Abstract
Wilson disease (WD) is an autosomal recessive disease of copper excess due to pathogenic variants in the ATP7B gene coding for a copper-transporting ATPase. We present a 5-year-old girl with the homozygous frame shift variant NM_000053.3: c.19_20del in exon 1 of ATP7B (consecutive exon numbering with c.1 as first nucleotide of exon 1), detected by whole-exome sequencing as a secondary finding. The variant leads to a premature termination codon in exon 2. The girl exhibited no WD symptoms and no abnormalities in liver biopsy. ATP7B liver mRNA expression was comparable to healthy controls suggesting that nonsense-mediated mRNA decay (NMD) could be bypassed by the mechanism of translation reinitiation. To verify this hypothesis, a CMV-driven ATP7B minigene (pcDNA3) was equipped with the authentic ATP7B 5′ untranslated region and a truncated intron 2. We introduced c.19_20del by site-directed mutagenesis and overexpressed the constructs in HEK293T cells. We analyzed ATP7B expression by qRT-PCR, northern and western blot, and examined protein function by copper export capacity assays. Northern blot, qRT-PCR, and western blot revealed that c.19_20del ATP7B mRNA and protein is expressed in size and amount comparable to wild-type. Copper export capacity was also comparable to wild-type. Our results indicate that c.19_20del in ATP7B is able to bypass NMD by translation reinitiation, demonstrating that the classification of truncating variants as pathogenic without additional investigations should be done carefully.
Similar content being viewed by others
Introduction
Wilson disease (WD, OMIM-P: 277900), an autosomal recessive disease of copper retention, is caused by bi-allelic variants negatively altering the function of the ATP7B gene (OMIM-G: 606882). ATP7B variants causing a (likely) altered function were classified as “(likely) pathogenic” according to the classification of the American College of Medical Genetics and Genomics (ACMG) [1]. For better readability, the ACMG nomenclature is used in the manuscript. ATP7B codes for a copper-transporting p-type ATPase of 157 kDa. Under physiological copper concentrations, ATP7B is localized in the trans-Golgi network membrane and transfers copper to ceruloplasmin which supplies extrahepatic organs with the metal [2]. However, ATP7B is able to translocate in the apical membrane to carry excessive copper out of the cell [3]. Lack of or non-functional ATP7B leads to an intracellular accumulation of copper mainly in liver and brain. Therefore, WD usually presents as liver disease or neuro-psychiatric disorder [4]. Phenotypes may differ considerably in view of organ involvement, severity, progression, and age of onset [4,5,6] but are generally not genotype-related [7]. WD is lethal without treatment, but timely therapy with a copper chelator such as penicillamine as initial agent for clinically affected patients or with zinc for presymptomatic patients or as maintenance therapy results in a normal life expectancy [4].
Here, we describe a 5-year-old girl with the homozygous frame shift variant c.19_20del in exon 1 of ATP7B (NM_000053.3, consecutive exon numbering with c.1 as first nucleotide of exon 1) leading to a premature termination codon (PTC) in exon 2 but without WD symptoms or abnormalities in liver biopsy. The variant was detected as secondary finding by whole-exome sequencing (WES) performed due to visual impairment and progressive hearing loss. Loudianos et al. [8] have reported on a 45-year-old man carrying the same frame shift variant compound heterozygous with the known pathogenic ATP7B variant c.-441_427del p.? who has also been asymptomatic. The ATP7B variant c.19_20del has also been detected on one of 400 mutant alleles in a cohort of 227 WD patients from Czech Republic and Slovakia [9]. Nine of the 227 patients have been described as asymptomatic. Unfortunately, it was not mentioned if any of those nine patients carried the ATP7B c.19_20del frame shift variant.
Nonsense and frame shift variants have usually been considered to be pathogenic as they lead to nonsense-mediated mRNA decay (NMD), except for variants located in the extreme 3′ end of the gene [1]. However, several studies revealed that also variants within the first bases downstream of the start codon are able to circumvent NMD [10,11,12]. NMD is a surveillance function degrading PTC-containing mRNA to prevent production of truncated proteins. According to the canonical model of NMD, the exon junction complex (EJC) model, NMD is harnessing exon–exon junctions to identify PTCs [13,14,15]. Approximately 24 nucleotides upstream of each exon–exon junction, a multiprotein complex, the EJC, is deposited during pre-mRNA splicing [16]. Subsequently, in the first “pioneer” round of translation, EJCs are removed by the ribosome. EJCs that are not removed by the ribosome due to PTCs are a signal for NMD. Compatible with this, bypassing of NMD occurs if a PTC is located <50 nt upstream of the last exon–exon junction as there is no possibility of a remaining EJC. There are several mechanisms how NMD can further be circumvented [12]. One of them is the reinitiation of translation at an in-frame start codon downstream of the PTC [10, 11]. This mechanism is likely, if the PTC is located in close proximity to the start codon, precisely within the first 200 nt [12]. This holds true for c.19_20del in ATP7B. The hypothetical mechanism of NMD and NMD bypass for c.19_20del is illustrated in Fig. 1a. Potential in-frame start codons in exon 2 of ATP7B are shown in Fig. 1b.
Hypothetical mechanism of nonsense-mediated mRNA decay (NMD) bypass for c.19_20del in ATP7B. a mRNA obtains exon junction complexes (EJCs) during pre-mRNA splicing. Ribosome removes EJCs in the pioneer round of translation. NMD: In case of a premature termination codon (PTC), EJCs remain on the mRNA which is subsequently degraded by NMD. No NMD: If translation can be reinitiated at a downstream in-frame AUG, all EJCs are removed and a protein with truncated N-terminus is expressed. b N-terminal ATP7B cDNA sequence: Color switch black/blue in lettering indicates exon–exon junctions. The original and potential new in-frame start codons are highlighted in green. Red TAA shows PTC generated by c.19_20del (marked in yellow). Gray indicates control variant c.915T>A p.(Cys305*). Reference transcript NM_000053.3, consecutive exon numbering with c.1 as first nucleotide of exon 1
We investigated how effectively c.19_20del is able to bypass NMD by analyzing ATP7B mRNA and protein levels as well as protein function to assess the variant’s pathogenicity.
Materials and methods
Patient samples
Samples from 15 patients with WD and 8 hepatically healthy donors were obtained from medically necessary liver biopsies or tissue of explanted livers. The study protocol conforms to the ethical guidelines of the Declaration of Helsinki (1964) and has been approved by the ethics committee of Hannover Medical School (No 3539-2017). As control patient data were anonymized prior to our study (and after patients or legal guardians had consented to the collection of biomaterial for research purposes) obtaining informed consent was not possible.
Sequencing
Genomic DNA was isolated from whole blood cells of the subject using the chemagic DNA Blood Kit special (PerkinElmer, Waltham, USA) according to the manufacturer’s protocol. Exonic regions were enriched using the SureSelect Human All Exon kit from Agilent followed by sequencing as 100 bp paired-end runs on a HiSeq2500 (Illumina, Inc., San Diego, CA, USA). Reads were aligned to the human reference genome (UCSC Genome Browser build hg19) using Burrows-Wheeler Aligner (v.0.7.5a). Detection of single-nucleotide variants and small insertions and deletions (indels) was performed with SAMtools (version 0.1.19). For analysis of rare bi-allelic variants, only variants with a minor allele frequency of less than 1% in the Munich in-house database were filtered [17]. Results were confirmed by Sanger sequencing. For this purpose exon 1 of ATP7B was amplified using forward primer GAATCATCCGTGTGAAGAGG and revers primer CAAGACATCCCTGGAGCTG. Sequencing was performed on ABI PRISM 3130xl Genetic Analyser (Applied Biosystems/Thermo Fisher, Brunswick, Germany). Data were analyzed using Sequence Pilot software (JSI medical systems, Ettenheim, Germany).
Multiplex ligation-dependent probe amplification (MLPA)
For MLPA, kit P098-C2 (MRC-Holland, Amsterdam; Netherlands) was used according to manufacturer’s instructions. Fragment analysis was performed on a CEQ 8000 Sequencer (Beckman Coulter, Krefeld, Germany). Data analysis was performed using Sequence Pilot software.
Vector constructs
ATP7B expression vectors were assembled using a gBlocks fragment (IDT, Leuven, Belgium) containing exon 1–3 of ATP7B (NM_000053.3, consecutive exon numbering with c.1 as first nucleotide of exon 1), including the 5′ untranslated region (UTR) and a truncated intron 2, and a PCR product containing exon 3–21 of ATP7B using vector pLB1080 (a gift from Ann Hubbard, Addgene plasmid # 37826) as template. The gBlocks fragment was introduced to pcDNA3 (Invitrogen/Life Technologies, Carlsbad, USA) using KpnI and XhoI restriction sites (pcDNA3_gBlocks). The PCR fragment was introduced to pcDNA3_gBlocks using XbaI and XhoI restriction sites.
Variants c.19_20del and c.915T>A p.(Cys305*) were introduced using Quick change site directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). For further details and sequence information see Supplementary Methods. All sequences of introduced fragments and variants were verified by Sanger sequencing.
For construction of a copper-sensitive firefly luciferase reporter vector, a sequence containing four tandem repeats of metal-responsive element d of the mouse metallothionein I [18] promotor and a TATA box was synthesized by genscript (Piscataway, NJ, USA) and introduced into pGL3 basic vector (Promega, Mannheim, Germany) using KpnI and NheI restriction sites. For sequence information see Supplementary Methods. The principle of the method has been described by van den Berghe et al. [19, 20].
Cell culture
HEK293T cell line was kindly provided by Johann Meyer (Institute of Experimental Hematology, Hannover Medical School, Hannover, Germany) and cultured in DMEM containing 10% FCS, 1% sodium pyruvat, and 100 U/mL penicillin/streptomycin at 37 °C and 5% CO2 in a humidified incubator.
Transfection
To overexpress ATP7B or its mutants for qRT-PCR measurements and western blot, 250,000 HLE cells per well were seeded 24 h before transfection in 6 well format. For Lipofectamine 2000 transfection (Thermo Fisher Scientific, Braunschweig, Germany), 300 ng of the respective pcDNA3 vector were filled up with MIGR1 (gift of Warren Pear, Addgene plasmid # 27490) to a total DNA amount of 2000 ng per well (10 µL Lipofectamine 2000 per well). The empty vector pcDNA3 served as negative control. After 24 h medium was changed. After additional 24 h RNA or whole-cell lysate was isolated.
mRNA analysis
RNA was extracted with RNeasy Mini Kit (Qiagen, Hilden, Germany). For RNA from snap-frozen liver, the tissue was previously crushed with pestle and mortar in liquid nitrogen. RNA was reverse transcribed using Revert Aid First Strand cDNA synthesis kit (Thermo Fisher Scientific). qRT-PCR was performed in technical duplicates or triplicates using hybridization probes (TIB MOLBIOL, Berlin, Germany). Cell culture samples were measured in biological triplicates. Primary liver material was measured in biological duplicates or only once due to limited liver tissue samples. For details see Supplementary Methods.
Northern blot was performed as described previously [21]. In brief, 3 µg of liver biopsy derived RNA or 7.5 µg of total RNA obtained from transfected HEK293T cells were separated in a denaturing 1% agarose gel and transferred to Biodyne B membrane (Pall, Dreieich, Germany) via capillary blot. Northern blots were hybridized with a radio labeled probe (Supplementary Methods) and incubated over night at 42 °C. Upon exposition, autoradiography was detected on X-ray films.
Western blot
Harvested cells were lysed in RIPA buffer. Protein was electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gel, and transferred onto a nitrocellulose membrane (GE Healthcare, Freiburg, Germany). The membrane was blocked for 1 h in non-fat milk. After incubation with primary antibody (binding at the C-terminus of ATP7B) over night at 4 °C and secondary antibody at room temperature for 1 h the membrane was developed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). β-Actin served as loading control. For details see Supplementary Methods.
Copper export capacity assay
For Lipofectamine 2000 transfection, 25 ng pGL3 reporter construct, 3 ng pRL-TK (Promega), and 25 ng of the respective pcDNA3 vector were filled up with MIGR1 to a total DNA amount of 250 ng and added to 9000 HEK293T cells seeded the day before (white 96 well plates, 9000 cells per well, 0.5 µL Lipofectamine 2000 per well). Co-transfected pRL-TK coding for renilla luciferase served for normalization. After 24 h cells were treated with 50 µM CuCl2 (Sigma-Aldrich). Luciferase activity was measured after additional 24 h with the DualGlo Luciferase Assay System (Promega) and normalized to renilla luciferase activity. Values were corrected for pcDNA3_GFP and the copper export capacity was determined by setting the extent of reduction for pcDNA3_wild-type at 1.
Data base entry
Patient and variant information can be found at Global Variome shared LOVD (https://databases.lovd.nl/shared/individuals with individual ID 00207500 and 00207521)
Results
Case report
We report on a girl presenting with visual impairment and progressive hearing loss at the age of five. She is the third child of consanguineous parents. One older brother was healthy and one presented with neurological symptoms after a severe metabolic crisis. WES revealed a riboflavin transporter deficiency (Brown–Vialetto–Van-Laere syndrome) in both affected children. As a secondary finding, a homozygous deletion of two base pairs in exon 1 of ATP7B was detected (NM_000053.3, consecutive exon numbering with c.1 as first nucleotide of exon 1; c.19_20del) in the girl. This variant is predicted to result in a frame shift p.(Gln7Aspfs*14) leading to a PTC in exon 2. The ATP7B variant was confirmed by Sanger sequencing. Since a genetic analysis of DNA from the parents was not possible the homozygous status of the ATP7B variant was confirmed by ruling out a heterozygous deletion by Multiplex Ligation-dependent Probe Amplification. The two brothers were found heterozygous for the ATP7B variant. Until the age of five the girl presented without hepatic WD symptoms and without abnormalities in liver biopsy (liver copper content at the age of five: 83 µg/g dry tissue, normal: 250 µg/g dry tissue; ceruloplasmin levels 297 mg/L, normal: >200 mg/L; ceruloplasmin is measured as a marker in WD. Non-copper bound apoceruloplasmin due to non-functional ATP7B is degraded faster [22]). After confirmation of the WES secondary finding at the age of five, but without knowledge of our current results, penicillamine therapy (15 mg/kg/d) was initiated for 17 months and then replaced by zinc acetate treatment (Wilzin® 2 × 25 mg/d, later raised to 2 × 50 mg/d) carried out up to now for our index patient. Zinc induces intestinal cell metallothionein, which binds copper and prevents its absorption. To address the riboflavin transporter deficiency life-long treatment with riboflavin (2 × 100 mg/d) was initiated simultaneously to penicillamine treatment. At the age of eight, the girl temporarily presented with neurological symptoms which had likely been due to the riboflavin transporter deficiency as cranial magnetic resonance imaging (cMRI) was without evidence of copper accumulation. The girl did not present any hepatological symptoms up to now. Presently, the girl is ten years old and is able to visit regular school. Figure S1 in Supplementary Results illustrates clinical details of the index patient in a timeline.
ATP7B liver mRNA expression in the patient with c.19_20del is unexpectedly high
As the deletion c.19_20del is located in close proximity to the start codon and two variant carriers, our index patient as well as the 45-year-old man described by Loudianos et al. [8], showed no hepatic WD symptoms, we investigated if the deletion causes NMD or is capable to escape. We therefore analyzed ATP7B mRNA expression levels by qRT-PCR in liver tissue from our index patient with homozygous c.19_20del, genetically and clinically proven WD patients, and controls without liver disease (Fig. 2a). RNA expression level in our patient with c.19_20del was found to be in the range of healthy controls and notably higher than in WD patients with other truncating variants in ATP7B. Northern blot investigations confirmed our qRT-PCR results (Fig. 2b). Furthermore, northern blot results showed no differences in height of the ATP7B gel bands indicating no effect on splicing of the tested variants.
ATP7B liver mRNA expression of patient with c.19_20del is unexpectedly high. Patients were grouped regarding their type of ATP7B variants. a Relative ATP7B mRNA expression in liver tissue of Wilson disease (WD) patients (all carrying pathogenic variants) and controls determined by qRT-PCR (TATA box-binding protein as reference gene). b Relative ATP7B mRNA expression in liver tissue of exemplarily chosen WD patients determined by northern blot (upper section) in comparison to qRT-PCR (lower section). Full size northern blot pictures are shown in Fig. S2 in Supplementary Results. Details of ATP7B variants of investigated patients are given in Tab. S1 in Supplementary Results. Biological replicates n = 1
c.19_20del mRNA and protein is comparable in size and amount to wild-type
To further investigate the impact of c.19_20del on ATP7B mRNA expression and protein level, we expressed the c.19_20del variant, the wild-type, and as a positive control for NMD, the known pathogenic nonsense variant c.915T>A p.(Cys305*) which is also located in exon 2 of the ATP7B gene in HEK293T cells. In order to be able to study NMD we also included the ATP7B 5′UTR as well as a truncated intron 2. The expected splicing leads to an introduction of EJCs necessary to investigate NMD. ATP7B mRNA expression level quantified by qRT-PCR was slightly lower for the c.19_20del variant compared to wild-type. However, the signal of the NMD positive control allele was 100 fold lower (Fig. 3a, upper section). These findings were confirmed by northern blot (Fig. 3a, lower section). Again, all bands occured at the same height in the gel which indicates no large differences regarding splicing of intron 2. Western blot analysis revealed that expression of the c.19_20del variant results in an ATP7B protein of about 150 kDa in size. It was undistinguishable from the amount and of slightly smaller size compared to the wild-type protein (157 kDa, UniProtKB - P35670, Isoform 1). In contrast, no protein was detected when the pathogenic variant c.915T>A p.(Cys305*) was expressed (Fig. 3b).
RNA and protein size and amount as well as copper export capacity of c.19_20del is comparable to wild-type. a, b HEK293T cells transfected with pcDNA3 empty vector or ATP7B expression vectors pcDNA3_wild-type, pcDNA3_c.19_20del, pcDNA3_915T>A for 48 h or left untreated. a Relative ATP7B mRNA expression in transfected HEK293T cells determined by qRT-PCR (TATA box-binding protein as reference gene) (upper section) and northern blot (lower section). b ATP7B protein expression determined by western blot. Full size northern and western blot pictures are shown in Fig. S3 and S4 in Supplementary Results. c HEK293T cells were co-transfected with pGL3_4MRE, pRL-TK and pcDNA3 empty vector, pcDNA3_GFP or ATP7B expression vectors pcDNA3_wild-type, pcDNA3_c.19_20del or pcDNA3_915T>A. After 24 h cells were treated with 50 µM CuCl2. Luciferase activity was measured after additional 24 h and normalized to renilla luciferase activity. Values were corrected for pcDNA3_GFP and the copper export capacity was determined by setting the extent of reduction for pcDNA3_wild-type at 1. Dependence of luciferase activity on copper concentration is shown in Fig. S5 in Supplementary Results. Statistics: one-way ANOVA/Bonferroni’s Multiple Comparison Test, ***P ≤ 0.001; ****P < 0.0001. Error bars show SD. Biological replicates n = 3
c.19_20del copper export capacity is comparable to wild-type
To analyze whether the ATP7B c.19_20del protein is able to export copper from the cytosol to the extracellular compartment, we performed copper export capacity assays. Copper export capacity was analyzed using firefly luciferase as reporter driven by a copper sensor. The copper sensor is based on metal-responsive elements of the mouse metallothionein-1 promotor cloned upstream of the firefly luciferase open reading frame as described by van den Berghe et al. [19, 20]. The copper export capacity was significantly lower when expressing the pathogenic variant c.915T>A p.(Cys305*), but undistinguishable when wild-type or c.19_20del alleles were expressed (Fig. 3c).
Discussion
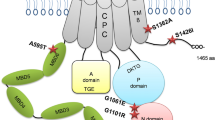
Our in vivo and in vitro analyses indicate that NMD for variant c.19_20del, which leads to a PTC in exon 2 of ATP7B, is at least in part circumvented (Figs. 2 and 3a). We were able to detect ATP7B protein for c.19_20del comparable in size and amount to wild-type ATP7B (Fig. 3b). As expected, control variant c.915T>A p.(Cys305*) showed only low mRNA levels and no protein was detectable, indicating that no full-length protein was built as the epitope for the ATP7B antibody is located at the C-terminus. Therefore, we suppose that for c.19_20del NMD is circumvented due to reinitiation of translation at one of the ten in-frame downstream ATGs in exon 2 (Fig. 1). We cannot completely rule out that the effect is due to a mechanism other than translation reinitiation, for instance, due to an aberrant splicing event which restores the reading frame. However, as shown in Fig. 2b and Fig. S2 in Supplementary Results no aberrant splicing event could be detected on total liver RNA from the patients. In addition, our minigene construct including intron 2 also did not show aberrant splicing (Fig. 3a and Fig. S3 in Supplementary Results) while Fig. 3b (and Fig. S4 in Supplementary Results) showed a slightly lower migrating protein band in the western blot arguing for reinititation. Reinitiation would result in a protein with truncated N-terminus. As the protein detected for c.19_20del in western blot was only slightly lower migrating compared to the wild-type (Fig. 3b and Fig. S4 in Supplementary Results), we hypothesize that one of the first downstream ATGs is the main starting point for reinitiation. Sizes of proteins resulting from the selection of the different ATGs are shown in Tab. S2 in Supplementary Results. The N-terminal part of ATP7B consists of six metal binding domains with the first one starting at amino acid (AA) position 56 [23].The first likely pathogenic (according to Richards et al. [1]) missense variant in ATP7B downstream of the original start codon is located at AA position 41 (c.122A>G p.(Asn41Ser)) [24]. Braiterman et al. [25] described that AAs 37–45 are required to retain the protein in the trans-Golgi network under low copper levels and target it apically in polarized hepatic cells under high copper levels. This indicates that a fully functional and trafficking capable ATP7B protein can only be built, if no more than the first 36 AAs are lacking. This likely restricts the new translation start site to the start codon at AA position 33 (Fig. 4). Expendability of the extreme N-terminal part of ATP7B is further supported by the fact that the first block of consecutive moderately conserved AAs downstream of the original start codon can be found at position 38–45, whereas lower numbered AAs are not conserved (aside from single moderately conserved AAs 25 and 34). In our copper export capacity assays (Fig. 3c), we could show that c.19_20del has a copper transport activity similar to the wild-type, showing that the protein is capable of transferring copper from one compartment to another. Our findings indicate that c.19_20del is able to escape NMD by the mechanism of translation reinitiation, with AA position 33 being the most probable new start site to allow the production of a fully functional and correctly localized protein. We therefore suggest to use p.(Met1_Ala32del). These findings are likely not only relevant for the variant investigated by us, but also for other variants located upstream of this variant in the ATP7B gene.
Potential translation reinitiation sites in ATP7B carrying c.19_20del. The N-terminal part of ATP7B consists of six metal binding domains (MBD, only four are fully shown) connected by linkers [23]. The first likely pathogenic, according to Richards et al. [1], missense variant in ATP7B downstream of the original start codon M1 is located at amino acid position 41 [c.122A>G p.(Asn41Ser)]. Amino acids 37–45 are required to retain the protein in the trans-Golgi network (TGN) under low copper levels and target it apically in polarized hepatic cells under high copper levels [25]. The new translation start site is likely restricted to the methionine (M) at amino acid position 33. Reference transcript NM_000053.3
Frame shift variant c.19_20del in ATP7B has also been detected in an asymptomatic 45-year-old man described by Loudianos et al. [8]. He carried the variant compound heterozygous with the known pathogenic variant c.-441_427del p.?. Clinical examinations of the man have revealed only slightly decreased ceruloplasmin levels (20 mg/dL, normal: 25–62 mg/dL) and steatosis, which could have been due to mild obesity (body mass index = 30). Liver chemistry as well as liver copper content (110 µg/g dry tissue, normal: 250 µg/g dry tissue) and urine copper content (basal: 10 µg/24 h, normal: <50 µg/24 h; penicillamine challenge: 600 µg/24 h, normal <1600 µg/24 h) have been within normal range. Brain magnet resonance imaging has been without abnormalities and no Kayser–Fleischer rings, caused by copper deposition in Descemet’s membrane in the eye [4], have been detected. Four years later the clinical picture has not changed although therapy was declined. This further supports no or only a mild pathogenic effect of frame shift variant c.19_20del.
With regard to our current results, an adjustment of therapy for our index patient is indicated to prevent a copper deficiency, which could result in hematological and neurological complications [26, 27], although the girl has been without hematological symptoms up to now. The therapy adjustment is planned as follows: bisection of zinc dose, with monthly and later 3-monthly clinical and laboratory monitoring. After twelve months without detected abnormalities, therapy will be terminated with continuing control of liver enzymes, urinary copper excretion, cMRI and liver histology in course (see also Fig. S1 in Supplementary Results).
Nonsense or frame shift variants are often considered as (likely) pathogenic. According to the ACMG standards and guidelines for the interpretation of sequence variants [1] also c.19_20del is classified as “likely pathogenic” [criteria PVS1: null variants, PM2: extremely low allele frequency, here: 0.014% (gnomAD)]. Several caveats have been mentioned for the criterion PVS1: genes, where loss of function is not a known disease mechanism, splice variants that are predicted to lead to exon skipping with leaving the remaining protein intact, the presence of multiple transcripts and location of the variant at the very 3′ end of a gene. The latter reflects the process of NMD bypassing if a PTC is located less than 50 nt upstream of the last exon–exon junction. However, other NMD bypassing mechanisms are not mentioned, although some of them have been shown to be not so rare at all. Lindeboom et al.[12] demonstrated on the basis of cancer genome data, that NMD efficiency is reduced to 35% if the PTC is located within the first 200 nt from the start codon whereas 93% NMD efficiency has been detected for PTCs > 200 nt from the start codon. This underlines that the classification of truncating variants in the first bases after the start codon should be done carefully without additional investigations, especially if the patient is asymptomatic regarding the gene-associated disease.
In 1.2% of exome or genome analyses, secondary findings, i.e., variants unrelated to the primary clinical diagnosis that triggered the genetic testing, are found [28]. At the moment, only few recommendations exist on how to handle secondary findings, with considerable variability concerning the list of genes that should be taken into account, e.g., from the ACMG [29]; the French Society of Predictive and Personalized Medicine (SFMPP) [30]; the European Society of Human Genetics (ESHG) [31], or the “100K Genomes Project” (https://www.genomicsengland.co.uk/taking-part/results/). As exome and genome sequencing are quickly becoming routinely used as a diagnostic tool, the question of how to handle secondary findings is becoming increasingly important. Especially in diseases such as WD, where clinical symptoms can be very subtle even in adults, it can be hard to define the clinical value of a genetic variant, as demonstrated here. This underlines the importance of supplementing high-throughput sequencing results with comprehensive pedigree and clinical data to better assess the clinical significance of the variant. Disclosure of secondary findings classified as “(likely) pathogenic” simply based on criteria as PVS1 and PM2 and without additional, in particular functional, data should be well considered.
In summary, our findings indicate that c.19_20del p.(Met1_Ala32del) is able to escape NMD by the mechanism of translation reinitiation underlining the importance of thorough classification of frame shift and nonsense variants, especially in cases of secondary findings.
References
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Ramos D, Mar D, Ishida M, et al. Mechanism of copper uptake from blood plasma ceruloplasmin by mammalian cells. PLoS ONE. 2016;11:e0149516.
Polishchuk R, Lutsenko S. Golgi in copper homeostasis: a view from the membrane trafficking field. Histochem Cell Biol. 2013;140:285–95.
Hedera P. Update on the clinical management of Wilson’s disease. Appl Clin Genet. 2017;10:9–19.
Ala A, Borjigin J, Rochwarger A, Schilsky M. Wilson disease in septuagenarian siblings: Raising the bar for diagnosis. Hepatology. 2005;41:668–70.
Kim JW, Kim JH, Seo JK, et al. Genetically confirmed Wilson disease in a 9-month old boy with elevations of aminotransferases. World J Hepatol. 2013;5:156–9.
Ferenci P. Phenotype-genotype correlations in patients with Wilson’s disease. Ann N Y Acad Sci. 2014;1315:1–5.
Loudianos G, Incollu S, Mameli E, Lepori MB. Wilson’s disease in an adult asymptomatic patient: a potential role for modifying factors of copper metabolism. Ann Gastroenterol. 2016;29:96–8.
Vrabelova S, Letocha O, Borsky M, Kozak L. Mutation analysis of the ATP7B gene and genotype/phenotype correlation in 227 patients with Wilson disease. Mol Genet Metab. 2005;86:277–85.
Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–33.
Neu-Yilik G, Amthor B, Gehring NH, et al. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA. 2011;17:843–54.
Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–8.
Thermann R, Neu-Yilik G, Deters A, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–94.
Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–83.
Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–97.
Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–9.
Kremer LS, Bader DM, Mertes C, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun. 2017;8:15824.
Stuart GW, Searle PF, Chen HY, Brinster RL, Palmiter RD. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci USA. 1984;81:7318–22.
van den Berghe PV, Folmer DE, Malingre HE, et al. Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem J. 2007;407:49–59.
van den Berghe PV, Stapelbroek JM, Krieger E, et al. Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology. 2009;50:1783–95.
Colombat M, Paradis V, Bieche I, et al. Quantitative RT-PCR in cirrhotic nodules reveals gene expression changes associated with liver carcinogenesis. J Pathol. 2003;201:260–7.
Holtzman NA, Gaumnitz BM. Studies on the rate of release and turnover of ceruloplasmin and apoceruloplasmin in rat plasma. J Biol Chem. 1970;245:2354–8.
Arioz C, Li Y, Wittung-Stafshede P. The six metal binding domains in human copper transporter, ATP7B: molecular biophysics and disease-causing mutations. Biometals. 2017;30:823–40.
Deguti MM, Genschel J, Cancado EL, et al. Wilson disease: novel mutations in the ATP7B gene and clinical correlation in Brazilian patients. Hum Mutat. 2004;23:398.
Braiterman L, Nyasae L, Guo Y, Bustos R, Lutsenko S, Hubbard A. Apical targeting and Golgi retention signals reside within a 9-amino acid sequence in the copper-ATPase, ATP7B. Am J Physiol Gastrointest Liver Physiol. 2009;296:G433–44.
Horvath J, Beris P, Giostra E, Martin PY, Burkhard PR. Zinc-induced copper deficiency in Wilson disease. J Neurol Neurosurg Psychiatry. 2010;81:1410–1.
Dziezyc K, Litwin T, Sobanska A, Czlonkowska A. Symptomatic copper deficiency in three Wilson’s disease patients treated with zinc sulphate. Neurol Neurochir Pol. 2014;48:214–8.
Hart MR, Biesecker BB, Blout CL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2018; e-pub ahead of print 5 October 2018; https://doi.org/10.1038/s41436-018-0308-x
Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55.
Pujol P, Vande Perre P, Faivre L, et al. Guidelines for reporting secondary findings of genome sequencing in cancer genes: the SFMPP recommendations. Eur J Hum Genet. 2018;26:1732–42.
van El CG, Cornel MC, Borry P, et al. Whole-genome sequencing in health care. Recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2013;21(Suppl 1):S1–5.
Acknowledgements
We thank Claudia Davenport for manuscript proofreading.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, Cluster of Excellence REBIRTH; ‘From Regenerative Biology to Reconstructive Therapy’ EXC 62).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Stalke, A., Pfister, ED., Baumann, U. et al. Homozygous frame shift variant in ATP7B exon 1 leads to bypass of nonsense-mediated mRNA decay and to a protein capable of copper export. Eur J Hum Genet 27, 879–887 (2019). https://doi.org/10.1038/s41431-019-0345-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-019-0345-1
This article is cited by
-
SRSF7 maintains its homeostasis through the expression of Split-ORFs and nuclear body assembly
Nature Structural & Molecular Biology (2020)
-
Molecular diagnostics of Mendelian disorders via combined DNA and RNA sequencing
Medizinische Genetik (2019)