint traces of introgression by jeanneae (loc. 1–3). The genome-average cline fitted along the southern transition accordingly received a large width w = 136.8 km (95% CI: 83.4–222.4 km), with a center c = 223 km (95% CI: 180.1–270.7 km) located within our sampling gap across the Guadiana River and surroundings (Fig. 4a).
The latter shows average population (map) and individual (barplots) ancestry to the clusters of galganoi (purple) and jeanneae (blue) (K = 2), combining individuals from two separate datasets of 1121 (library 1) and 4748 SNPs (library 2), respectively. The large rivers discussed in the text are indicated on the map. The inset map shows the distribution of each taxon (adapted from Dufresnes 2019). Photo: D. g. galganoi (CD).
a D. g. jeanneae/galganoi along an east–west transect in S-Iberia. b D. pictus/scovazzi along an east-west transect in N-Morocco. c P. punctatus/hespericus along an east–west transect in NE-Spain. d P. atlanticus/ibericus along a west-east transect in S-Portugal. The crosses show the average ancestry at each sampled site, encircled proportionally to sample sizes. Photos (from the top left, clockwise): D. g. galganoi (CD), D. scovazzi (F. Deschandol), P. ibericus (CD), P. punctatus (CD).
This wide nuclear transition broadly coincides with the distribution of alleles sequenced with mitochondrial primers, and assigned to mtDNA haplotypes in previous studies (Vences et al. 2014). Individuals bearing double peaks at these sequences, and thus possibly affected by nuclear pseudogenes or heteroplasmy (see “Discussion” section), were almost exclusively sampled near the area of contact (Fig. 3). For 16 S, base ambiguities reflected the co-amplification of the typical galganoi and jeanneae haplotypes (Table S1), since no intraspecific polymorphism was found at the short fragment analyzed. For cyt-b, diagnostic SNPs were also involved in most ambiguities, but not necessarily all of them at once, and some intraspecific SNPs were heterozygous in some individuals as well.
In contrast, admixture was almost absent between D. pictus and D. scovazzi (794 SNPs, Fig. 5). All frogs from the Moulouya valley belong to D. pictus (loc. 11–17), while those from the rest of Morocco were fully assigned to D. scovazzi (loc. 1–10) (Figs. 5 and S3). The cline fitted across the populations sampled along the coast expectedly tended towards zero, since no hybrids were detected (w = 0.1 km, 95% CI: 0.1–69.7 km; c = 93.0 km, 95% CI: 78.9–133.2 km; Fig. 4b). Assuming that the two species do form a hybrid zone within the ~60-km sampling gap spanning between loc. 10 (pure scovazzi) and loc. 12 (pure pictus), the corresponding cline would be 17.4 km wide at maximum (analytical calculation by approximating Q = 0.999 at the edges of the gap). Further inland, a weak signal of introgression was suggested in an isolated population of D. pictus from the upper Moulouya River (loc. 11; Figs. 5 and S3). The nuclear and mitochondrial distributions were identical (Fig. 5).
The latter shows average population (map) and individual (barplots) ancestry to the clusters of scovazzi (yellow) and pictus (red) (K = 2), based on 794 SNPs. The Moulouya River is indicated. The inset map shows the distribution of each taxa (adapted from Dufresnes 2019). Photo: D. scovazzi (F. Deschandol).
Contact zones in Pelodytes
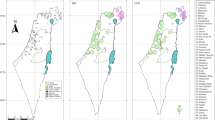
Introgression patterns remarkably differed between the Pelodytes hybrid pairs as well. Based on our RAD analyses (3788 SNPs), most populations in Catalonia west of Barcelona are a mixture of P. punctatus and P. hespericus (Figs. 6 and S4). This transition was quantified by a cline width w = 148.1 km (95% CI: 97.8–254.3 km), and a center predicted near the town of Lleida (c = 207.6, 95% CI: 183.2–247.5 km) (Fig. 4c). The species mitotypes were syntopic at only two localities in the middle of the hybrid zone (loc. 25, 27) (Fig. 6).
The latter shows average population (map) and individual (barplots) ancestry to the clusters of punctatus (blue) and hespericus (green) (K = 2), based on 3788 SNPs. The main regions/towns mentioned in the text are indicated on the map. The inset map shows the distribution of each taxa (adapted from Dufresnes 2019). Photo: P. punctatus (CD).
In Southwest Iberia, populations were assigned to either P. atlanticus, in most of Portugal, or P. ibericus in southeastern Portugal and Andalusia (3396 SNPs; Figs. 7 and S4). The transition identified was narrow, with a steep nuclear cline of w = 12.3 km (95% CI: 1.9–38.6 km) and c = 26.7 km (95% CI: 14.1–37.5 km), located close to the village of Colos (Fig. 4d). We did sample both mitochondrial haplotypes in loc. 12, a flooded field near Colos, but could not sequence nuclear loci due to insufficient DNA quality. All southern P. atlanticus appear slightly introgressed by P. ibericus, and the mtDNA of the latter was detected about 25 km away from the center of the contact zone (Por8).
The latter shows average population (map) and individual (barplots) ancestry to the clusters of atlanticus (purple) and P. ibericus clusters (orange) (K = 2), based on 3396 SNPs. The inset map shows the distribution of each taxa (adapted from Dufresnes 2019). Photo: P. atlanticus (CD).
Discussion
Insights on the evolution of Discoglossus and Pelodytes
The diversification of Discoglossus frogs has been a model to test competing biogeographic scenarios for allopatric speciation in the Western Mediterranean, from mtDNA (Fromhage et al. 2004), allozymes (Zangari et al. 2006), and nuclear sequences at individual genes (Pabijan et al. 2012). Our genome-average phylogeny was fully concordant with previous work, thus supporting their conclusions on the alternative chains of geotectonic events proposed for the vicariance events (discussed in Pabijan et al. 2012). All our divergence time estimates were remarkably close to those obtained by Fromhage et al. (2004) under the corresponding calibration scenario of their mitochondrial and protein clocks (scenario I). Whether the Messinian Salinity Crisis was responsible for the simultaneous disconnection of Iberian-N-African (D. galganoi/scovazzi) and Sicilian-Tyrrhenian ranges (D. pictus/sardus) remains an open question (Pabijan et al. 2012), but this appears plausible given the similar divergences of these populations (Zangari et al. 2006, Pabijan et al. 2012) and the prevalence of this phylogeographic pattern in the Mediterranean herpetofauna (Ehl et al. 2019). Finally, we also agree with previous studies (Martínez-Solano 2004; Vences et al. 2014) that the dramatic climate oscillations and sea-level subsidence linked to the glaciations may have triggered the Plio-Pleistocene divergences found in Iberia (D. g. galganoi/jeanneae) and between Sicily and the E-Maghreb (D. p. pictus/auritus), respectively (Dawson 1992; Wilson et al. 1999). The latter parallels the split between the Sicilian and African populations of the green toad Bufotes boulengeri (2.5 My; Dufresnes et al. in 2019b), suggesting common events of post-Messinian oversea dispersal at times when the low sea levels possibly brought the coasts as close as 50 km (see also Stöck et al. 2016).
Range isolation during glacial periods was also invoked for the cryptic Pelodytes radiation, where extant lineages hypothetically diverged in separate Iberian microrefugia (Díaz‐Rodríguez et al. 2015, 2017). One drawback in the reconstruction of the evolutionary history of Pelodytes is the absence of accurately dated events to calibrate the tree (Díaz‐Rodríguez et al. 2017). However, the oldest fossils (~3.5 My) are just slightly older (~2 My) than the age of the tree root estimated with a mitochondrial clock (Díaz‐Rodríguez et al. 2017 and reference therein), so there is little doubt on the Plio-Pleistocene origin of this group. Moreover, our analyses highlighted a possible conflict regarding the phylogenetic position of P. atlanticus, which appears genetically closer to P. ibericus, but was placed with P. punctatus/hespericus in previous analyses, notably the mitochondrial tree (with moderate node support; Díaz‐Rodríguez et al. 2015, 2017). One explanation is that P. atlanticus, P. ibericus and the ancestor of P. punctatus/hespericus all diverged simultaneously, blurring phylogenetic inferences drawn from a few markers. Alternatively, the discordance may stem from ancestral hybridization, following recurrent episodes of range contractions and secondary contacts, in which case we cannot exclude that past gene flow affected the nuclear phylogeny of Pelodytes. These hypotheses could be specifically tested in future phylogenetic frameworks, including data from the Caucasian relative P. caucasicus as outgroup.
Integrating hybrid zone studies in taxonomy
The contrasting patterns of admixture documented at phylogeographic transitions emphasize how the probability to speciate can be roughly predicted by the relative divergence between hybridizing versus non-hybridizing pairs of taxa in a given radiation, as recently shown in other vertebrates (Singhal and Moritz 2013; Dufresnes et al. 2019a, 2019b). Here, the lesser diverged D. g. galganoi/jeanneae and P. punctatus/hespericus admix across wide hybrid zones, indicative of little barriers to gene flow, while their elders D. pictus/scovazzi and P. ibericus/atlanticus feature restricted and localized introgression, consistent with stronger selection against hybrids.
The alternative hypothesis that limited dispersal prevents admixture among north-African Discoglossus, and southern Iberian Pelodytes is unlikely. The Moulouya valley is a prominent phylogeographic break for the Maghreb (Beddek et al. 2018) but does not constitute an ecological barrier for Discoglossus according to species distribution modeling (Vences et al. 2014). Although their ranges could be regionally disrupted at present (see Beukema et al. 2013), which would explain our ~60-km sampling gap in the studied transition (between loc. 10 and 12, Fig. 5), D. pictus successfully colonized the western side of the Moulouya, and admixture with D. scovazzi in its upper stretch testifies of past and/or present opportunities for gene flow. More generally, painted frogs are known to expand very rapidly, and thus, to be efficient dispersers (Veith and Martens 1987). In southern Portugal, the distribution of Pelodytes is continuous, and the landscape appears favorable to amphibian movements (CD pers. obs.). Under these conditions, the documented P. atlanticus/ibericus hybrid zone would be unrealistically recent if it was not mediated by reproductive isolation. Applying a model of neutral diffusion and assuming a dispersal rate of 1 km per generation (Barton and Gale 1993; based on previous estimates in a similar sized Anura), it would only take 24 generations to exceed the narrow cline width documented in our study. Hence, these transitions may even fit the definition of tension zones, where genes are selectively filtered across species boundaries.
If they exist, phenotypic characters related to assortative mate choice between the investigated taxa are not obvious. From current knowledge, the four Pelodytes species show little phenotypic differentiation, including breeding calls (Díaz‐Rodríguez et al. 2017), the major cue of anuran sexual selection. In fact, some bioacoustic differences were found only between P. hespericus and other Iberian Pelodytes, but were apparently insufficient to avoid massive hybridization with P. punctatus. In Discoglossus, the most divergent species have distinct mating calls (Glaw and Vences 1991), but temporal and spectral call variables remain nearly identical among Iberian (D. g. galganoi/jeanneae) and African taxa (D. pictus/scovazzi), which can be considered truly cryptic (Glaw and Vences 1991; Vences and Glaw 1996). Therefore, the contrasting patterns of admixture in these two groups should mostly reflect how DMIs have affected post-zygotic isolation rather than pre-mating barriers, even if other traits potentially affecting mate choice (e.g., odors) remain to be investigated.
Combining hybrid zone and phylogenetic analyses, we thus dissected the gray zones of speciation in Discoglossus and Pelodytes. According to the hybrid zone approach, we recommend to maintain D. scovazzi and D. pictus as distinct species, but D. g. galganoi and D. g. jeanneae as conspecific. The Sicilian endemic D. p. pictus and the North-African D. p. auritus should also keep their subspecific status, given that they diverged at approximately the same time than D. g. galganoi/jeanneae (Figs. 1 and S1). In Pelodytes, the novel taxa recently described by Díaz‐Rodríguez et al. (2017) received contrasting evaluations: P. hespericus belongs to the same species as P. punctatus, while the restricted introgression between P. atlanticus and P. ibericus supports species distinctiveness. Since P. atlanticus may not form a monophyletic clade with P. punctatus, and given their respective amounts of divergence, we thus recommend to recognize P. ibericus, P. atlanticus, and P. punctatus as three valid species, the latter including the subspecies P. p. punctatus and P. p. hespericus.
One major lesson from this comparative framework is that our taxonomic recommendations would have differed without the hybrid zone evidence. Built from our nuclear SNP phylogenies, the coalescent models overestimated the number of species in both groups, which is not surprising given the fully resolved trees and the exclusion of hybrid samples from the analysis (Leaché et al. 2014). Although powerful to reliably identify evolutionary significant units, the multispecies coalescent appears inappropriate for ranking their taxonomy, since the approach may interpret monophyletic genetic differentiation as speciation (Sukumuran and Knowles 2017, Campillo et al. 2019). Paradoxically, this oversplitting issue should disproportionally affect genomic datasets, which now disentangle divergences far beyond species boundaries. Enhancements in phylogenetic resolution will thus inevitably widen the gap between species delimitations obtained from the general lineage (De Queiroz 2007) versus the biological species concept (Mayr 1942).
We thus advise caution when drawing systematic conclusions based solely on phylogenetic inferences. Beyond model-based evidence, many claims for new species rely on mitochondrial divergence and private alleles at non-diagnostic nuclear genes (e.g., Ghielmi et al. 2016). In the genomic era, such data should qualify as too preliminary to call for taxonomic decisions, because mtDNA does not necessarily reflect the true evolutionary relationships of lineages, and whether the reported genetic distances are consistent with reproductive isolation is rarely addressed specifically. As cryptic species are increasingly considered, many biologists now integrate independently evolving populations in biodiversity assessments, by using their own delimitation criteria (often biased toward distance-based methods), hence causing a paradigm shift of the species concept (Fišer et al. 2018). Acknowledging this shift, and in an effort to develop multicriteria approaches of species delimitation under integrative taxonomy (Padial et al. 2010), we recommend to implement robust nuclear phylogenies together with hybrid zone analyses as a standard practice in molecular systematics, whenever feasible.
On thresholds of speciation
When no hybrid zone exists between candidate lineages, a classical approach of preliminary species delimitation consists in applying thresholds of divergence. Although disputable as it aims at dividing the continuous process of speciation, the rationale of thresholds remains consistent with the evolutionary view that reproductive isolation accelerates quickly once DMIs have broken down enough co-adapted gene combinations, and start to significantly reduce hybrid fitness (Orr and Turelli 2001; Matute et al. 2010; Moyle and Nakazato 2010). Hence, little divergence is expected to separate the extreme situations of lineage fusion, when speciation reverses (Seehausen 2006), from restricted gene flow, which may further create conditions favorable to reinforcement (Servedio and Noor 2003; Butlin and Smadja 2018). We accordingly noticed a sudden increase of hybrid zone steepness with divergence time in species-rich anuran radiations, and thus relatively narrow gray zones e.g., <1 My in Pelobates (Dufresnes et al. 2019a) and Bufotes (Dufresnes et al. 2019b). Which estimates are most suitable to capture this stepwise progression of speciation? Could thresholds be transposed across taxonomic groups if hybrid zones are not available/affordable to analyze? In the following, we provide a brief account of the current state of research, with some recommendations.
Early mentions of the threshold concept are found in Maxson and Maxson (1979), who hypothesized that an allozyme distance of Nei’s D > 0.15 corresponds to a genetic isolation of ~2 My in salamanders—later considered enough to complete the speciation process (Highton 2000, but see Vences and Wake 2007). Ever since, DNA divergence at mitochondrial genes has often been used as a proxy, because their resolution allows to discriminate young monophyletic lineages when single nuclear genes cannot. In an influential study on bufonid toads, Malone and Fontenot (2008) built upon a vast number of hybridization experiments to relate mtDNA divergence (12 S and 16 S), with reproductive incompatibility, and found evidence for full post-zygotic isolation in some lineages differing by ca. 3% sequence divergence, and in almost all lineages differing by >8%. Applying an approach introduced by Good and Wake (1992) and Wynn and Heyer (2001), Fouquet et al. (2007) showed that 16 S sequence divergences below 2% mostly reflect geographical isolation by distance in Neotropical frogs, whereas the relation is no longer significant with divergences >5%. Along these lines, Vieites et al. (2009) set up a threshold of 3% of 16 S divergence to define candidate species in Malagasy anurans. A 3% divergence cutoff is also used in bacteria, based on the prokaryotic 16 S rRNA gene (corresponding to mitochondrial 12 S rRNA) (Stackebrandt and Goebel 1994).
Despite their common usage, we refrain to recommend thresholds built exclusively from mitochondrial genes, because of their heterogeneous rates of evolution between lineages (e.g., Irisarri et al. 2012), the possible presence of pseudogenes (see below), as well as the frequent occurrence of cyto-nuclear discordances (Toews and Brelsford 2012; Bonnet et al. 2017), altogether making them unreliable molecular witnesses of evolutionary processes (e.g., Bernardo et al. 2019). Here, a >2% divergence threshold at the 16 S gene (Fouquet et al. 2007) would lead us to accept D. g. jeanneae as a species (2.2% from D. g. galganoi) but reject the valid P. atlanticus (0.8% from P. ibericus). A reassessment in the light of the gray zone, and in comparison with other estimates, shall inform on the accuracy of different sets of mitochondrial genes for future prospects.
Genome-wide nuclear divergence should offer more biologically informative metrics. Bacterial taxonomists are advised to perform full genome comparison to ascertain species status for isolates below the 16 S threshold (Stackebrandt and Goebel 1994; Stackebrandt and Ebers 2006). This remains difficult in eukaryotes, even if sequence divergence, from conserved genomic regions such as transcriptomes, offers some alternatives (Roux et al. 2016). Given the rising success of RAD methods and the associated analysis tools in phylogeography (e.g., Leaché et al. 2014), divergence at concatenated RAD tags could make a promising solution. Yet, comparisons between radiations are problematical because the loci sequenced in a RAD library are specific to the protocol applied (notably the enzymes used), the set of lineages analyzed, and the filtering parameters. In this study, incipient species bear 0.34% (D. pictus/scovazzi) and 0.39% (P. atlanticus/ibericus) of net RAD divergence, which roughly corresponds to the values computed for taxa meeting in comparatively narrow contact zones, and analyzed with the same protocol in our previous work on European anurans: 0.36% for Pelobates balcanicus/syriacus (Dufresnes et al. 2019a), 0.41% for Bufotes boulengeri/balearicus (Dufresnes et al. 2019b), and 0.37% for Hyla savignyi/felixarabica (Dufresnes et al. 2019c). There are outliers, however, e.g., only 0.16% for the quasi-isolated pair Pelobates fuscus/vespertinus (Dufresnes et al. 2019a), but up to 0.28% for the widely hybridizing P. punctatus/hespericus (this study). While it would be tempting to set up a RAD threshold of >0.30% from these examples, this would only pertain to our RAD protocol, and might still overlook some valid species. The same applies to other customized genomic protocols, such as target capture methods (e.g., Wielstra et al. 2019).
Divergence time, which correlates with genetic divergence, and by extension, reproductive isolation, could make a better proxy, provided resolved phylogenies, and comparable calibrations (Ehl et al. 2019). One advantage of molecular dating thresholds is their application to all kinds of DNA sequences. Based on Messinian calibrations, narrow amphibian hybrid zones (<30 km) are only found between taxa older than 3 My in Pelobates (Dufresnes et al. 2019a), but not younger than 5 My in Discoglossus (this study) and Hyla (Dufresnes et al. 2018), and 6 My in Bufotes (Dufresnes et al. 2019b). From these observations, we recently proposed 3 My as a low-bound yardstick for European anurans (Dufresnes et al. 2019a). Although some remarkably consistent timeframes of speciation stand out from global comparative analyses (2–3 My as the average species age across the tree of life, Hedges et al. 2015), the heterogeneous rates of diversification among vertebrate groups and between biogeographic regions (e.g., Cattin et al. 2016) obviously hinder the use of any universal checkpoint to delimit cryptic species.
To conclude, while proposed cutoff values may provide a preliminary indication of the level of distinctiveness of newly discovered lineages, the strong variability in both the timeframe of speciation and our ability to quantify it from current molecular estimates stresses the need to customize thresholds specifically for the radiation under focus. Implementing the hybrid zone approach of species delimitation will thus remain a necessary luxury in systematics. Even then, researchers may never agree on how narrow hybrid zones should be to reflect speciation, i.e., when hybridization and genetic introgression still occur (Harrison and Larson 2014). Rather than the amount of divergence and introgression, thresholds derived from the genetic architecture of this introgression—such as the proportion of barrier loci accumulating with reproductive isolation—may provide an indication of “whole-genome cohesiveness” in the face of gene flow (the “genic view” of speciation, Wu 2001), and thus, contribute to solving the century-old debate about the taxonomy of cryptic evolutionary lineages. Finally, thresholds may not be implemented as dichotomous decision tools but rather as probabilistic indicators of species delimitation, from which increasing excesses translate into higher probabilities of completed speciation.
On the “mitochondrial” polymorphism in Discoglossus
Finally, a puzzling result of our study is the co-amplification of different “mtDNA” haplotypes within single D. g. galganoi/jeanneae individuals (Vences et al. 2014). The most likely explanation is the presence of mutated pseudo-mtDNA copies in the nuclear genome (numts) (Hazkani-Covo et al. 2010), which can skew mtDNA-driven systematics (Dubey et al. 2009). All mtDNA genes tested in this genus yielded heterozygous chromatogram profiles in several individuals at least, including 16 S and cyt-b (as reported), but also ND1, ND4, ND5, and COI (IMS, unpublished data). Vences et al. (2016) confirmed this result for the 16 S gene using Illumina amplicon sequencing, for which D. g. galganoi/jeanneae samples (notably from the hybrid zone) received high read variation compared to D. pictus//scovazzi. The fact that heterozygous individuals mostly originate from the area of contact suggests a link with the hybridization events, leading to what could be called “cyto-numt” discordances. For instance, pure D. g. jeanneae from loc. Ib10–12 (on the basis of nuclear markers), which are heterozygotes at pseudo-mtDNA amplicons, could possess jeanneae-like numts (transposed from their original jeanneae mtDNA), but galganoi mitochondrial genomes were acquired through recent hybridization. Consequently, we cannot reliably infer the mitochondrial transition between these taxa.
Alternatively, heteroplasmy, i.e., the co-occurrence of more than one mtDNA copy in a single cell, could also explain our observations, although this phenomenon is exceedingly rare in vertebrates (Duan et al. 2018). Heteroplasmy results from paternal leakage (the transfer of sperm mitochondria into the egg), which was actually associated to hybridization in birds (Kvist et al. 2003; Gandolfi et al. 2017). Whether the sperm of Discoglossus hybrids is “leaking” mtDNA would be a fascinating question to address in future research, and illustrates how the continuum of divergence from population to speciation offers rich evolutionary frameworks to study the consequences of hybridization.
Data availability
References
Adams M, Raadik TA, Burridge CP, Georges A (2014) Global biodiversity assessment and hyper‐cryptic species complexes: more than one species of elephant in the room? Syst Biol 63:518–533
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Avise J, Walker D, Johns G (1998) Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc R Soc Lond B 265:1707–1712
Avise J (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge, MA
Barton NH (1983) Multilocus clines. Evolution 37:454–471
Barton N, Gale KS (1993) Genetic analysis of hybrid zones. In: Harrison R (ed) Hybrid zones and the evolutionary process. Oxford University Press, New York, NY, p 13–45
Beddek M, Zenboudji-Beddek S, Géniez P, Fathalla R, Sourouille P, Arnal V et al. (2018) Comparative phylogeography of amphibians and reptiles in Algeria suggest common causes for the east-west phylogeographic breaks in the Maghreb. PLoS ONE 13:e0201218
Bernardo PH, Sánchez-Ramírez S, Sánchez-Pacheco S, Álvarez-Castañeda ST, Aguilera-Miller EF, Mendez-de la Cruz FR et al. (2019) Extreme mito-nuclear discordance in a peninsular lizard: the role of drift, selection, and climate. Heredity 123:359–370
Beukema W, de Pous P, Donaire-Barroso D, Bogaerts S, Garcia-Porta J, Escoriza D et al. (2013) Review of the systematics, distribution, biogeography and natural history of Moroccan amphibians. Zootaxa 3661:1–60
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K et al. (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155
Bolnick DI, Fitzpatrick BM (2007) Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst 38:459–487
Bonnet T, Leblois R, Rousset F, Crochet P-A (2017) A reassessment of explanations for discordant introgressions of mitochondrial and nuclear genomes. Evolution 71:2140–218
Bouckaert RR, Heled J (2014) DensiTree 2: seeing trees through the forest. https://doi.org/10.1101/012401
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C‐H, Xie D et al. (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537
Bouckaert RR, Drummond AJ (2017) bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol 17:42
Brelsford A, Dufresnes C, Perrin N (2016) High‐density sex‐specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116:177–181
Busack SD (1986) Biochemical and morphological differentiation in Spanish and Moroccan populations of Discoglossus and the description of a new species from Southern Spain (Amphibia, Anura, Discoglossidae). Ann Carnegie Mus 55:41–61
Butlin R, Smadja CM (2018) Coupling, reinforcement, and speciation. Am Nat 191:155–172
Campillo LC, Barley AJ, Thomson RC (2019) Model-based species delimitation: are coalescent species reproductively isolated? Syst Biol https://doi.org/10.1093/sysbio/syz072
Catchen J, Hohenlohe P, Bassham S, Amores A, Cresko W (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Cattin L, Schuerch J, Salamin N, Dubey S (2016) Why are some species older than others? A large-scale study of vertebrates. BMC Evol Biol 16:90
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates Inc, Sunderland
Dawson AG (1992) Ice age earth: late-quaternary geology and climate. Routledge, London
De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
Derryberry EP, Derryberry GE, Maley JM, Brumfield RT (2014) HZAR: hybrid zone analysis using an R software package. Mol Ecol Resour 14:652–663
Díaz‐Rodríguez J, Gonçalves H, Sequeira F, Sousa-Neves T, Tejedo M, Ferrand N et al. (2015) Molecular evidence for cryptic candidate species in Iberian Pelodytes (Anura, Pelodytidae). Mol Phylogenet Evol 83:224–241
Díaz‐Rodríguez J, Gehara M, Márquez R, Vences M, Gonçalves H, Sequeira F et al. (2017) Integration of molecular, bioacoustical and morphological data reveals two new cryptic species of Pelodytes (Anura, Pelodytidae) from the Iberian Peninsula. Zootaxa 4243:1–41
Duan M, Tu J, Lu Z (2018) Recent advances in detecting mitochondrial DNA heteroplasmic variations. Molecules 23:323
Dubey S, Michaux J, Brünner H, Hutterer R, Vogel P (2009) False phylogenies on wood mice due to cryptic cytochrome-b pseudogene. Mol Phylogenet Evol 50:633–641
Dufresnes C (2019) Amphibians of Europe, North-Africa and the Middle East. Bloomsbury, London
Dufresnes C, Mazepa G, Rodrigues N, Brelsford A, Litvinchuk SN, Sermier R et al. (2018) Genomic evidence for cryptic speciation in tree frogs from the Apennine Peninsula, with description of Hyla perrini sp. nov. Front Ecol Evol 6:144
Dufresnes C, Strachinis I, Suriadna N, Mykytynets G, Cogălniceanu D, Székely P et al. (2019a) Phylogeography of a cryptic speciation continuum in Eurasian spadefoot toads (Pelobates). Mol Ecol 28:3257–3270
Dufresnes C, Mazepa G, Jablonski D, Caliari Oliveira R, Wenseleers T, Shabanov DA et al. (2019b) Fifteen shades of green: the evolution of Bufotes toads revisited. Mol Phylogenet Evol 141:106615
Dufresnes C, Mazepa G, Jablonski D, Sadek RA, Litvinchuk SN (2019c) A river runs through it: tree frog genomics supports the Dead Sea Rift as a rare phylogeographical break. Biol J Linn Soc 128:130–137
Earl DA, vonHoldt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Edward SV (2009) Is a new and general theory of molecular systematics emerging? Evolution 63:1–19
Ehl S, Vences M, Veith M (2019) Reconstructing evolution at the community level: a case study on Mediterranean amphibians. Mol Phylogenet Evol 134:211–225
Fišer C, Robinson CT, Malard F (2018) Cryptic species as a window into the paradigm shift of the species concept. Mol Ecol 27:613–635
Fouquet A, Gilles A, Vences M, Marty C, Blanc M, Gemmell NJ (2007) Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS ONE 2:e1109
Fromhage L, Vences M, Veith M (2004) Testing alternative vicariance scenarios in Western Mediterranean discoglossid frogs. Mol Phylogenet Evol 31:308–322
Gandolfi A, Crestanello B, Fagotti A, Simoncelli F, Chiesa S, Girardi M et al. (2017) New evidences of mitochondrial DNA heteroplasmy by putative paternal leakage between the rock partridge (Alectoris graeca) and the chukar partridge (Alectoris chukar). PLoS ONE 12:e0170507
García-París M, Jockusch EL (1999) A mitochondrial DNA perspective on the evolution of Iberian Discoglossus (Amphibia: Anura). J Zool Lond 248:209–218
Garrick RC, Banusiewicz JD, Burgess S, Hyseni C, Symula RE (2019) Extending phylogeography to account for lineage fusion. J Biogeogr 46:268–278
Gavrilets S (2004) Fitness landscapes and the origin of species. Princeton University Press, Princeton
Ghielmi S, Menegon M, Marsden SJ, Laddaga L, Ursenbacher S (2016) A new vertebrate for Europe: the discovery of a range-restricted relict viper in the western Italian Alps. J Zool Syst Evol Res 54:161–173
Glaw F, Vences M (1991) Bioacoustic differentiation in Painted frogs (Discoglossus). Amphib-Reptilia 12:385–394
Good DA, Wake DB (1992) Geographic variation and speciation in the torrent salamanders of the genus Rhyacotriton (Caudata: Rhyacotritonidae). Univ Calif Publ Zool 126:1–91
Gourbiere S, Mallet J (2009) Are species real? The shape of the species boundary with exponential failure, reinforcement and the ‘missing snowball’. Evolution 64:1–24
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Grummer JA, Bryson RW, Reeder TW (2013) Species delimitation using Bayes factors: simulations and application to the Sceloporus scalaris species group (Squamata: Phrynosomatidae). Syst Biol 63:119–133
Gutiérrez-Rodríguez J, Salvi D, Geffen E, Gafny S, Martínez-Solano I (2014) Isolation and characterisation of novel polymorphic microsatellite loci in Iberian painted frogs (Discoglossus galganoi and D. jeanneae), with data on cross-species amplification in Discoglossus and Latonia (Alytidae). Herpetol J 24:261–265
Harrison RG, Larson EL (2014) Hybridization, introgression, and the nature of species boundaries. J Hered 105:795–809
Hazkani-Covo E, Zeller RM, Martin W (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6:e1000834
Hedges SB, Marin J, Suleski M, Paymer M, Kumar S (2015) Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32:835–845
Highton R (2000) Detecting cryptic species using allozyme data. In: Bruce RC, Jaeger RG, Houck LD (eds) The Biology of Plethodontid Salamanders, Kluwer Academic/Plenum Publishers, New York, p 215–241
Irisarri I, San Mauro D, Abascal F, Ohler A, Vences M, Zardoya R (2012) The origin of modern frogs (Neobatrachia) was accompanied by acceleration in mitochondrial and nuclear substitution rates. BMC Genomics 13:e626
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Krijgsman W, Hilgen FJ, Raffi I, Sierro FJ, Wilson DS (1999) Chronology, causes and progression of the Messinian salinity crisis. Nature 400:652–655
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analyses across computing platforms. Mol Biol Evol 35:1547–1549
Kvist L, Martens J, Nazarenko AA, Orell M (2003) Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol Biol Evol 20:243–247
Leaché AD, Fujita MW, Minin VN, Bouckaert RR (2014) Species delimitation using genome-wide SNP data. Syst Biol 63:534–542
Leaché AD, Bouckaert RR (2018) Species trees and species delimitation with SNAPP: a tutorial and worked example. http://evomicsorg.wpengine.netdna-cdn.com/wp-content/uploads/2018/01/BFD-tutorial-1.pdf
Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV (2009) Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol 53:320–328
Malone JH, Fontenot BE (2008) Patterns of reproductive isolation in toads. PLoS ONE 3:e3900
Martínez‐Solano I (2004) Phylogeography of Iberian Discoglossus (Anura: Discoglossidae). J Zool Syst Evol Res 42:298–305
Matute DR, Butler IA, Turissini DA, Coyne JA (2010) A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329:1518–1521
Maxson LR, Maxson RD (1979) Comparative albumin and biochemical evolution in plethodontid salamanders. Evolution 33:1057–1062
Mayr E (1942) Systematics and the origin of species from the viewpoint of a zoologist. Harvard University Press, Cambridge, MA
Mendelson T, Inouye B, Rausher M (2004) Quantifying patterns in the evolution of reproductive isolation. Evolution 58:1424–1433
Moyle LC, Nakazato T (2010) Hybrid incompatibilities “snowballs” between Solanum species. Science 329:1521–1523
Orr H (1995) The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139:1805–1813
Orr HA, Turelli M (2001) The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55:1085–1094
Pabijan M, Crottini A, Reckwell D, Irisarri I, Hauswaldt SJ, Vences M (2012) A multigene species tree for Western Mediterranean painted frogs (Discoglossus). Mol Phylogenet Evol 64:690–696
Padial JM, Miralles A, de la Riva I, Vences M (2010) The integrative future of taxonomy. Front Zool 7:16
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Real R, Barbosa AM, Martínez-Solano I, García-París M (2005) Distinguishing the distributions of two cryptic frogs (Anura: Discoglossidae) using molecular data and environmental modeling. Can J Zool 83:536–545
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rosenblum E, Sarver B, Brown J, Des Roches S, Hardwick K, Hether T et al. (2012) Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol Biol 39:255–261
Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N (2016) Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol 14:e20000234
Seehausen O (2006) Conservation: losing biodiversity by reverse speciation. Curr Biol 16:334–347
Servedio MR, Noor MAF (2003) The role of reinforcement in speciation: theory and data. Annu Rev Ecol Evol Syst 34:339–364
Singhal S, Moritz C (2013) Reproductive isolation between phylogeographic lineages scales with divergence. Proc R Soc B 280:20132246
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Stöck M, Grifoni G, Armor N, Scheidt U, Sicilia A, Novarini N (2016) On the origin of the recent herpetofauna of Sicily: comparative phylogeography using homologous mitochondrial and nuclear genes. Zool Anz 261:70–81
Sukumaran J, Knowles LL (2017) Multispecies coalescent delimits structure, not species. PNAS 114:1607–1612
Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21:3907–3930
Veith M, Martens H (1987) Discoglossus pictus. In: Atlas of Amphibians and Reptiles in Europe. Societas Europaea Herpetologica, Museum National d’Histoire Naturelle, Bonn and Paris, p 104–105
Vences M, Glaw F (1996) Further investigations on Discoglossus bioacoustics: relationships between D. galganoi galganoi, D. g. jeanneae and D. pictus scovazzi. Amphib-Reptilia 17:333–340
Vences M, Lyra ML, Perl RGB, Bletz MC, Stankovic D, Lopes et al. (2016) Freshwater vertebrate metabarcoding on Illumina platforms using double-indexed primers of the mitochondrial 16S rRNA gene. Conserv Genet Resour 8:323–327
Vences M, de Pous P, Nicolas V, Diaz-Rodriguez J, Donaire D, Hugemann K et al. (2014) New insights on phylogeography and distribution of painted frogs (Discoglossus) in northern Africa and the Iberian Peninsula. Amphib-Reptilia 35:305–320
Vences M, Wake DB (2007) Speciation, species boundaries and phylogeography of amphibians. In: Heatwole HH, Tyler M (eds) Amphibian Biology, vol. 6, Systematics. Surrey Beatty & Sons, Chipping Norton, Australia, pp 2613–2669
Vieites DR, Wollenberg KC, Andreone F, Köhler J, Glaw F, Vences M (2009) Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proc Natl Acad Sci USA 106:8267–8272
Wielstra B, McCartney-Melstad E, Arntzen JW, Butlin RK, Shaffer HB (2019) Phylogenomics of the adaptive radiation of Triturus newts supports gradual ecological niche expansion towards an incrementally aquatic lifestyle. Mol Phylogenet Evol 133:120–127
Wilson RCL, Drury SA, Chapman JL (1999) The great ice age. The Open University, London, New York, NY
Wollenberg Valero KC, Marshall JC, Bastiaans E, Caccone A, Camargo A et al. (2019) Patterns, mechanisms and genetics of speciation in reptiles and amphibians. Genes 10i:E646
Wu CI (2001) The genic view of the process of speciation. J Evol Biol 14:851–865
Wynn A, Heyer WR (2001) Do geographically widespread species of tropical amphibians exist? An estimate of genetic relatedness within the Neotropical frog Leptodactylus fuscus (Schneider 1799) (Anura Leptodactylidae). Trop Zool 14:255–285
Zangari F, Cimmaruta R, Nascetti G (2006) Genetic relationships of the western Mediterranean painted frogs based on allozymes and mitochondrial markers: evolutionary and taxonomic inferences (Amphibia, Anura, Discoglossidae). Biol J Linn Soc 87:515–536
Zink RM, Barraclough G (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17:2107–2121
Acknowledgements
We thank G. Lavanchy and G. Fabre for their help in the lab, as well as R. Butlin for his useful comments in the paper. J.W. Arntzen, M. Barbosa, D. Buckley, D. Donaire, H. El Mouden, D. Escoriza, M. García-París, P. Galan, I. Garín, D. Gómez, A. Gosá, J. Gutiérrez, P. Hernández Sastre, H. Iglesias, U. Joger, D. Martínez, A. Montori, R. Pereira, E. Recuero, T. Slimani, A. Sánchez Vialas and G. Velo-Antón provided samples or assisted with field work. We appreciated the help of I. Rey and B. Álvarez (Tissue and DNA collection in MNCN) and P. Geniez (CNRS-EPHE BEV collection in CEFE) in facilitating access to samples under their care. The support and constructive remarks of three anonymous reviewers were also decisive to improve this paper. Our study was funded by the Swiss National Science Foundation, under grant 31003A_166323 to NP and fellowship P2LAP3_171818 to CD. BA was supported by a fellowship from the Fyssen Foundation, and MV by the Deutsche Forschungsgemeinschaft (grant VE247/16-1—HO 3492/6-1) in the framework of the “TaxonOmics” priority program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dufresnes, C., Pribille, M., Alard, B. et al. Integrating hybrid zone analyses in species delimitation: lessons from two anuran radiations of the Western Mediterranean. Heredity 124, 423–438 (2020). https://doi.org/10.1038/s41437-020-0294-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-020-0294-z
This article is cited by
-
Multiple contact zones and karyotypic evolution in a neotropical frog species complex
Scientific Reports (2024)
-
Easternmost distribution of Bufo bufo (Linnaeus, 1758) in Türkiye: implications for the putative contact zone between B. bufo and B. verrucosissimus
Genetica (2023)
-
Physical and ecological isolation contribute to maintain genetic differentiation between fire salamander subspecies
Heredity (2021)
-
The effect of phylogeographic history on species boundaries: a comparative framework in Hyla tree frogs
Scientific Reports (2020)