Abstract
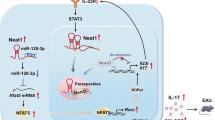
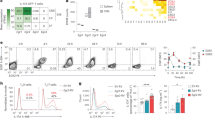
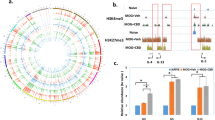
Circular RNAs (circRNAs) regulate gene expression and participate in various biological and pathological processes. However, little is known about the effects of specific circRNAs on T helper cell 17 (Th17) differentiation and related autoimmune diseases, such as multiple sclerosis (MS). Here, using transcriptome microarray analysis at different stages of experimental autoimmune encephalomyelitis (EAE), we identified the EAE progression-related circINPP4B, which showed upregulated expression in Th17 cells from mice with EAE and during Th17 differentiation in vitro. Silencing of circINPP4B inhibited Th17 differentiation and alleviated EAE, characterized by less demyelination and Th17 infiltration in the spinal cord. Mechanistically, circINPP4B served as a sponge that directly targeted miR-30a to regulate Th17 differentiation. Furthermore, circINPP4B levels were associated with the developing phases of clinical relapsing-remitting MS patients. Our results indicate that circINPP4B plays an important role in promoting Th17 differentiation and progression of EAE by targeting miR-30a, which provides a potential diagnostic and therapeutic target for Th17-mediated MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data are available in the main text or the supplementary materials.
References
Dolati S, Babaloo Z, Jadidi-Niaragh F, Ayromlou H, Sadreddini S, Yousefi M. Multiple sclerosis: therapeutic applications of advancing drug delivery systems. Biomed Pharmacother. 2017;86:343–53.
Popescu BF, Lucchinetti CF. Pathology of demyelinating diseases. Annu Rev Pathol. 2012;7:185–217.
Duffy SS, Lees JG, Moalem-Taylor G. The contribution of immune and glial cell types in experimental autoimmune encephalomyelitis and multiple sclerosis. Mult Scler Int. 2014;2014:285245.
Ramaglia V, Rojas O, Naouar I, Gommerman JL. The Ins and Outs of Central Nervous System Inflammation-Lessons Learned from Multiple Sclerosis. Annu Rev Immunol. 2021;39:199–226.
Matsui M. [Immunology for understanding the pathogenesis of multiple sclerosis]. Rinsho Shinkeigaku. 2013;53:898–901.
Moser T, Akgün K, Proschmann U, Sellner J, Ziemssen T. The role of TH17 cells in multiple sclerosis: therapeutic implications. Autoimmun Rev. 2020;19:102647.
Qu X, Han J, Zhang Y, Wang X, Fan H, Hua F, et al. TLR4-RelA-miR-30a signal pathway regulates Th17 differentiation during experimental autoimmune encephalomyelitis development. J Neuroinflammation. 2019;16:183.
Brucklacher-Waldert V, Stuerner K, Kolster M, Wolthausen J, Tolosa E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain: J Neurol. 2009;132:3329–41.
Paradowska A, Maślińiski W, Grzybowska-Kowalczyk A, Łacki J. The function of interleukin 17 in the pathogenesis of rheumatoid arthritis. Arch Immunol Ther Exp (Warsz.) 2007;55:329–34.
Quinn JL, Kumar G, Agasing A, Ko RM, Axtell RC. Role of TFH Cells in Promoting T Helper 17-Induced Neuroinflammation. Front Immunol. 2018;9:382.
Qu X, Han J, Zhang Y, Wang Y, Zhou J, Fan H, et al. MiR-384 Regulates the Th17/Treg Ratio during Experimental Autoimmune Encephalomyelitis Pathogenesis. Front Cell Neurosci. 2017;11:88.
Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, et al. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci. 2014;11:810–8.
Dolati S, Marofi F, Babaloo Z, Aghebati-Maleki L, Roshangar L, Ahmadi M, et al. Dysregulated Network of miRNAs Involved in the Pathogenesis of Multiple Sclerosis. Biomed Pharmacother. 2018;104:280–90.
Qu X, Zhou J, Wang T, Han J, Ma L, Yu H, et al. MiR-30a inhibits Th17 differentiation and demyelination of EAE mice by targeting the IL-21R. Brain Behav Immun. 2016;57:193–9.
Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44.
Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13:669–80.
Zhang C, Hu J, Yu Y. CircRNA Is a Rising Star in Researches of Ocular Diseases. Front Cell Dev Biol. 2020;8:850.
Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661–80.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
D’Ambra E, Capauto D, Morlando M. Exploring the Regulatory Role of Circular RNAs in Neurodegenerative Disorders. Int J Mol Sci. 2019;20:5477.
Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166.
Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18:133.
Fischer MT, Sharma R, Lim JL, Haider L, Frischer JM, Drexhage J, et al. NADPH oxidase expression in active multiple sclerosis lesions in relation to oxidative tissue damage and mitochondrial injury. Brain:J Neurol. 2012;135:886–99.
Casella G, Rasouli J, Boehm A, Zhang W, Xiao D, Ishikawa L, et al. Oligodendrocyte-derived extracellular vesicles as antigen-specific therapy for autoimmune neuroinflammation in mice. Sci Transl Med. 2020;12:eaba0599.
Ishihara A, Ishihara J, Watkins EA, Tremain AC, Nguyen M, Solanki A, et al. Prolonged residence of an albumin-IL-4 fusion protein in secondary lymphoid organs ameliorates experimental autoimmune encephalomyelitis. Nat Biomed Eng. 2021;5:387–98.
Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19.
Kumar L, Shamsuzzama, Haque R, Baghel T, Nazir A. Circular RNAs: the Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol Neurobiol. 2017;54:7224–34.
Shen L, Bai Y, Han B, Yao H. Non-coding RNA and neuroinflammation: implications for the therapy of stroke. Stroke Vasc Neurol. 2019;4:96–8.
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865–80. e821
Iparraguirre L, Muñoz-Culla M, Prada-Luengo I, Castillo-Triviño T, Olascoaga J, Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum Mol Genet. 2017;26:3564–72.
Paraboschi EM, Cardamone G, Soldà G, Duga S, Asselta R. Interpreting Non-coding Genetic Variation in Multiple Sclerosis Genome-Wide Associated Regions. Front Genet. 2018;9:647.
Zurawska A, Mycko MP, Selmaj KW. Dominant role of circular RNA in miRNA circuit in multiple sclerosis. Mult Scler J. 2018;24:P1074.
Zurawska A, Mycko MP, Selmaj KW. Circular RNAs as a novel layer of regulatory mechanism in multiple sclerosis. J Neuroimmunol. 2019;334:576971.
Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, et al. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORgammat and STAT3. J Immunol. 2014;192:5599–609.
Mycko MP, Cichalewska M, Cwiklinska H, Selmaj KW. miR-155-3p Drives the Development of Autoimmune Demyelination by Regulation of Heat Shock Protein 40. J Neurosci: Off J Soc Neurosci. 2015;35:16504–15.
Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9.
Wu R, He Q, Chen H, Xu M, Zhao N, Xiao Y, et al. MicroRNA-448 promotes multiple sclerosis development through induction of Th17 response through targeting protein tyrosine phosphatase non-receptor type 2 (PTPN2). Biochem Biophys Res Commun. 2017;486:759–66.
Liu R, Ma X, Chen L, Yang Y, Zeng Y, Gao J, et al. MicroRNA-15b Suppresses Th17 Differentiation and Is Associated with Pathogenesis of Multiple Sclerosis by Targeting O-GlcNAc Transferase. J Immunol. 2017;198:2626–39.
Liu R, Li Y, Zhou H, Wang H, Liu D, Wang H, et al. OIP5-AS1 facilitates Th17 differentiation and EAE severity by targeting miR-140-5p to regulate RhoA/ROCK2 signaling pathway. Life Sci. 119108 (2021).
Guan D, Li Y, Cui Y, Guo Y, Dong N, Li G, et al. Down-regulated miR-374c and Hsp70 promote Th17 cell differentiation by inducing Fas expression in experimental autoimmune encephalomyelitis. Int J Biol Macromol. 2020;154:1158–65.
Zhao M, Sun D, Guan Y, Wang Z, Sang D, Liu M, et al. Disulfiram and Diphenhydramine Hydrochloride Upregulate miR-30a to Suppress IL-17-Associated Autoimmune Inflammation. J Neurosci: Off J Soc Neurosci. 2016;36:9253–66.
Gao Y, Han D, Feng J. MicroRNA in multiple sclerosis. Clin Chim Acta; Int J Clin Chem. 2021;516:92–9.
Schiavinato J, Haddad R, Saldanha-Araujo F, Baiochi J, Araujo AG, Santos Scheucher P, et al. TGF-beta/atRA-induced Tregs express a selected set of microRNAs involved in the repression of transcripts related to Th17 differentiation. Sci Rep. 2017;7:3627.
Yang Q, Pan W, Qian L. Identification of the miRNA-mRNA regulatory network in multiple sclerosis. Neurol Res. 2017;39:142–51.
Quintana E, Ortega FJ, Robles-Cedeño R, Villar ML, Buxó M, Mercader JM, et al. miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult Scler. 2017;23:1716–26.
Teymoori-Rad M, Mozhgani SH, Zarei-Ghobadi M, Sahraian MA, Nejati A, Amiri MM, et al. Integrational analysis of miRNAs data sets as a plausible missing linker between Epstein-Barr virus and vitamin D in relapsing remitting MS patients. Gene. 2019;689:1–10.
Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology. 1983;33:1444–52.
Acknowledgements
We thank the Affiliated Hospital of Xuzhou Medical University for the clinical samples. We thank the Majorbio Cloud Platform for microarray data analysis. This research was supported by the National Natural Science Foundation of China (81771337, 81271345), The National Key R&D Program of China (2017YFA0104202), The Natural Science Foundation of Jiangsu Province (BK20161174), the 333 Project of Jiangsu Province, The Xuzhou Basic Research Science and Technology Project (KC19059) and Xuzhou Medical University Scientific Research Fund for Talents.
Author information
Authors and Affiliations
Contributions
JJH: designed the research, performed the experiments, analyzed the data, and wrote the paper. WZ: performed the experiments and analyzed the data. WHF: performed the experiments and analyzed the data. FXD: performed the experiments and analyzed the data. FH: designed the research and analyzed the data. RQY: designed the research and analyzed the data. XBQ designed the research, performed the experiments, analyzed the data, and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, J., Zhuang, W., Feng, W. et al. The circular RNA circINPP4B acts as a sponge of miR-30a to regulate Th17 cell differentiation during progression of experimental autoimmune encephalomyelitis. Cell Mol Immunol 18, 2177–2187 (2021). https://doi.org/10.1038/s41423-021-00748-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-021-00748-y
Keywords
This article is cited by
-
Brain alarm by self-extracellular nucleic acids: from neuroinflammation to neurodegeneration
Journal of Biomedical Science (2023)
-
Circ_0000479 promotes proliferation, invasion, migration and inflammation and inhibits apoptosis of rheumatoid arthritis fibroblast-like synoviocytes via miR-766/FKBP5 axis
Journal of Orthopaedic Surgery and Research (2023)
-
MiR-30a-centered molecular crosstalk regulates Th17 differentiation
Cellular & Molecular Immunology (2022)