Abstract
Species’ geographical ranges are often restricted due to niche limitation resulting in geographical isolation and reduced population size at range margins. Under the “abundant center” paradigm, static marginal populations are thus expected to show higher genetic differentiation and lower genetic diversity than core populations. Low mate availability may also drive shifts toward higher propensity for selfing in geographically marginal populations. However, these predictions remain to be validated for contemporary range shifts occurring under current environmental change. This study is devoted to bridging this gap and assesses the spatial patterns of genetic structure and mating system across the geographical range of two coastal plant species characterized by contrasting contemporary range dynamics: the receding myrmecochorous Dune pansy (Viola tricolor subsp. curtisii) and the widespread expanding hydrochorous Rock samphire (Crithmum maritimum). Both species exhibited high propensity for selfing, with indications of inbreeding depression acting at early life stages. In Dune pansy, a biogeographical break was observed between core and marginal populations, with trailing-edge populations showing higher levels of genetic differentiation, reduced genetic diversity, and higher levels of selfing estimated through progeny arrays. In contrast, genetic structuring was weak in Rock samphire and no clear spatial trends were observed in genetic diversity nor in mating system, likely the result of efficient long-distance seed dispersal by sea-surface currents. Our study highlights that key species differences in life-history traits related to dispersal and/or mate limitation modify the expectations of genetic diversity loss and mating system shift in contemporary range-expanding populations, as compared with historical core populations.
Similar content being viewed by others
Introduction
Numerous ecological and evolutionary processes shape the geographical distribution of species, including climate features, the strength of biotic interactions, habitat and resource availability, anthropogenic habitat modifications, and key life-history traits such as mating system variation or dispersal abilities (Gaston 2009; Eckert et al. 2010; Grossenbacher et al. 2015; Moeller et al. 2017). Static species’ range boundaries often stem from declining environmental suitability with increasing distance from the species’ range core (e.g., Jump et al. 2003). Under the “abundant center” paradigm, static equilibrium range limits might take place, with marginal populations prone to extinction events or bottlenecks, as they occur in less suitable habitats with low density of conspecifics (Brown 1984; Sagarin et al. 2006; Phillips et al. 2010). Over the past decades, many empirical studies indeed showed increased genetic drift, shifts in mating system, and potentially reduced adaptive potential in marginal populations as compared with core populations (Eckert et al. 2008). These studies often involved surveys of historical range shifts since the ice retreat of last glacial maximum or rapidly expanding invasive species (e.g., Hewitt 2000; Hampe and Petit 2005; Colautti et al. 2010; Leys et al. 2014; Hopper et al. 2019). Nonetheless, ongoing human-driven environmental changes trigger current geographical expansion or retraction of species’ geographical ranges and local adaptation to changing conditions through the evolution of key life-history traits related to dispersal and reproductive traits (Jump and Peñuelas 2005; Colautti et al. 2010; Steinbauer et al. 2018; Vilà-Cabrera et al. 2019). In this respect, empirical evidence of concomitant changes in mating system and patterns of genetic structure for contemporary shifting ranges in a changing world is still lacking, especially in sessile organisms such as plant species for which gene flow within and among populations only occurs through seed and pollen dispersal (Hargreaves and Eckert 2014; Auffret et al. 2017). This study aims at bridging this gap and focuses on spatial patterns of genetic structure and mating system across two plant species’ geographical ranges characterized by ongoing contrasting dynamics of expansion and contraction.
Understanding the spatial genetic structure across species’ ranges using core populations and expanding or contracting range populations is motivated by evolutionary and management/conservation concerns (Vilà-Cabrera et al. 2019). Populations located at range margins are indeed generally geographically isolated within a matrix of inhospitable habitats and often of low population size (Kawecki 2008). When compared with core populations, the levels of genetic diversity are expected to be lower in marginal populations because of magnified genetic drift effects through reduced effective population size and founder effects (e.g., Hewitt 2000; Lönn and Prentice 2002; Leys et al. 2014; reviewed in Eckert et al. 2008). At the same time, an increase in among-population genetic differentiation is also expected owing to reduced levels of gene flow at range margins compared with core populations (e.g., Griffin and Willi 2014; Samis et al. 2016). Although both types of effects on genetic structure are supported by a large body of empirical work for static range boundaries (Eckert et al. 2008; López-Villalobos and Eckert 2018), differences in genetic structure between core and currently expanding or receding marginal populations still need to be investigated.
Theoretical studies showed that, in addition to drift and founder effects, ongoing extinction and recolonization processes are also an important determinant of genetic structure (Slatkin 1977; Whitlock and McCauley 1990). At range margins, the turnover of local populations can enhance opportunities for both genetic drift and gene flow, depending on the number of colonists relative to the number of recurrent migrants, and on whether the colonists arise from a single source (propagule-pool model) or many sources (migrant-pool model, see Pannell and Charlesworth 2000 for review). Therefore, counterintuitive patterns of genetic structure may arise for contemporary range shifts. First, marginal populations may be larger and more genetically diverse than core populations if the geographical range of the species is not constrained by habitat suitability (Sagarin et al. 2006; Eckert et al. 2008). Second, whereas lower levels of genetic diversity are expected in leading-edge populations, higher genetic diversity may persist in remnant trailing-edge populations (Hampe and Petit 2005; Arenas et al. 2012; Vilà-Cabrera et al. 2019). Third, asymmetrical long-distance dispersal may increase the levels of genetic diversity at the leading edge of the distribution and lower the among-population genetic differentiation (Bialozyt et al. 2006; Eckert et al. 2008; Leys et al. 2014). Overall, population genetic differentiation can increase or decrease depending on the source of colonists, the rate of colonization, and the rate of gene flow among static marginal populations (Whitlock and McCauley 1990; Le Corre and Kremer 1998; Pannell and Charlesworth 2000). These patterns remain to be investigated in native species for which geographical ranges are currently shifting.
Mating system variation can also play a role in successful colonization of marginal habitats characterized by low densities of conspecifics and/or poor pollination opportunities (Jain 1976; Grossenbacher et al. 2015; Pannell 2015; Moeller et al. 2017). Provided that inbreeding depression is low and/or mildly deleterious alleles are purged, self-fertilization is thought to facilitate geographical expansion (Baker 1955; Pannell 2015). Theoretical models predict that populations located at range margins should evolve toward selfing (Pannell and Barrett 1998; Cheptou and Massol 2009; Rodger et al. 2018), and many empirical studies suggest that the propensity for selfing is higher in leading-edge populations (Darling et al. 2008; Griffin and Willi 2014; López-Villalobos and Eckert 2018; see however Herlihy and Eckert 2005; Colautti et al. 2010). In trailing-edge populations, mate limitation may also select for higher selfing rates, which may be detrimental for long-term species’ persistence in a warming world due to increased genetic drift and resulting depletion in adaptive standing genetic variation (Hampe and Petit 2005; Levin 2012; Hargreaves and Eckert 2014; Vilà-Cabrera et al. 2019).
Broadly speaking, newly established populations at expanding leading edges are expected to show low effective population sizes and moderate levels of genetic diversity, high selfing rates, and possibly low incoming gene flow through long-distance dispersal events relative to core populations. On the other hand, trailing-edge populations are expected to face habitat fragmentation, triggering high genetic differentiation among populations. However, these predictions mostly hold for historical range shifts that left genetic footprint of postglacial recolonization or retraction. Empirical support of concomitant changes in mating system and genetic structure for ongoing contracting and expanding ranges are still lacking (Hargreaves and Eckert 2014).
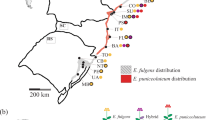
To bridge these gaps, we analyzed the genetic structure and mating system of two diploid native plant species with contrasting life-history traits: the widespread Rock samphire (Crithmum maritimum, Apiaceae), currently expanding in northwestern Europe, and the endangered Dune pansy (Viola tricolor subsp curtisii, Violaceae) which is receding in northern France (Fig. 1). Both species are perennial and self-compatible and have a strictly coastal distribution, which facilitates categorization into central (core) and marginal populations. These two focal species differ in life-history traits, notably in seed dispersal mode (hydrochory and likely associated long-distance dispersal events through marine currents for Rock samphire, short-distance dispersal events through myrmecochory for Dune pansy). We investigated the large-scale spatial patterns of genetic structure of a comprehensive sampling of populations collected from the range center to the leading (Rock samphire) or trailing edge (Dune pansy) of their geographical distribution. We specifically addressed the following questions related to contemporary expanding or receding geographical ranges:
-
(1)
Do currently shifting marginal populations display lower census and effective population size? If so, are they more genetically depauperate than stable historical core populations? If so, are they more genetically depauperate than core populations? We expect low genetic diversity in expanding colonizing populations of Rock samphire. However, a genetic legacy of high genetic diversity at the contemporary trailing edge of the species’ range could be expected for the retracting populations of Dune pansy.
-
(2)
Do expanding or retracting populations show higher levels of genetic differentiation compared with stable historical core populations and if so, to what extent? In spite of the mitigating effect of long-distance dispersal events through hydrochory, we expect stronger levels of genetic differentiation for Rock samphire populations at expanding range margins due to sequential founder effects. We also expect that biogeographical legacy should attenuate this pattern among currently shifting trailing-edge populations of Dune pansy.
-
(3)
Can we detect fine-scale genetic discontinuities mirroring contemporary expansion and retraction shifts? We expect asymmetrical gene flow from core (source) to contemporary expanding or retracting populations (sinks).
-
(4)
Is there a shift toward a higher propensity for selfing in marginal populations of currently shifting species, as predicted by Baker’s law for stable historical range limits? We predict no significant changes in selfing rates for these species, owing to the time lag necessary to approach genetic equilibrium (Pannell 2015).
Species’ geographical distribution (gray squares) and geographical locations of sampled populations (black stars) for Crithmum maritimum (a) and Viola tricolor subsp. curtisii (b). Sources of distribution data: http://siflore.fcbn.fr and http://www.gbif.org/
Methods
Study species, sampling, and genotyping
Rock samphire (Crithmum maritimum L., Apiaceae) is an insect-pollinated perennial halophyte and heliophilous coastal plant found on rocky shores in Western Europe (Fig. 1a). The species is restricted to a thin strip along the shoreline, and produces small corky achenes which can drift through marine currents, with seeds remaining viable over several months (Ridley 1930). Self-fertilization occurs, but strictly requires a pollination vector (Latron et al. unpublished results). Patterns of within-population gene flow in Rock samphire may mostly occur over short spatial distance through barochory and through pollen dispersal owing to local pollinator foraging. For the past decades, its geographical distribution has been expanding northwards in man-made habitats along the coastlines of northern France, Belgium, and the Netherlands, most often on the south facing parts of dikes, pontoons, pilings, riprap, and other artificial structures (Lambinon and Verloove 2012). Sampling included populations from the core of the range, on the Atlantic coast, up to the leading edge of the species distribution in the Netherlands (Fig. 1a).
Dune pansy (Viola tricolor subsp. curtisii [E. Forst] Syme, Violaceae) is an endangered insect-pollinated perennial species commonly found on semistabilized sand dunes on the coasts of the English Channel and North Sea (Fig. 1b). As for Rock samphire, self-fertilization obligately requires a pollinator (Latron et al. unpublished results). Determinants of among-population dispersal events are not documented in Dune pansy: seeds are mostly dispersed over very short distances, a few meters generally, by myrmecochory (Warren et al. 2010; Rix 2014). Over the past decades, Dune pansy lost habitats in northern France, due to urbanization and to dune fixation in peri-urban areas, and is now protected. We thus sampled populations of Dune pansy from the trailing edge of its distribution in northern France, to more central populations located in the Netherlands (Fig. 1b).
Rock samphire is a widespread species located around the Mediterranean and Atlantic coasts, up to the British Islands. Rock samphire sampling only covered the western part of continental Atlantic coast, excluding the UK (Fig. 1a). Dune pansy sampling covered the southern part of the species’ geographical distribution (Fig. 1b). For both species, we sampled marginal populations as exhaustively as possible, by screening every accessible habitat near range edges, while we collected well-distributed populations in the center of their distribution ranges (Fig. 1; Tables 1 and 2). In the core of each species’ distribution, populations occurred at almost every site with suitable habitat. Toward the edge of the distribution, the number of suitable habitats declines, fewer are occupied, and populations tend to be smaller. For Rock samphire, sampling was almost exhaustive from Den Helder (DH) to Breezanddijk (BREE; the Netherlands), and scattered in the more southern parts of the distribution range. For Dune pansy, population sampling was exhaustive from Stella-Plage (STEL) to Gravelines (GRAV, France), and scattered in the more northern parts of its geographic distribution. We also assessed the number of individuals in each population to get a census population size (Tables 1 and 2).
Overall, we sampled 22 Rock samphire populations and 19 Dune pansy populations, with sampling size varying from 3 to 39 individuals (24.3 ± 11.1) and from 23 to 30 individuals (29.6 ± 1.6), for a total of 508 and 563 fresh leaf samples, respectively (Fig. 1, Tables 1 and 2). WGS 84 coordinates of each individual were recorded. Coastal geographical distances among populations were calculated on the basis of the southernmost core population for Rock samphire (JSM) and the northernmost core population for Dune pansy (EZ).
Leaves were dried for 3 days at 50 °C, then DNA was extracted with the NucleoSpin 96 Plant II kit (Macherey Nagel, Duren, Germany) according to the manufacturer’s recommendations. All sampled individuals were genotyped at 21 and 10 nuclear microsatellite loci (Table S1) for Rock samphire and Dune pansy, respectively, as described in Latron et al. (2018). PCR products were sized using an ABI PRISM 3130 Sequencer and the software GeneMapper version 5 (Applied Biosystems). All genotypes were independently checked by two operators. Amplification failure was scarce (0.4% for Rock samphire and 0.9% for Dune pansy).
Levels of genetic diversity, genetic differentiation, and effective population size (Ne)
To identify rangewide spatial trends in population genetic structure, we calculated the following statistics using SPAGeDI version 1.5 (Hardy and Vekemans 2002): allelic richness (Ar) per locus and population, observed heterozygosity (HO), genetic diversity (HE), intrapopulation fixation index (FIS; Weir and Cockerham 1984) and mean levels and pairwise population genetic differentiation (FST). Allelic richness was estimated using a rarefaction procedure based on a minimal sample size of 11 and 23 individuals for Rock samphire and Dune pansy, respectively. Significance of FIS and FST estimates were assessed using 10 000 permutations of alleles within populations and of multilocus genotypes among populations, respectively. We chose to use FST since alternative measures of population differentiation tend to overestimate differentiation when mutation is high relative to migration rate, as is likely for microsatellite loci (Whitlock 2011). To estimate whether individual level of inbreeding increased for marginal populations, we computed the homozygosity by locus (HL, Aparicio et al. 2006), using the R package “genhet” (Coulon 2010). Likewise, to test whether contemporary effective population size (Ne) decreased toward the range edges, we estimated Ne based on levels of within-population linkage disequilibrium, as implemented in NeEstimator v2.1 (Do et al. 2014), for populations with at least ten sampled individuals.
Population genetic affiliation
To identify broadscale and cryptic geographical variation in allele frequencies depicting population boundaries, we performed a spatial principal component analysis (sPCA) as described in Jombart et al. (2008) and implemented in the R package “adegenet” (Jombart 2008). sPCA makes no assumptions with regard to linkage disequilibrium or departures from Hardy–Weinberg (HW) equilibrium. This analysis identifies independent synthetic components: those associated with the highest eigenvalues reflect positive spatial autocorrelation (“global structure”) while those associated with the lowest eigenvalues depict repulsion patterns (Jombart et al. 2008).
As a complementary approach, we tested whether the genetic differentiation among sampled locations mirrored the occurrence of distinct genetic pools by carrying out a non spatially explicit Bayesian genetic clustering, as implemented in the software InStruct (Gao et al. 2007). In contrast to the seminal procedure of Pritchard et al. (2000), individual assignments to the most likely number K of clusters do not assume random mating and a mean selfing rate can be estimated for each inferred cluster. We ran 35 Markov chain Monte Carlo (MCMC) chains for each value of K, ranging from 1 to the actual number of sampled populations for each species. Each run consisted in 200 000 initial burn-in iterations followed by 106 MCMC iterations. We used the ad hoc statistic ΔK (Evanno et al. 2005) to assess the most likely number of clusters.
Large- and fine-scale spatial genetic structure
To assess the occurrence of within or among-population spatial genetic structure, we studied the relationship between genetic relatedness and geographical distance among individuals through spatial correlograms based on pairwise individual kinship coefficients (Fij, Loiselle et al. 1995). Standard errors of Fij estimations were assessed using a multilocus jackknife estimator and 95% confidence intervals were generated using 10 000 permutations of individual locations. To compare the strength of spatial genetic structure among populations, we calculated the Sp statistic, which is independent of the sampling scheme, and described in Vekemans and Hardy (2004).
Assessment of recent migration rates among populations
To assess whether recent migration events among populations were asymmetrical from historical core populations to marginal populations, we used Bayesian inferences implemented in BayesAss (Wilson and Rannala 2003). This software estimates relative rates of gene flow over the last few generations, without requiring genetic equilibrium. We launched several runs of analysis including 20 million MCMC iterations with a burn-in of 4 million iterations and mixing parameters of 0.3 (0.6) for allele frequencies and 0.5 (0.7) for inbreeding coefficients in Dune pansy (respectively in Rock samphire).
Inference of mating system variation
To determine whether selfing rates increased toward the edges of geographical distribution ranges, we estimated within-population selfing rates (s) using three independent methods, one based on the within-population genotypic structure (sFIS), the second being derived from the multilocus correlation structure (standardized identity disequilibrium, sID), and the third relying on a progeny-array design (sPA). While sPA quantifies the amount of self-fertilized seeds, sFIS and sID estimate the amount of selfing at later life stages that undergone natural selection thinning.
sFIS was estimated from the classical relationship s = 2FIS/(1 + FIS) assuming inbreeding equilibrium in an infinite population for which the mating system is thought to be the most salient source of inbreeding (Hedrick 2011). sID was derived from maximum likelihood estimates based on the distribution of multilocus heterozygosity (identity disequilibrium; David et al. 2007). sFIS and sID estimates are thus based on a population structure approach. sPA was based on a progeny-array analysis using a set of maternal plants and offspring genotypes. Mating system parameters were estimated at the population level using a maximum-likelihood approach under a mixed-mating system model, using the MLTR v3.2 software (Ritland 2002). sFIS and sID were estimated for all populations, but sPA was only determined for five or six populations in each species (Tables 1 and 2). We chose to estimate sPA for a small number of large progeny arrays to better estimate individual selfing rates. 6 ± 1.58 sampled individuals per population were used as mother plants with 12.5 ± 2.1 seeds per plant, for a grand total of 506 and 375 genotyped seedlings for Rock samphire and Dune pansy, respectively. Finally, we also obtained an estimate of selfing rates at the level of the individual clusters inferred by Instruct.
Results
Levels of genetic diversity, effective population size, and genetic differentiation
In Rock samphire, moderate levels of genetic diversity were observed across loci (Table 1): mean allelic richness of 1.86 ± 0.47, mean He of 0.22 ± 0.11, and Ho of 0.13 ± 0.08). Linkage-based estimates of Ne were generally low (2 to 47, mean = 13.5, s.d. = 12.6). None of these estimates of genetic diversity significantly correlated with geographical location along the distribution range of this species (Figs. 2a, b and S1B, D).
Geographical distribution of genetic diversity in populations of Crithmum maritimum (a, b) and Viola tricolor subsp. curtisii (c, d). a, c allelic richness (Ar); b, d expected heterozygosity (HE). Insets show the estimates of genetic diversity with respect to the coastline geographical distance to central (core) populations; i.e., from the southernmost population for Crithmum maritimum and the northernmost population for Viola tricolor ssp. curtisii. For significant correlations, dashed lines indicate the 95% confidence interval
Dune pansy exhibited higher levels of genetic diversity (Table 2 and Fig. 2c, d): mean allelic richness of 2.94 ± 1.01, mean He of 0.38 ± 0.16, and Ho of 0.31 ± 0.14. All of these estimates significantly decreased from central to marginal populations (Figs. 2c, d and S2B; all at P < 0.05). All individuals from the trailing-edge population GRAV shared the same fully homozygous genotype. Yet, the most marginal population STEL had levels of genetic diversity similar to those found in central populations. Estimates of Ne varied greatly among populations (1–330, mean = 50; s.d. = 78), with no clear geographical trend (Fig. S2D).
Populations were highly differentiated in both species: mean multilocus FST were 0.37 ± 0.20 in Rock samphire and 0.33 ± 0.20 in Dune pansy. In rock samphire, FST ranged from 0.03 to 0.90, and among 190 pairwise FST values, only two were not significant. The same held for Dune pansy: pairwise FST estimates ranged from 0.04 to 0.90 with only one nonsignificant pairwise FST after Bonferroni correction, indicating substantial genetic differentiation among almost all populations (Fig. 4a, c). While pairwise FST values did not correlate with geographical distances in Rock samphire (rz = 0.024, Mantel test, P > 0.05), they did in Dune pansy (rz = 0.500, Mantel test, P < 0.001).
Population genetic affiliation and large- and fine-scale spatial genetic structure
Contrasting geographical partitions of populations were observed for the two focal species. Rock samphire showed a significant global structure (G-test = 0.11, P = 0.024) and four spatially structured groups were identified by the first three axes of the sPCA (Moran’s I = 0.72, 0.59, and 0.49 with var = 0.29, 0.23, and 0.17, respectively; Figs. 3a, S3): (i) southern populations from the Atlantic coast, (ii) neighboring populations from Normandy, (iii) scattered populations from northern France, (iv) isolated marginal Dutch populations. The same four groups were obtained when analyzing pairwise FST values among neighboring populations (Fig. 4a), with similar within-group mean pairwise FST (Fig. 4b). Bayesian clustering accounting for partial selfing identified three main clusters that did not correspond to well-defined geographically delimited genetic entities, and that showed substantial selfing rates ranging from 0.65 to 0.78 (Fig. S4).
Population genetic affiliations using sPCA analysis for Crithmum maritimum (a) and for Viola tricolor subsp. curtisii (b). Population colors reflect their coordinates on the first three (Crithmum maritimum) or two (Viola tricolor subsp. curtisii) axes of the sPCA. Coordinates along these three (a) or two (b) axes were assigned to the Red, Green, and Blue channels (only red and green were thus used in b). For Crithmum maritimum, the main sea surface currents are also shown (taken from Lazure and Desmare 2012) as well as breaks between genetic clusters (black arrows)
Pairwise FST estimates for Crithmum maritimum (a, b) and Viola tricolor subsp. curtisii (c, d). a, c: matrices of pairwise FST values for both species. Populations are listed from north to south in both cases. Black lines indicate population clusters, as determined by comparison of pairwise FST among neighboring populations. High FST values among two neighboring populations defined a break among clusters. Cluster names are given above the matrices. b, d: distribution of pairwise FST values within each cluster
Dune pansy showed striking geographical structuring with two differentiated lineages (Fig. 3b) supported by the sPCA global test (G-test = 0.14, P < 0.001). The first two sPCA axes accounted for most of the spatial genetic patterning in the data (Moran’s I = 0.88 and 0.69 with var = 0.77 and 0.18, respectively), with a genetic break between northern and southern populations occurring between populations MON and OVO, and a weak genetic discontinuity located more southerly, between populations WULF and GRAV (Fig. S5). This suggested admixed ancestry for the southernmost Dutch populations and northernmost French populations. Bayesian clustering clearly confirmed these two clusters and consistently inferred mixed ancestry for all individuals of the southernmost Dutch population OKA (Fig. S6). Selfing rate was of 0.34 and of 0.69 for the northern and southern lineages, respectively. Besides, this analysis also suggested a mixed affiliation of the southernmost French population STEL, which accordingly showed low pairwise genetic differentiation with core Dutch populations (Fig. 4c). Core Dutch populations were less differentiated than marginal populations (mean pairwise FST estimates of 0.20 and 0.54, respectively; P < 0.01; Fig. 4d).
Both species displayed large-scale significant spatial genetic structure with a clear decrease in genetic similarity among individuals with increasing geographical distance (Fig. 5a, c). Positive spatial autocorrelation only occurred at the intrapopulation level in Rock samphire (Fig. 5a). When performing this analysis at the level of genetic clusters identified above, a stronger signal of isolation by distance was observed for both species (Fig. 5b, d). Spatial genetic structure was clearly stronger for trailing-edge populations than for core populations in Dune pansy (Fig. 5d). Within populations, both species showed a significant pattern of isolation by distance in 63% of populations of Rock samphire and in 78% of Dune pansy populations, as indicated by the Sp statistic. Sp did not show any geographical trend for either species (Tables 1 and 2).
Spatial autocorrelograms depicting the relationship between pairwise kinship coefficients Fij (Loiselle et al. 1995) among individuals of Rock samphire (a, b) or Dune pansy (c, d), and the logarithm of coastal geographical distances. a, c: among all individual samples; b, d: within spatial clusters determined by the sPCA analysis. Solid lines correspond to the most marginal cluster, and dashed and dotted lines to the more central clusters. On all autocorrelograms, black symbols indicate values of Fij that are significantly different from zero. The Sp statistic is given for each autocorrelogram (all at P < 0.05)
Recent migration rates
Estimated migration rates among populations were generally low: the proportion of nonmigrants ranged from 67 to 88% in both species (Tables S2 and S3). For Dune pansy, a mean of 19% immigration events was observed, with the central populations WAS, MEIJ, and MONP contributing to emigration to diverse central populations (Table S2). The trailing-edge population CAL was inferred to send migrants to close neighboring populations. Surprisingly, populations OKA and STEL, which appeared of mixed ancestry, were not inferred to send nor to receive large proportions of migrants. For Rock samphire, inferred migration rates were slightly larger, with a mean incoming gene flow of 22% and strong evidence for asymmetrical gene flow coming from Normandy (Table S3). Indeed, populations from the Normandy peninsula (HER, LEVI, BRF, GOU, and BRV) sent migrants to most other populations, with dispersal events occurring randomly, irrespective of population location. The smallest expanding range-edge populations did not show higher immigration rates than core populations (20–23% immigrants in the four population clusters identified by the sPCA; Table S3).
Geographical variation in mating system along the distribution range
In Rock samphire, significant departures from Hardy–Weinberg equilibrium were detected in all but one population, with substantial FIS estimates ranging from 0.14 to 0.88 (0.41 ± 0.22, all at P < 0.05; Table 1 and Fig. S1A). The only population that did not show significant departures from HW expectations was PLOUHA (FIS = 0.17), probably due to sampling effects: only 11 individuals were sampled out of a much larger population of inaccessible plants on a cliff. Individual estimates of homozygosity by locus (HL) were high, ranging from 0.66 to 1 (0.82 ± 0.11; Table 1), but did not show any spatial trends (Fig. S1C). Selfing rate estimates supported a predominant self-fertilization mating system, regardless of the statistical method used to infer them. Mean selfing rate over populations was 0.57 ± 0.20, 0.53 ± 0.26, and 0.84 ± 0.10 using sFIS, sID, and sPA metrics, respectively (Fig. 6a and Table 1). Selfing rates based on FIS and on identity disequilibrium were strongly correlated (r2 = 0.78, P < 0.001; Fig. S7A). However, selfing rates estimated from progeny array analyses did not correlate with and were generally much higher than the former two estimates (Fig. 6 and S7A). Although selfing rates based on FIS and on identity disequilibrium did not show any significant geographical trends, selfing rates derived from progeny arrays indicated a marginal trend for increasing selfing rates in leading-edge populations (r2 = 0.54, P = 0.097; Fig. S7B).
Dune pansy showed a genotypic structure suggestive of a more mixed-mating system with a lower propensity for selfing. Significant departures from HW expectations were observed in 17 out of 19 populations, with FIS values ranging from 0.09 to 0.53 (0.20 ± 0.11) but no geographical trend emerged (Table 2 and Fig. S2A). One population (OKA) that was previously identified as being strongly admixed was characterized by a significant heterozygote excess, with a mean multilocus FIS value of −0.46 (P < 0.001). Overall, these results are consistent with relatively high individual HL estimates, varying from 0.47 to 1.00 (0.64 ± 0.16; Table 2) and significantly increasing from central to trailing-edge populations (r2 = 0.44, P = 0.002; Fig. S2C). Selfing rates estimated by FIS and identity disequilibrium methods were moderately correlated (r2 = 0.23, P = 0.06; Fig. S8A) and indicated a mixed-mating system (mean sFIS = 0.27 ± 0.17, sID = 0.14 ± 0.13), but did not show any significant geographical patterning from core to edge populations (Figs. 6b and S8B). Nonetheless, the progeny-based selfing rate estimations (mean sPA = 0.42 ± 0.25) showed a significant increase in selfing rates toward the receding range margin (P < 0.01; Figs. 6b and S8B).
Discussion
Variation of genetic diversity and effective population size along the geographical range
Due to geographical isolation and reduced census population size, lower levels of genetic diversity and effective population size, and higher levels of genetic differentiation are expected within and among static marginal populations relative to historical core populations (Le Corre and Kremer 1998; Pannell and Charlesworth 2000; Eckert et al. 2008). These expected patterns of genetic structure have been well established for invasive species or species that underwent historical range shifts due to postglacial range expansion (e.g., Hewitt 2000; Jump et al. 2003; Griffin and Willi 2014; Samis et al. 2016; López-Villalobos and Eckert 2018). However, empirical investigations of genetic structure of dynamic native species’ range over contemporary time scales are still lacking, especially for trailing edges of currently receding species (Hampe and Petit 2005; Arenas et al. 2012; Vilà-Cabrera et al. 2019).
Interestingly, no evidence of spatial trends in genetic diversity or Ne was observed across the range-wide distribution of Rock samphire. Ne estimates were particularly low, a finding expected for perennial species with a large propensity for selfing (Hamrick and Godt 1996; Griffin and Willi 2014; Ellegren and Galtier 2016). The lack of spatial trends in genetic diversity may be attributable to efficient seed dispersal by hydrochory (Kudoh and Whigham 1997; Favre-Bac et al. 2016). Occasional long-distance dispersal events mediated by seeds drifting along major sea currents can partially offset the effects of geographical isolation of marginal populations (Fievet et al. 2007; Leys et al. 2014). In line with these confounding effects, no significant pattern of isolation by distance was observed among Rock samphire populations, and a significant spatial genetic structure only occurred at the intrapopulation level. Finally, the lack of reduced Ne at leading range margins suggested that newly established populations are likely to be founded by several individuals. The mode of colonization has theoretically been recognized as a major factor influencing the level of genetic diversity and the amount of population differentiation, especially at range margin (Pannell and Charlesworth 2000). Long-distance seed dispersal by hydrochory may thus compensate for spatially restricted pollen flow and lead to departures from the common view of reduced genetic diversity that applies for historical leading-edge populations. This is predicted under a migrant-pool model where colonists are drawn from a random sample of the whole metapopulation (Whitlock and McCauley 1990). Empirical support for a migrant-pool model was observed for historical cytoplasmic variation in sea beet, a coastal plant species with similar life-history traits (Leys et al. 2014). Our study demonstrates that efficient long-distance dispersal abilities may attenuate contemporary changes in genetic structure expected at expanding leading edges owing to genetic drift (see also Bialozyt et al. 2006 for theoretical expectations).
In contrast, under a relaxed propagule-pool model where new populations are founded from few sources, genetic differentiation is always expected to increase relative to an island model (Slatkin 1977; Whitlock and McCauley 1990). Dune pansy seeds are dispersed over very short spatial distances, first by fruit explosion then by myrmecochory (Oostermeijer 1989; Rix 2014). Consistently, Dune pansy showed a strong pattern of isolation by distance. This suggests spatially restricted dispersal enhancing genetic drift effects, in line with patterns of genetic structure observed in various plant species with similar life-history traits (Hamrick and Godt 1996; Lönn and Prentice 2002; Hirai et al. 2012; Auffret et al. 2017). As observed for historical trailing edges (e.g., Griffin and Willi 2014), a significant decrease in genetic diversity was observed toward the trailing edge of the Dune pansy. One population in particular exemplified this pattern, because all sampled individuals shared the same fully homozygous genotype (population GRAV). However, the southernmost Dune pansy population (STEL), still exhibited levels of genetic diversity comparable to core populations. This population may benefit from being situated in an unusually large unmanaged dune complex, and may have retained its former rear-edge polymorphism (see Hampe and Petit 2005; Vilà-Cabrera et al. 2019). However, we did not observe any private alleles in this population, as would be expected for static remnant marginal populations (Hampe and Petit 2005). Beyond this legacy of past genetic diversity and of past population connectedness, another nonexclusive hypothesis may involve genetic admixture, e.g., through human-mediated sand transport. Overall and beyond a snapshot of neutral genetic variation, understanding adaptive differentiation will help design efficient management strategies to decide which dune pansy populations should be used as seed sources for translocation, supplementation, or assisted migration to preserve currently receding trailing-edge populations (Vilà-Cabrera et al. 2019).
Large- and fine-scale spatial genetic structure
Pollen and seed dispersal distance are crucial in determining population genetic connectedness in light of ongoing rapid environmental changes and alteration (Vekemans and Hardy 2004; Favre-Bac et al. 2016; Auffret et al. 2017). Myrmecochores are typically poor dispersers and ant-mediated seed dispersal does not facilitate range expansion (Warren et al. 2010). In a retracting and shortly-dispersed species such as Dune pansy, striking biogeographical patterns are thus expected (Hamrick and Godt 1996; Hewitt 2000; Vekemans and Hardy 2004). The recent migration rates among populations indeed indicated low levels of contemporary gene flow restricted to the nearest neighboring populations through a classical stepping-stone model. Moreover, a genetic discontinuity separated core Dutch populations from marginal French populations, with admixed ancestry for an intermediate population located in between. Finally, pairwise FST estimates suggested stronger level of genetic differentiation among trailing-edge populations than among core populations, consistent with theoretical and empirical expectations of increasing spatial isolation of marginal receding populations, especially when range contraction is slow (Arenas et al. 2012; Samis et al. 2016). In contrast, gene flow was not impeded among central populations benefiting from nearly continuous suitable dune habitats, with migration events occurring over long distances. Consistent with increasing geographical isolation, spatial genetic structure increased toward southern marginal populations. Altogether, patterns of contemporary genetic structure for this species perfectly mirrored what is expected for historical range contraction (Hampe and Petit 2005; Griffin and Willi 2014).
A somewhat different pattern of large-scale spatial genetic structure was observed in Rock samphire. Rock samphire is an entomophilous species with spatially-restricted pollen flow mostly occurring within populations. As such, a significant spatial genetic structure due to kinship structure was observed within most populations. However, no clear biogeographical patterns related to either recent range expansion or postglacial recolonization were identified using nonspatially explicit Bayesian clustering. Our results thus contrasted to most studies classically based on the ‘abundant center’’ hypothesis or on spreading invasive species (Jump et al. 2003; Eckert et al. 2008; Griffin and Willi 2014; Samis et al. 2016). This lack of global structure is consistent with theoretical expectations under a migrant-pool model and with relatively high estimated levels of contemporary rangewide migration events (Whitlock and McCauley 1990; Moeller et al. 2011). However, sPCA analyses with spatial priors allowed us to depict broad genetic breaks, matching main marine currents trajectories along the Atlantic, English channel, and North sea coastlines, as observed in hydrochorous coastal plant species with similar habitat requirements (Ridley 1930; Kadereit et al. 2005; Leys et al. 2014). A first break separated southern populations from Brittany and Normandy, consistent with long-term sea-surface current forking. Populations from Normandy appeared more genetically related with moderate pairwise genetic differentiation, which may suggest efficient connectivity through swirling currents (Salomon and Breton 1993; Fievet et al. 2007). Altogether, our results suggest that contemporary long-distance seed drifting, together with high seed longevity, partly erased the patterns of genetic variation expected under a simple isolation by distance model under drift/gene flow equilibrium for historical range shifts (Le Corre and Kremer 1998; Bialozyt et al. 2006; Moeller et al. 2011).
Mating system across the geographical range
Geographical variation in mating system may result from a diverse array of selective pressures, and determine opportunities for further adaptive evolution in rapidly changing human-altered environments (Jain 1976; Eckert et al. 2010; Grossenbacher et al. 2015; Pannell 2015; Moeller et al. 2017). Both Rock samphire and Dune pansy showed mixed-mating systems, as demonstrated by high mean FIS values and homozygosity by locus. Accordingly, both direct (progeny-array based) and indirect (population-structure based) estimates of selfing rate point to substantial selfing, especially in Rock samphire. In contrast to what is observed in mixed-mating species such as Dune pansy, predominant selfing increases the among-population genetic variance, enhances local inbreeding level, and decreases pollen flow. Therefore, Rock samphire populations were strongly genetically structured (mean FST = 0.37), harbored significant intrapopulation kinship structure and did not follow a large-scale isolation by distance pattern reflecting a gene flow/drift equilibrium. Along with reduced Ne, such findings are typical of predominantly selfing perennial species (Jain 1976; Hamrick and Godt 1996; Vekemans and Hardy 2004; Griffin and Willi 2014; Ellegren and Galtier 2016).
In Rock samphire, selfing rates estimated from population genetic structure all exceeded 25% and reached up to 94%. In a colonizing species, low mate availability and pollen limitation may favor selfers in expanding leading-edge populations (Eckert et al. 2008; Pannell 2015; Moeller et al. 2017). Nonetheless, indirect estimates of selfing rates did not increase toward leading-edge populations, as expected for newly established invasive species or historical expansion front following the ice retreat after the last glacial maximum (e.g., Griffin and Willi 2014; see however Colautti et al. 2010). Our results thus suggested attenuated founder effects during ongoing colonization. Indeed, the possibility of long-distance seed dispersal through marine hydrochory makes this species a potentially efficient colonizer (Ridley 1930; Kadereit et al. 2005). Multiple colonization events from diverse sources may offset drift effects and mate limitation, as theoretically expected under a migrant-pool model (Baker 1955; Whitlock and McCauley 1990; Bialozyt et al. 2006; Pannell 2015). Thus, contemporary expanding species may not always follow the expected pattern of increased selfing in stable leading-edge populations, either because (i) some traits related to dispersal make them efficient colonizers with large populations near the leading range limits (e.g., Herlihy and Eckert 2005), (ii) population expansion is not limited by the availability of pollinators and compatible mates (Colautti et al. 2010; Moeller et al. 2012), or (iii) colonization events are old enough for plants to have shifted back toward a more open mating system (Hargreaves and Eckert 2014; Pannell 2015). For the above reasons, Rock samphire populations did not show the expected pattern of increased selfing rates toward the leading edge of the species’ distribution.
Owing to urbanization and to diverse dune management strategies along the coast of Northern France, trailing-edge Dune pansy populations are more geographically isolated and generally smaller than core populations. In this mixed-mating species, mate limitation is expected in trailing-edge populations facing anthropogenic disturbances, which would drive an evolutionary shift towards more selfed progeny, as observed in other species, including coastal Dune plants (Darling et al. 2008; Eckert et al. 2010; Griffin and Willi 2014; López-Villalobos and Eckert 2018). Direct estimates based on progeny arrays indeed showed increased selfing rates toward currently receding trailing-edge populations, indicating pollen limitation and seed discounting in harsh marginal environments, and suggesting small Ne, as often observed for historical rear edges (Hampe and Petit 2005; Gaston 2009; Griffin and Willi 2014). However, caution has to be taken: indirect estimates of selfing rates based on population genetic structure did not show this expected increase in selfing rates in contemporary receding trailing-edge populations of Dune pansy. This highlights the need to use both direct and indirect estimators of selfing rates to avoid over-interpretation (Bürkli et al. 2017).
Selfing rate estimates using progeny arrays were also consistently higher than those estimated using population structure. Beyond departures from a pure inbreeding equilibrium (Hedrick 2011), this may be due to the time lag necessary to approach genetic equilibrium in receding or expanding populations (Pannell 2015). Alternatively, this may suggest the existence of some level of inbreeding depression acting at early developmental stages in both species, with selfed offspring showing lower survival than outcrossed offspring. Thus, thinning effects may explain why the difference in estimated selfing rates between central and marginal populations diminishes along life stages. In line with the conclusions of Bürkli et al. (2017), our results suggest that direct and indirect selfing rate estimates are not necessarily correlated, because they capture selfing rates at different life stages. To draw correct inferences on mating system, selfing rates need to be estimated at different life stages using both direct and indirect approaches, especially for populations with contemporary dynamic geographical ranges.
Conclusion
Patterns of genetic structure and mating system are rarely studied for native species undergoing retracting or expanding range dynamics at contemporary time scales. A reduction in genetic diversity and an increase in genetic differentiation in marginal populations, expected under the ‘abundant center’’ paradigm, was only observed near the trailing edge of the receding Dune pansy. Dune pansy and Rock samphire did not show the expected increase in selfing rates in marginal populations when considering estimates based on population genetic structure. However, progeny arrays suggested that most seeds were sired by selfing in currently shifting marginal populations, which may indicate the occurrence of some degree of inbreeding depression.
Altogether, this study lends support to the importance of life-history traits related to dispersal abilities in shaping contemporary genetic structure in a changing world. Lower genetic diversity, higher selfing rate, and higher genetic differentiation can reasonably be expected in marginal populations of fragmented and short-dispersed taxa. Nonetheless, these assumptions do not necessarily hold for self-compatible species with efficient dispersal capabilities, such as Rock samphire (reviewed in Grossenbacher et al. 2015; Moeller et al. 2017).
Data availability
Genotypes are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3m6917t.
References
Aparicio JM, Ortego J, Cordero PJ (2006) What should we weigh to estimate heterozygosity, alleles or loci? Mol Ecol 15:4659–4665
Arenas M, Ray N, Currat M, Excoffier L (2012) Consequences of range contractions and range shifts on molecular diversity. Mol Biol Evol 29:207–218
Auffret AG, Rico Y, Bullock JM, Hooftman DAP, Pakeman RJ, Soons MB et al. (2017) Plant functional connectivity—integrating landscape structure and effective dispersal. J Ecol 105:1648–1656
Baker HG (1955) Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution 9:347–349
Bialozyt R, Ziegenhagen B, Petit RJ (2006) Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. J Evol Biol 19:12–20
Brown JH (1984) On the relationship between abundance and distribution of species. Am Nat 124:255
Bürkli A, Sieber N, Seppälä K, Jokela J (2017) Comparing direct and indirect selfing rate estimates: when are population-structure estimates reliable? Heredity 118:525–533
Cheptou P-O, Massol F (2009) Pollination fluctuations drive evolutionary syndromes linking dispersal and mating system. Am Nat 174:46–55
Colautti RI, White NA, Barrett SCH (2010) Variation of self‐incompatibility within invasive populations of purple loosestrife (Lythrum salicaria L.) from Eastern North America. Int J Plant Sci 171:158–166
Le Corre V, Kremer A (1998) Cumulative effects of founding events during colonisation on genetic diversity and differentiation in an island and stepping-stone model. J Evol Biol 11:495–512
Coulon A (2010) GENHET: an easy-to-use R function to estimate individual heterozygosity. Mol Ecol Resour 10:167–169
Darling E, Samis KE, Eckert CG (2008) Increased seed dispersal potential towards geographic range limits in a Pacific coast dune plant. New Phytol 178:424–435
David P, Pujol B, Viard F, Castella V, Goudet J (2007) Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol 16:2474–2487
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimatorv2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Eckert CG, Kalisz S, Geber MA, Sargent R, Elle E, Cheptou PO et al. (2010) Plant mating systems in a changing world. Trends Ecol Evol 25:35–43
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nat Rev Genet 17:422–433
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Favre-Bac L, Mony C, Ernoult A, Burel F, Arnaud J-F (2016) Ditch network sustains functional connectivity and influences patterns of gene flow in an intensive agricultural landscape. Heredity 116:200–212
Fievet V, Touzet P, Arnaud JF, Cuguen J (2007) Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: do marine currents shape the genetic structure? Mol Ecol 16:1847–1864
Gao H, Williamson S, Bustamante CD (2007) A Markov Chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176:1635–1651
Gaston KJ (2009) Geographic range limits: achieving synthesis. Proc R Soc B Biol Sci 276:1395–1406
Griffin PC, Willi Y (2014) Evolutionary shifts to self-fertilisation restricted to geographic range margins in North American Arabidopsis lyrata. Ecol Lett 17:484–490
Grossenbacher D, Runquist RB, Goldberg EE, Brandvain Y (2015) Geographic range size is predicted by plant mating system. Ecol Lett 18:706–713
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Hamrick J, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc B 351:1291–1298
Hardy OJ, Vekemans X (2002) Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hargreaves AL, Eckert CG (2014) Evolution of dispersal and mating systems along geographic gradients: implications for shifting ranges. Funct Ecol 28:5–21
Hedrick PW (2011) Genetics of populations, 4th edn. Jones & Bartlett Publishers, Inc., Sudbury, MA
Herlihy C, Eckert C (2005) Evolution of self-fertilization at geographical range margins? A comparison of demographic, floral, and mating system variables in central vs. peripheral populations of Aquilegia canadensis (Ranunculaceae). Am J Bot 92:744–751
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hirai M, Kubo N, Ohsako T, Utsumi T (2012) Genetic diversity of the endangered coastal violet Viola grayi Franchet et Savatier (Violaceae) and its genetic relationship to the species in subsection Rostratae. Conserv Genet 13:837–848
Hopper JV, McCue KF, Pratt PD, Duchesne P, Grosholz ED, Hufbauer RA (2019) Into the weeds: Matching importation history to genetic consequences and pathways in two widely used biological control agents. Evol Appl 12:773–790
Jain SK (1976) The evolution of inbreeding in plants. Annu Rev Ecol Syst 7:469–495
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Dufour AB, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103
Jump AS, Peñuelas J (2005) Running to stand still: adaptation and the response of plants to rapid climate change. Ecol Lett 8:1010–1020
Jump AS, Woodward FI, Burke T (2003) Cirsium species show disparity in patterns of genetic variation at their range-edge, despite similar patterns of reproduction and isolation. New Phytol 160:359–370
Kadereit JW, Arafeh R, Somogyi G, Westberg E, Westberg E, Kadereit JW (2005) Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale. Taxon 54:861–876
Kawecki TJ (2008) Adaptation to marginal habitats. Annu Rev Ecol Evol Syst 39:321–342
Kudoh H, Whigham DF (1997) Microgeographic genetic structure and gene flow in Hibiscus moscheutos (Malvaceae) populations. Am J Bot 84:1285–1293
Lambinon J, Verloove F (2012) Nouvelle flore de la Belgique, du G.-D. de Luxembourg, du nord de la France et des régions voisines, 6th edn. Jardin Botanique National de Belgique, Meise, Belgium
Latron M, Arnaud J-F, Ferla H, Godé C, Duputié A (2018) Polymorphic nuclear markers for coastal plant species with dynamic geographic distributions, the rock samphire (Crithmum maritimum) and the vulnerable dune pansy (Viola tricolor subsp. curtisii). Mol Biol Rep 45:203–209
Lazure P, Desmare S (2012) Caractéristiques physiques et état écologique - Manche - Mer du Nord - Etat physique et chimique, caractéristiques physiques, courantologie. Ifremer/Agence des aires marines protégées. https://www.ifremer.fr/sextant_doc/dcsmm/documents/Evaluation_initiale/MMN/EE/MMN_EE_06_Courantologie.pdf. (Accessed Sep 13 2019)
Levin DA (2012) Mating system shifts on the trailing edge. Ann Bot 109:613–620
Leys M, Petit EJ, El-Bahloul Y, Liso C, Fournet S, Arnaud JF (2014) Spatial genetic structure in Beta vulgaris subsp. maritima and Beta macrocarpa reveals the effect of contrasting mating system, influence of marine currents, and footprints of postglacial recolonization routes. Ecol Evol 4:1828–1852
Loiselle BA, Sork VL, John N, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lönn M, Prentice HC (2002) Gene diversity and demographic turnover in central and peripheral populations of the perennial herb Gypsophila fastigiata. Oikos 99:489–498
López-Villalobos A, Eckert CG (2018) Consequences of multiple mating-system shifts for population and range-wide genetic structure in a coastal dune plant. Mol Ecol 27:675–693
Moeller DA, Briscoe Runquist RD, Moe AM, Geber MA, Goodwillie C, Cheptou PO et al. (2017) Global biogeography of mating system variation in seed plants. Ecol Lett 20:375–384
Moeller DA, Geber MA, Eckhart VM, Tiffin P (2012) Reduced pollinator service and elevated pollen limitation at the geographic range limit of an annual plant. Ecology 93:1036–1048
Moeller DA, Geber MA, Tiffin P (2011) Population genetics and the evolution of geographic range limits in an annual plant. Am Nat 178:S44–S57
Oostermeijer JGB (1989) Myrmecochory in Polygala vulgaris L., Luzula campestris (L.) DC. and Viola curtisii Forster in a Dutch dune area. Oecologia 78:302–311
Pannell JR (2015) Evolution of the mating system in colonizing plants. Mol Ecol 24:2018–2037
Pannell JR, Barrett SCH (1998) Baker’s law revisited: reproductive assurance in a metapopulation. Evolution 52:657–668
Pannell JR, Charlesworth B (2000) Effects of metapopulation processes on measures of genetic diversity. Philos Trans R Soc B Biol Sci 355:1851–1864
Phillips BL, Brown GP, Shine R (2010) Life-history evolution in range-shifting populations. Ecology 91:1617–1627
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ridley (1930) The dispersal of plants throughout the world. L. Reeve & Company, Limited, Ashford, Kent
Rix M (2014) Viola tricolor subsp. curtisii (Violaceae). Curtis’s Bot Mag 31:17–25
Rodger JG, Landi P, Hui C (2018) Heterogeneity in local density allows a positiveevolutionary relationship between self-fertilisation and dispersal. Evolution 72:1784–1800
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 47:35–52
Sagarin RD, Gaines SD, Gaylord B (2006) Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends Ecol Evol 21:524–530
Salomon JC, Breton M (1993) An atlas of long-term currents in the Channel. Oceano Acta 16:439–448
Samis KE, López-Villalobos A, Eckert CG (2016) Strong genetic differentiation but not local adaptation toward the range limit of a coastal dune plant. Evolution 70:2520–2536
Slatkin M (1977) Gene flow and genetic drift in a species subject to frequent local extinctions. Theor Popul Biol 12:253–262
Steinbauer MJ, Grytnes JA, Jurasinski G, Kulonen A, Lenoir J, Pauli H et al. (2018) Accelerated increase in plant species richness on mountain summits is linked to warming. Nature 556:231–234
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Vilà-Cabrera A, Premoli AC, Jump AS (2019) Refining predictions of population decline at species’ rear edges. Glob Chang Biol 25:1549–1560
Warren RJ, Giladi I, Bradford MA (2010) Ant-mediated seed dispersal does not facilitate niche expansion. J Ecol 98:1178–1185
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC (2011) G’ST and D do not replace FST. Mol Ecol 20:1083–1091
Whitlock MC, McCauley DE (1990) Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1724
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Acknowledgements
We are grateful to Vincent Comor, Chloé Ponitzki, and Eric Schmitt for their help in population sampling, and to Peter Maas and Casper Zuyderduyn, Harrie van der Hagen, Mark van Til, Tycho Hoogstrate, and Jan IJff for facilitating access to Dune pansy populations in the Netherlands. The authors thank the French Ministry for Higher Education and Research, the Hauts-de-France Regional Council (AREoLAiRe project), and the European Regional Development Fund (CLIMIBIO project) for their financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Latron, M., Arnaud, JF., Ferla, H. et al. Effects of contemporary shifts of range margins on patterns of genetic structure and mating system in two coastal plant species. Heredity 124, 336–350 (2020). https://doi.org/10.1038/s41437-019-0269-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-019-0269-0