Abstract
The Integrator complex subunit 1 (INTS1) is a component of the integrator complex that comprises 14 subunits and associates with RPB1 to catalyze endonucleolytic cleavage of nascent snRNAs and assist RNA polymerase II in promoter-proximal pause-release on protein-coding genes. We present five patients, including two sib pairs, with biallelic sequence variants in INTS1. The patients manifested absent or severely limited speech, an abnormal gait, hypotonia and cataracts. Exome sequencing revealed biallelic variants in INTS1 in all patients. One sib pair demonstrated a missense variant, p.(Arg77Cys), and a frameshift variant, p.(Arg1800Profs*20), another sib pair had a homozygous missense variant, p.(Pro1874Leu), and the fifth patient had a frameshift variant, p.(Leu1764Cysfs*16) and a missense variant, p.(Leu2164Pro). We also report additional clinical data on three previously described individuals with a homozygous, loss of function variant, p.(Ser1784*) in INTS1 that shared cognitive delays, cataracts and dysmorphic features with these patients. Several of the variants affected the protein C-terminus and preliminary modeling showed that the p.(Pro1874Leu) and p.(Leu2164Pro) variants may interfere with INTS1 helix folding. In view of the cataracts observed, we performed in-situ hybridization and demonstrated expression of ints1 in the zebrafish eye. We used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 to make larvae with biallelic insertion/deletion (indel) variants in ints1. The mutant larvae developed typically through gastrulation, but sections of the eye showed abnormal lens development. The distinctive phenotype associated with biallelic variants in INTS1 points to dysfunction of the integrator complex as a mechanism for intellectual disability, eye defects and craniofacial anomalies.
Similar content being viewed by others
Introduction
The integrator complex, INT, is composed of 14 integrator subunits that associate with the C-terminal domain of Rbp1, the largest subunit of ribonucleic acid (RNA) polymerase II (RNAPII) [1, 2]. In D. melanogaster, the integrator complex catalyzes the endonucleolytic cleavage of nascent small nuclear RNAs (snRNAs) at their 3′ ends and disruption of almost any integrator subunit causes snRNA misprocessing [1, 3]. There is still a paucity of information regarding the individual subunits of INT and their impact on human disease. The proteins show relatively little resemblance to other proteins, although the predicted motifs include domains that are suggestive of protein interactive surfaces, such as α-helical repeats (armadillo-like repeats (ARM), tetratricopeptide repeats (TPR), von Willebrand type A like (VWA), and HEAT domains) [1, 2, 4].
The largest integrator subunit in humans, INTS1, is composed of 2190 amino acids and has a molecular weight of 244 kDa [1]. INTS1 is conserved amongst many metazoans, including humans, rat, mouse, zebrafish and fruit fly, suggesting a specialized and non-redundant function. Targeted disruption of Ints1 in the mouse results in embryonic lethality at the early blastocyst stage for embryos with a pure genetic background [5]. The mutant mice have increased levels of unprocessed, primary U2 snRNA, suggestive of aberrant snRNA processing [5]. In Oryzias latipes, ints1 is strongly expressed in the brain and detectable in many other tissues at 6 days post fertilization (dpf) [6]. The awabancha (awb) mutant contains a splice site loss of function variant in ints1 (then called kiaa1440) and displays defective development of the thymic anlage and small eyes at 1.25 dpf and 2.5 dpf [6]. These mutants also had reduced liver and gall bladder size at 6 dpf, absence of the spleen at 7 dpf and died prior to hatching [6]. A zebrafish ints1 mutant has not yet been reported.
Despite the murine and medaka models for ints1 loss of function, a human phenotype associated with compromised INTS1 function was only recently described. Three unrelated patients, all of Dutch ancestry, demonstrated moderate to severe intellectual disability, lack of speech development, impaired mobility, cataracts, and a consistent pattern of dysmorphic features comprising dolichocephaly, a prominent forehead, wide-spaced eyes, pectus deformities and irregularly implanted and overlapping toes [7]. Findings noted in single patients were hypoplasia of the cerebellar vermis, renal dysplasia, cleft lip and palate and congenital heart disease. All three patients were homozygous for the same nonsense variant in INTS1, c.[5351C>A];[5351C>A], predicting p.[(Ser1784*)];[(Ser1784)] (NM_001080453.2). The INTS1 nonsense variant led to strong reduction of the mRNA expression in skin fibroblasts, consistent with a loss of function model of pathogenicity [7].
Herein we report an additional five children with biallelic sequence variants in INTS1 who manifest a similar pattern of clinical features comprising developmental delays, juvenile cataracts and craniofacial findings. We add on detailed clinical information regarding the three previously described patients and review the phenotype. We also provide data from in-situ hybridization studies and a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 model of loss of ints1 function in zebrafish (Danio rerio) that confirm the importance of this gene in eye development.
Material and Methods
Case reports
Clinical details and pedigree information were obtained from all patients as per standard clinical practice. The parents of patients 1 and 2 provided consent for publication of photographs that did not include recognizable facial features; the parents of patients 3 and 4 did not provide additional consent for publication of photographs. The parents of patient 5 provided consent for publication of facial photographs.
Exome sequencing
For patients 1 and 2, genomic DNA libraries were prepared (SeqCap EZ Human Exome Library v3.0; Roche Nimblegen, Madison, WI, USA) and sequenced on a HiSeq 2000 (Illumina, San Diego, CA, USA) for pair-end 100 cycles. Sequencing reads were aligned to the hg19 reference genome using the BurrowsWheeler Alignment tool (BWA v0.5.9) using the default parameters. Variants were subsequently called using Genome Analysis Toolkit v 1.3–21-gcb284ee [8, 9]. Variants were filtered with the recommendations listed in the Best Practice Variant Detection with GATK v3 with ŒQD < 2.0¹, ŒMQ < 40.0¹, ŒFS > 60.0¹, ŒHaplotypeScore > 13.0¹, ŒMQRankSum < −12.5¹, ŒReadPosRankSum < −8.0¹ for SNPs and ŒQD < 2.0¹, ŒReadPosRankSum < −20.0¹, ŒInbreedingCoeff < −0.8¹, ŒFS > 200.0¹ for Indels. The filtered list was then annotated with both ANNOVAR 2012Mar08 version [10] and snpEff v2.0.5 in GRCh37.64 [11]. The potential deleteriousness of novel sequence variants was assessed using PolyPhen-2 [12], Sorting Intolerant from Tolerant [13] and MutationTaster [14]. We utilized VarSifter [15] to explore the dataset and the INTS1 sequence variants were verified using Sanger sequencing.
For patients three and four, WES sequencing was performed as part of the Care4Rare consortium as previously described [16]. Variants were filtered to those that were homozygous in both patients three and four, and had a minor allele frequency of <0.005 in both of the 1000 Genomes or the exome variant server (EVS) databases, and absent or with a low frequency in an in-house database. The remaining variants were assessed using PolyPhen-2, SIFT, and MutationTaster. For patient five, whole exome sequencing was performed as a clinical test (GeneDx, Inc.) using the IDT xGen Exome Research Panel v1.0. Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19, and analyzed for sequence variants using a custom-developed analysis tool as previously described [17]. Exome sequencing of patient 6, 7, and 8 was previously described [7].
Clinical data and variants were submitted to the gene variant database at www.LOVD.nl/INTS1 (patient IDs 00164989–00164991).
Structure modeling
We used HHpred server [18] to identify structures of homologous proteins. HHpred identified homologs with low sequence identity of ~12% only for the C-terminal part of the protein (residues 1661–2184). We used MODELLER [19] to generate structural models for this part based on the identified homologs (PDB 2db0, 2vgl, and 4uqi).
In-situ hybridization
All animal experiments were performed under a protocol approved by the Institute for Animal Care and Use Committee (IACUC) at UCSF (AN108657–02C). EKW strain zebrafish were maintained at 28 °C with a 14 h (h) light/10 h dark cycle. We used primers to amplify the full-length ints1 transcript from wildtype Danio rerio cDNA (XM_003198152.5; Table S1). Database searches showed no significant cross-hybridization between the amplified cDNA and other Danio rerio genes. The cDNA fragment was cloned into pCRII-TOPO (Invitrogen/ThermoScientific, San Francisco, CA) and cDNA orientation was verified by Sanger sequencing. Digoxigenin-labeled sense and antisense RNA probes were prepared as previously described (DIG RNA labeling kit; Roche, Indianapolis, IN) [20]. In-situ hybridization was performed according to standard protocols on wildtype, whole zebrafish embryos at 48 h post fertilization (hpf) [20].
CRISPR/Cas9 targeting of ints1 in Danio rerio
We chose not to use antisense morpholinos due to the possibility of non-specific effects from this methodology on eye size and development. For CRISPR/Cas9, we used a modification of a previously described method [21]. We designed a CRISPR small guide (sg)RNA to target exon 8 of ints1 that is upstream of an annotated domain of unknown function (DUF3677) spanning exons 8–10 of this gene (Table S2). We co-injected sgRNA and Cas9 protein into wildtype zebrafish eggs at the one cell stage and used Sanger sequencing at 24–48 hpf to assess for mutagenesis in batches of 10–20 larvae (for primers, see Table S3). FO larvae with insertion/deletion (indel) variants were raised and fin-clipped at 6–8 weeks of age in batches of 10–20 larvae for further genotyping with Sanger sequencing to determine the exact indel variants present. We selected founders with small indels introducing a frameshift or eliminating a splice site in ints1, thus predicting premature protein truncation and nonsense-mediated decay (Table S4). These founders were outcrossed to wildtype Danio rerio and the F1 progeny were raised and genotyped to identify heterozygous fish with germline transmission of an indel variant. F1 fish that were heterozygous for indel variants in ints1 were incrossed to obtain homozygous or compound heterozygous F2 larvae that had biallelic ints1 variants predicting loss of function.
Larvae obtained from incrosses of heterozygous fish with ints1 indel variants were examined for eye size and lens abnormalities at 3 to 7 dpf using light microscopy. Tails of larvae from incrosses were genotyped by Sanger sequencing and the corresponding heads of larvae with biallelic ints1 variants were then sectioned at 6 dpf so that the eyes could be studied in more detail relating genotype to hematoxylin and eosin staining and immunohistochemistry. For hematoxylin and eosin staining, slides were immersed in hematoxylin (VWR International, Brisbane, CA) for three minutes, washed with tap water, dipped twice in Scott’s water and then placed in 75% ethanol for 15 s. Slides were stained with eosin (VWR International, Brisbane, CA) for 15 s and then dipped in 75% ethanol prior to mounting. For immunohistochemistry, larval heads were fixed in 4% paraformaldehyde, washed with phosphate buffered saline (PBS) and placed in 25% (w:v) sucrose followed by 35% (w:v) sucrose. Cryosections were made according to standard techniques. Sections were dried overnight and stored at −80 °C. For staining, sections were washed three times in PBS with 0.1% Tween 20 (PBST) and incubated in blocking buffer containing 4% normal goat serum, 2% normal sheep serum and 0.2% Tween (all v-v) in PBS for 4 h at 4 °C. Slides were incubated with a 1 in 500 dilution of zl-1 primary antibody (Zebrafish International Resource Center) in blocking buffer overnight at 4 °C, washed in PBST, and then incubated overnight with a 1:500 dilution of antimouse IgG labeled with Texas red (M32017; Invitrogen, Carlsbad, CA) in blocking buffer as a secondary antibody at 4 °C. Sections were mounted with Abcam fluoroshield mounting medium containing DAPI (Abcam, Burlingame, CA) as a counterstain and imaged with a fluorescent microscope.
Lastly, in view of the lethality of the mouse model of Ints1 loss of function at the stage of gastrulation, we used light microscopy to photograph the progeny of incrossed, heterozygous F1 adult fish at 1 h, 2 h, 4 h, and 24 hpf. After photographs were taken, DNA was extracted and the larvae were genotyped by Sanger sequencing, so that progress through gastrulation and eye morphology could be correlated to ints1 genotype for wildtype, heterozygous, and compound heterozygous larvae.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
We used qRT-PCR to determine if there was altered expression of other INT complex genes between wildtype larvae and larvae from an incross of heterozygous, ints1 indel fish. 1 ng cDNA was amplified using gene-specific primers (Table S5) at final concentrations of 2.5 μM and Universal SybrGreen mastermix containing Rox (Roche, Pleasanton, CA) at a final concentration of 2.5 μM. Reactions were run on a StepOne cycler and analyzed with StepOne™ Software and ExpressionSuite software (Thermo Fisher Scientific, San Francisco, CA) according to the ΔΔCt method. We used gapdh (NM_001115114.1) or eef1a1l1(NM_131263.1) as internal control genes. All experiments were performed on three independent samples from wildtype larvae and larvae resulting from an incross of heterozygous, ints1 indel fish. Each reaction was run in triplicate.
Results
Case reports
Clinical details from the eight cases are summarized in Table 1, Figs. 1–3, and Fig. S1. Further clinical information is available in Supplemental Data. Similar to the previously reported patients with biallelic INTS1 variants [7], all patients demonstrated hypotonia and gait disturbances, with several patients taking independent steps at or after 2 years of age (patients 1 and 3). The mobility of the second patient deteriorated at 5 years of age after he was independently ambulant at 14 months of age, but he retained the ability to walk with assistance. All patients had significant developmental delays, with absent or minimal speech. Three were given a diagnosis of autism (patients 2, 3, and 4) and one carried a diagnosis of autism spectrum disorder (patient 5).
Biallelic variants in INTS1 are associated with facial dysmorphic features. a Front view of patient 2, the younger sibling from family 1, demonstrating a broad nasal tip, downturned corners of the mouth, low-set ears and a pectus deformity. b Front view of patient 1, the older sibling from family 1, demonstrating downturned corners of the mouth and low-set ears. c Front view of patient 5, demonstrating frontal bossing, upslanting palpebral fissures and a wide nasal tip. d Left foot from patient 1, demonstrating overlapping toes with clinodactyly. e Oral region of patient 1, demonstrating a wide diastema between the upper two incisor teeth. f Chest and abdomen of patient 1, demonstrating a pectus deformity and G-tube insertion site
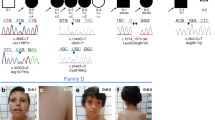
Phenotype findings in patients homozygous for p.[(Ser1784*)] in INTS1 Upper row: patient 6 at the age of 6 years; Middle row: patient 7 at the age of 19 years; Lower row: patient 8 at the age of 2 years (first two pictures) and 6 years (3rd picture). All patients share the same homozygous p.[(Ser1784*)];[(Ser1784*)] variant, but are unrelated
All patients developed cataracts between one and 3 years of age, as found in the first three reported patients, thus confirming the ocular involvement in patients with INTS1 variants [7]. Three patients were diagnosed with myopia and the siblings from the first family had additional ophthalmological findings of microphthalmia, microcornea and colobomas (Table S6).
Four of the new patients shared dysmorphic features similar to those noted in the first three reported cases. Features described in more than half of the eight individuals with biallelic INTS1 variants include frontal bossing, small palpebral fissures that are slanted, wide-spaced eyes, full cheeks, a broad nasal bridge and a broad or bulbous nasal tip, low-set ears, down-turned corners of the mouth and a small chin (Table 1). Additional distinctive findings in the majority included a wide diastema between the upper incisor teeth, short neck, pectus deformities and broad halluces with overlapping toes (Table 1; Fig. 2). Other craniofacial features that were less commonly observed included brachycephaly (previous patients were reported to have dolichocephaly) [7], epicanthus and dysplastic ears (Fig. 1). Overall, the facial features and skeletal changes showed striking similarities to those of previously reported patients (Fig. 3) [7].
Brain magnetic resonance imaging (MRI) findings are summarized in Table S7 and ranged from normal (patients 7 and 8) to demonstrating non-specific abnormalities, with small anterior and hippocampal commissures with mildly reduced volume of the splenium of the corpus callosum and adenohypophysis in one child (patient 2) and hypoplasia of the inferior vermis of the cerebellum (patient 8; Table S7). Even among the three patients with the identical, homozygous nonsense variant, p.[(Ser1784*)], brain MRI abnormalities were not consistent, with only patient 6 showing hypoplasia of the inferior vermis of the cerebellum (Fig. S1). Patient 6 has also been treated for epilepsy.
The two siblings from the first family have had heights on the 5th and 1st centiles and all patients with the p.(Ser1784*) variant have significant growth retardation. The first sibling pair also demonstrated abnormal thumbs; one brother had both a multicystic, dysplastic kidney and mild prolapse of the anterior leaflet of the mitral valve. Although the second pair of siblings did not have renal ultrasound scans or echocardiograms, patient 5 had renal pelviectasis and patient 7 showed a horseshoe kidney. Patient 2 had distinctive skeletal findings, with loss of the thoracic kyphosis and lumbar lordosis, levocurvature of the lumbar spine, mild coxa valga, shortened tibiae and a leg length discrepancy. His hand radiographs demonstrated hypoplasia of the first metacarpal and phalanges of the right hand with absence of the trapezium. Congenital heart disease was only observed in patients 7 and 8 [7].
Exome sequencing
Exome sequencing showed that all patients had biallelic variants in INTS1 (Table 2). The first sib pair were each compound heterozygous for a missense variant and a frameshift variant, c.[229C>T];[5398dup], predicting p.[(Arg77Cys)];[(Arg1800Profs*20)] in INTS1 (NM_001080453.2). The frameshift variant was predicted to be deleterious, but the missense variant was present in 0.025% of individuals in a control database (ExAC Browser, Table 2) and has been seen in the homozygous state in one individual (http://gnomad.broadinstitute.org/gene/ENSG00000164880; Table 2), suggesting that it is likely to be a hypomorphic variant or associated with incomplete penetrance. It is possible that p.(Arg77Cys) may not lead to clinical manifestations when present in the homozygous state, but that it could have sufficient effect on the protein to cause disease when inherited with a more severe sequence variant on the second allele. Protein structure data for this variant is unavailable. It is also noteworthy that the homozygous individual in gnomAD is of Hispanic ethnicity, the same ethnicity as the siblings with this variant, and that the frequency of the variant is highest in the Hispanic population at 0.13% (http://gnomad.broadinstitute.org/variant/7–1542657-G-A). However, the clinical findings in these patients are similar to those in others with INTS1 variants and we still consider this the causative gene. Both of the siblings from the second family were homozygous for a missense variant c.[5621C>T];[5621C>T], predicting p.[(Pro1874Leu)];[(Pro1874Leu)] that was also predicted to be deleterious and was absent from human genetic variation databases (Table 2). The fifth child also had biallelic INTS1 variants—c.[5290del];[6491T>C], predicting p.[(Leu1764Cysfs*16)];[(Leu2164Pro)]. This child’s healthy brother carried the c.[6491T>C] variant only. Splicing prediction programs did not demonstrate altered splicing with any of the variants (data not shown). Several of the variants would be expected to result in loss of function of the protein, consistent with the reported homozygosity for a nonsense variant in INTS1 in the first three reported patients [7]. Modeling studies of the C-terminal end of the protein showed that the missense variants p.(Pro1874Leu) and p.(Leu2164Pro) may interfere with helix folding (Fig. S2). Specifically, the p.(Pro1874Leu) variant is likely to disrupt the helix because it is located immediately after the helix in our model. The substitution of proline with leucine, which has a high helix propensity [22] cancels the helix break and disrupts the protein fold. The p.(Leu2164Pro) variant is expected to have an opposite effect. Leu2164 is located in the middle of the helix; the change to Pro breaks the helix. Protein structure data to model the p.(Arg77Cys) variant was not available.
In-situ hybridization
In-situ hybridization demonstrated expression of ints1 in the developing zebrafish eye and brain at 48 hpf (Fig. S3). This is consistent with the small eyes observed in the awb medaka model of ints1 loss of function and the eye involvement observed in the patients reported here.
CRISPR/Cas9 model of ints1 loss of function
We successfully targeted the single, ints1 transcript in Danio rerio and generated fish that were heterozygous for two separate indel variants predicting loss of function—c.1020delinsGG, predicting p.(Met341Aspfs*19), and c.1020_1021insG, predicting p.(Met341Hisfs*18) (XM_003198152.4; Table S4; Fig. S4). After incrossing a female that was heterozygous for c.1020_1021insG in ints1 and a male that was heterozygous for ints1 c.1020delinsGG, genotyping at 6 dpf revealed that inheritance of the variants was present in expected Mendelian ratios, with 5/30 (16.7%) larvae wildtype for ints1, 8/30 (26.7%) larvae heterozygous for the c.1020insG variant in ints1, 10/30 (33.3%) larvae heterozygous for the c.1020delinsGG variant in ints1, and 7/30 (23.3%) larvae that were compound heterozygous for both the c.1020insG and c.1020delinsGG variants in ints1. We did not detect any external abnormalities in the larvae that were compound heterozygous for the c.1020insG and c.1020delinsGG variants in ints1 at up to 6 dpf with light microscopy and development through gastrulation appeared to progress typically (Figs. S5 and S6).
Sectioning and staining of the eyes of larvae at 6 dpf showed grossly normal eye formation in larvae with biallelic ints1 variants (Fig. 4g–i) compared to wildtype (Fig. 4a–c) and heterozygous larvae (Fig. 4d–f). However, there was a subtle increase in lens thickness that was observed in the compound heterozygous larvae (Fig. 4l, m) compared to wildtype larvae (Fig. 4j) that was also detectable as an increase in staining with zl-1 antibody that labels lens fibers.
Compound heterozygous ints1 indel mutant larvae show abnormal lens morphology at 6 days post fertilization. Immunohistochemistry performed with zl-1 antibody and visualized with Texas Red demonstrates a thickened and less transparent lens appearance with reduced zl-1 staining in ints1 compound heterozygous indel larvae compared to wildtype controls. a–c Larva wildtype for ints1 (XM_003198152.4), demonstrating normal lens formation. d–f Larva heterozygous for ints1 targeted allele. g–i Larva that are compound heterozygous for variants c.[1020delinsGG];[c.1020_1021insG], predicting p.[(Met341Aspfs*19)];[(Met341Hisfs*18)] in ints1. The outer layer of the lenses is thickened and exhibits increased staining with zl-1 antibody that binds to lens fibers. Staining with hematoxylin and eosin revealed mildly thickened lenses in larvae with biallelic ints1 variants. j. Larva wildtype for ints1. k–l Larva heterozygous for ints1 targeted allele. m–n Larvae that are compound heterozygous for c.[1020delinsGG];[c.1020_1021insG], predicting p.[(Met341Aspfs*19)];[(Met341Hisfs*18)] in ints1, showing mild thickening of the outer layer of the ocular lens
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
We used qRT-PCR at 3 dpf to examine the effects of reduced ints1 expression on genes from the INT complex on RNA obtained from wildtype larvae and RNA obtained from an incross of fish that were heterozygous for an ints1 indel variant. A limitation of this experiment was that the incross would be expected to result in only 25% of larvae that were compound heterozygous for both ints1 indel variants. We were unable to phenotype the larvae from the heterozygous indel incross using a phenotypic marker that was visible by light microscopy due to their typical external appearance at 3 dpf; we could also not genotype and extract RNA from single larvae at this developmental stage due to larval size. We performed qRT-PCR for ints1, ints3 (NM_001044932.1), ints8 (NM_001020756.2), ints9 (NM_001077270.1), ints11 (NM_001020621.1) and ints12 (NM_001002161.3) in control and incrossed larvae. The results showed significant downregulation of ints1 as expected (p = 0.027; Fig. S7), but also downregulation of ints9 (p = 0.032), ints11 (p = 0.069), ints8 (p = 0.086), ints3 (p = 0.163), and ints12 (p = 0.176). These findings suggest that deficiency of ints1 could lead to dysregulation of additional genes in the INT complex. Interestingly, the results also suggested upregulation of ints8, although this was not statistically significant (p = 0.086) (Fig. S7).
Discussion
Herein we present two sibling pairs and a fifth patient with clinical features of autism and severe speech delay, hypotonia and gait disturbances, cataracts and dysmorphic craniofacial findings that included frontal bossing, small and slanted palpebral fissures, wide-spaced eyes, a broad nasal bridge with a broad or bulbous nasal tip, low-set ears, down-turned corners of the mouth, a wide diastema between the upper incisor teeth and a small chin. Other distinctive findings included stature at or below the 5th centile, short neck, pectus deformities and abnormal thumbs and toes. All patients had biallelic variants in INTS1. The overlap of clinical findings with previously reported patients who were homozygous for the p.(Ser1874*) variant in INTS1 implicate this gene in the pathogenesis of the phenotypic features of these patients. Our data suggest that the clinical findings associated with INTS1 variants may also include the eye findings of microphthalmia, microcornea and colobomas.
To date, there has been no clear phenotype-genotype correlation for INTS1 variants and the first sib pair (patients 1 and 2) that have a missense variant of unclear pathogenicity, p.(Arg77Cys), share similar clinical findings to the three previously described patients (patients 6, 7, and 8) who are homozygous for the nonsense variant, p.(Ser1874*) (Table 2). However, the nonsense variant is also predicted to create a new splice donor site that is stronger than the endogenous splice donor site (Fig. S8) that would delete 30 nucleotides if used to result in an in-frame deletion, p.(Ser1784_Ser1793del), thus this variant may also be a hypomorphic allele (data not shown). The missense variants in patients 3 and 4, p.(Pro1874Leu), and in patient 5, (p.Leu2164Pro), were predicted to interfere with helix folding (Fig. S2) and thus may affect the ability of INTS1 to bind to other members of the integrator complex.
We used whole mount in-situ hybridization to show that the ints1 gene is expressed in the developing Danio rerio eye (Fig. S3) and targeted the ints1 gene using CRISPR/Cas9 in zebrafish to produce larvae with biallelic indel variants affecting ints1 that predicted loss of function for this gene. Examination of the external appearance of larvae with biallelic ints1 variants by light microscopy revealed the fish passed through gastrulation normally with no obvious differences in external appearance (Figs. S5 and S6). However, staining of sections of the eyes with hematoxylin and eosin at 6 dpf showed slightly thickened lenses in the larvae that were genotyped to show biallelic ints1 variants (Fig. 4). This finding was also present with immunohistochemistry using zl-1 antibody that is expressed in lens fibers, when a broader band of zl-1 staining in larvae that were compound heterozygous for ints1 indels was observed compared to wildtype larvae at 6 dpf. The increased staining is in accordance with early markers of adult onset cataracts that include deeper staining of fiber cells, scattered cortical opacities and fiber cell disruption [23]. These findings, together with the cataracts noted in all patients with biallelic INTS1 variants reported to date, strongly support the involvement of this gene in eye and lens development. However, the relationship between the lens thickening and the development of cataracts is not known and the zebrafish model has limited utility to examine the other features found in patients with biallelic INTS1 variants, such as developmental delays and dysmorphic features.
In the mouse, the Ints1 protein has 2222 amino acids and predominantly localizes to the nucleus, with marginal cytoplasmic expression [5]. Homozygous null embryos (−/−) for Ints1 demonstrated abnormal blastocyst morphology as early as embryonic day (E)3.5 [5]. In a mixed background, depletion of Ints1 in homozygous null embryos is lethal at the morula stage [5]. After 8–10 backcrosses with the ICR strain, embryos were produced in the expected Mendelian ratios, but the blastoceles of homozygous null embryos did not expand and hatching from the zona pellucida and implantation did not occur, demonstrating developmental arrest. In vitro culture of homozygous null embryos showed increased apoptosis of the inner cell matrix and caspase-3 and caspase-7 activation [5]. Similar findings with abnormal gastrulation were not observed in our Danio rerio model of reduced ints1 function, with compound heterozygous larvae passing through gastrulation with typical morphology, suggesting functional redundancy for this gene in zebrafish compared to mouse.
To date, there are few details on how ints1 interacts with the other integrator complex subunits. INTS1 has been shown to interact with INTS12 which is involved in small nuclear (sn)RNA splicing and regulation of translation [3, 24]. In the mouse, Ints1 may directly bind to Ints2 [5, 25] and these proteins act as scaffolding for the assembly of the entire integrator complex. Disruption to Ints1 could thus compromise the function of the entire integrator complex [5]. Ints11 is a predominantly nuclear protein with sequence similarity to the putative β-lactamase catalytic domain (RNA-specific endonuclease) [26]. INTS9 and INTS11 are paralogues of the cleavage and polyadenylation specificity factor subunits CPSF100 and CPSF73, respectively, which form the endonuclease factor responsible for cleavage of pre-mRNA at the polyA site [7]. Knockdown of INTS1 and INTS11 specifically abrogated the response of immediate early genes (IEGs) to epidermal growth factor (EGF) stimulation in human cells [27].
We performed qRT-PCR to determine if the genes for the remaining INT subunits were dysregulated in the larvae resulting from an incross of ints1 heterozygous indel fish compared to an incross of wildtype fish. We found significant downregulation of ints1 as expected, but also downregulation of ints9, ints11, ints3, and ints12, suggesting that reduced ints1 expression could affect the expression of other INT genes. In contrast, ints8 may be upregulated in the incrossed larvae (Fig. S7). This is intriguing in view of the recent demonstration of biallelic variants in INTS8 in patients with intellectual disability and neuronal migration defects [7]. In HEK293T cells stably expressing wildtype or mutant c.2917_2925(ΔEVL) INTS8, FLAG-INTS8-ΔEVL eluates contained nearly undetectable amounts of INTS1, INTS12 and RPB1, reduced levels of associated INTS4, INTS9, and INTS11, but similar levels of INTS5 and INTS3, compared to wildtype FLAG-INTS8, and thus the ΔEVL variant may impair the ability of INTS8 to associate with members of the integrator complex, including INTS1 [7].
In summary, we present five patients with biallelic variants in INTS1 and provide further clinical details for three previously reported patients. All patients demonstrated severely compromised expressive speech, an abnormal gait, cataracts, and distinctive craniofacial dysmorphic features that included frontal bossing, wide-spaced eyes, a broad nasal bridge and nasal tip and downturned corners of the mouth. We were successfully able to target the zebrafish ints1 gene using CRISPR/Cas9. Larvae that were compound heterozygotes for deleterious ints1 variants showed no obvious external defects, but examination of their eyes revealed abnormal lens histology with thickening of the lenses and increased staining for zl-1 in lens fibers at 6 dpf. Our work provides additional data on the clinical features associated with biallelic INTS1 variants in humans and confirms abnormal lens development in a zebrafish model of reduced ints1 expression.
References
Baillat D, Wagner EJ. Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci. 2015;40:257–64.
Rienzo M, Casamassimi A. Integrator complex and transcription regulation: recent findings and pathophysiology. Biochim Biophys Acta. 2016;1859:1269–80.
Chen J, Waltenspiel B, Warren WD, et al. Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J Biol Chem. 2013;288:4867–77.
Chen J, Wagner EJ. snRNA 3’ end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–7.
Hata T, Nakayama M. Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim Biophys Acta. 2007;1773:1039–51.
Iwanami N, Okada M, Hoa V, et al. Ethylnitrosurea-induced thymus-defective mutants identify roles of KIAA1440, TRRAP, and SKIV2L2 in teleost organ development. Eur J Immunol. 2009;39:2606–16.
Oegema R, Baillat D, Schot R, et al. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13:e1006809.
Depristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491498.
Mills RE, Pittard WS, Mullaney JM, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21:830839
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucl Acids Res. 2010;38:e164.
Cingolani P. SnpEff: variant effect prediction. http://snpeff.sourceforge.net (2012).
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Schwarz JM, Rödelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6.
Teer JK, Green ED, Mullikin JC, et al. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600.
Hartley T, Wagner JD, Warman-Chardon J, et al. Whole-exome sequencing is a valuable diagnostic tool for inherited peripheral neuropathies: Outcomes from a cohort of 50 families. Clin Genet. 2018;93:301–9.
Tanaka AJ, Cho MT, Millan F, et al. Mutations in SPATA5 are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am J Hum Genet. 2015;97:457–64.
Zimmerman L, Stephens A, Nam SZ, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHPRED server at its core. J Mol Biol. 2017;S0022-2836:30587–9.
Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815.
Chao R, Nevin L, Agarwal P, et al. A male with unilateral microphthalmia reveals a role for TMX3 in eye development. PLoS ONE. 2010;5:e10565.
Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–5.
Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–7.
Al-Ghoul KJ, Lane CW, Taylor VL, et al. Distribution and type of morphological damage in human nuclear age-related cataracts. Exp Eye Res. 1996;62:237–51.
Kheirallah AK, de Moor CH, Faiz A, et al. Lung function associated gene Integrator Complex subunit 12 regulates protein synthesis pathways. BMC Genom. 2017;18:248.
Nagase T, Ishikawa K, Kikuno R, et al. Prediction of the coding sequence of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:337–45.
Dominski Z, Yang XC, Purdy M, et al. Differences and similarities between Drosophila and mammalian 3’end processing of histone pre-mRNAs. RNA. 2005;11:1835–47.
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009;25:2078–9.
Acknowledgements
We are grateful to the families for their participation. This work was supported by National Eye Institute, National Institutes of Health [grant number R21EY022779–01 to A.S.]; Erasmus Medical Center [grant Mrace #104673 to G.M.]. This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Genomics Institute, Ontario Research Fund, Génome Québec, and Children’s Hospital of Eastern Ontario Foundation. We also wish to acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada. R.E.L. and A.M.I. would like to thank Mary Anderson, Dr. Francois Bernier and Dr. Jillian Parboosingh for clinical and technical support, as well as helpful discussions.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
R.E.S. is an employee of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. The remaining authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Krall, M., Htun, S., Schnur, R.E. et al. Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur J Hum Genet 27, 582–593 (2019). https://doi.org/10.1038/s41431-018-0298-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0298-9
This article is cited by
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex
Nature Reviews Molecular Cell Biology (2023)
-
BRAT1 links Integrator and defective RNA processing with neurodegeneration
Nature Communications (2022)
-
Inositol hexakisphosphate is required for Integrator function
Nature Communications (2022)
-
Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder
Nature Communications (2021)
-
Biallelic INTS1 Mutations Cause a Rare Neurodevelopmental Disorder in Two Chinese Siblings
Journal of Molecular Neuroscience (2020)