Abstract
Glioblastoma is one of the most lethal malignant cancers, displaying striking intratumor heterogeneity, with glioblastoma stem cells (GSCs) contributing to tumorigenesis and therapeutic resistance. Pharmacologic modulators of ubiquitin ligases and deubiquitinases are under development for cancer and other diseases. Here, we performed parallel in vitro and in vivo CRISPR/Cas9 knockout screens targeting human ubiquitin E3 ligases and deubiquitinases, revealing the E3 ligase RBBP6 as an essential factor for GSC maintenance. Targeting RBBP6 inhibited GSC proliferation and tumor initiation. Mechanistically, RBBP6 mediated K63-linked ubiquitination of Cleavage and Polyadenylation Specific Factor 3 (CPSF3), which stabilized CPSF3 to regulate alternative polyadenylation events. RBBP6 depletion induced shortening of the 3’UTRs of MYC competing-endogenous RNAs to release miR-590-3p from shortened UTRs, thereby decreasing MYC expression. Targeting CPSF3 with a small molecular inhibitor (JTE-607) reduces GSC viability and inhibits in vivo tumor growth. Collectively, RBBP6 maintains high MYC expression in GSCs through regulation of CPSF3-dependent alternative polyadenylation, providing a potential therapeutic paradigm for glioblastoma.
Similar content being viewed by others
Introduction
Glioblastoma is the most prevalent and lethal primary brain tumor, with a median survival time of less than two years. Current standard-of-care for glioblastoma includes maximal surgical resection, combined radiotherapy and chemotherapy, followed by adjuvant chemotherapy, offering only palliation1. Glioblastoma displays intratumoral heterogeneity with self-renewing glioblastoma stem cells (GSCs) at the hierarchical apex2. GSCs are functionally characterized by their capacities of self-renewal and tumor initiation with additional contributions to tumor angiogenesis, radio-resistance, and chemoresistance3.
Glioblastomas were one of the first cancers to under comprehensive genetic analysis, which revealed frequent alterations leading to dysregulated signaling, including those involving epigenetic alterations, transcriptional regulators, and posttranscriptional regulation3,4,5. However, precision medicine based on genetic lesions has had a limited impact on the clinical management of glioblastoma patients, suggesting that alternative molecular targets may offer therapeutic benefits. Ubiquitination is one of the most common post-translational modifications that regulate the fate and/or function of substrate proteins. The ubiquitin‒proteasome pathway maintains the homeostasis of most proteins6,7. Ubiquitin molecules are added to target proteins through a catalytic cascade involving E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes, which can be reversed by deubiquitinating enzymes (DUBs)8. Prior studies have linked ubiquitin pathways to glioblastoma growth, in general, and GSC maintenance, specifically. For example, glioblastomas can overexpress USP15, which binds to the SMAD7-SMAD specific E3 ubiquitin protein ligase 2 (SMURF2) complex to deubiquitinate and stabilize the type I TGF-β receptor (TβR-I), enhancing TGF-β signaling9. In GSCs, USP9x abrogates aldehyde dehydrogenase 1A3 (ALDH1A3) polyubiquitination to stabilize ALDH1A3 and maintain GSCs10. Treatment of GSCs with the USP9x inhibitor WP1130 induces ALDH1A3 degradation and inhibits GSC growth10. Further, USP13 and FBXL14 reciprocally modulate MYC polyubiquitination in GSCs11. Therefore, deubiquitinases and ubiquitin ligases may be potential therapeutic targets in glioblastoma.
CRISPR/Cas9-based knockout (KO) high-throughput screening has been proven to be an effective platform for the discovery of target genes in cancer therapy. Previous studies of CRISPR screens in glioblastoma have largely focused on in vitro cultured cell models that do not adequately represent the complex effects of the tumor microenvironment, an approach that may limit the success of drug development. Here, we adapted a more physiologically relevant in vivo CRISPR screen to identify essential ubiquitin ligases and deubiquitinases for GSC maintenance and tumor formation.
Results
In vitro and in vivo CRISPR screens revealed novel regulators of GSC-derived tumor growth
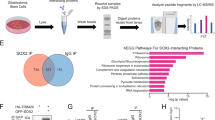
To systematically interrogate the functional contributions of DUBs and ubiquitin ligases to GSC, we performed parallel in vitro and in vivo CRISPR screens to identify DUBs and ubiquitin ligases essential for GSC survival and tumor formation. We designed and constructed a lentivirus-based CRISPR/Cas9 KO library targeting 387 human ubiquitin ligases and 76 deubiquitinases. The library contained 8 different sgRNAs for each targeted gene and 100 nontargeting, control sgRNAs (Supplementary Table S1). We then lentivirally transduced the library into two patient-derived GSCs (GSC468 and GSC738) and then performed puromycin selection. GSCs with CRISPR/Cas9-mediated KO were split into two groups: (1) in vitro cultures in stem cell conditions, and (2) an in vivo screening group intracranially implanted into immunocompromised NSG (NOD.Cg-Prkdc scid Il2rg tm1Wjl /SzJ) mice (Fig. 1a). We analyzed changes in sgRNA abundance in vivo and in vitro using MAGeCK, a bioinformatics pipeline for analysis of pooled genetic screening data. To identify potential therapeutic targets in glioblastoma, we focused on the depleted sgRNAs in our CRISPR screen results. Thirteen genes were hit common to both GSCs in vitro, while 29 genes were hit common to both GSCs in vivo. We next identified 6 highly confident hits common to both the in vitro and in vivo screens. These 6 hits included: RB Binding Protein 6, Ubiquitin Ligase (RBBP6), Ubiquitin Specific Peptidase 5 (USP5), Ubiquitin Specific Peptidase 7 (USP7), Ubiquitin Like with PHD And Ring Finger Domains 1 (UHRF1), BRCA1 Associated Protein (BRAP), and HECT, UBA And WWE Domain Containing E3 Ubiquitin Protein Ligase 1 (HUWE1) (Fig. 1b, c).
a Workflow of the CRISPR screens to identify potential targets for glioblastoma therapy. b Dot plot represents the CRISPR screen results. c Venn diagram showing the overlap between the in vivo CRISPR screen and in vitro CRISPR screen results in GSCs. d RBBP6 mRNA expression levels in the TCGA GBM dataset (U133A). Statistical significance was assessed using a t-test. e The mRNA level of RBBP6 in GSCs and matched DGCs. Statistical significance was assessed using a t-test, ***P < 0.001, n = 3. f RBBP6 mRNA expression level in a panel of 44 GSCs and 9 NSCs. Statistical significance was assessed using a t-test. g H3K27ac signal at the RBBP6 locus in three GSCs, three DGCs and four NSCs.
To prioritize hits for further study, we examined the expression of these genes in The Cancer Genome Atlas (TCGA)-glioblastoma dataset. Of the 6 shared genes, only the mRNA expression of RBBP6 was upregulated in glioblastoma tissue compared to normal brain tissue (Fig. 1d; Supplementary Fig. S1a). Furthermore, RNA sequencing (RNA-seq) and H3K27ac (Histone 3 lysine 27 acetyl, a marker of active enhancer and promoter regions) chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) data from our group and other published datasets12,13 showed that RBBP6 was preferentially expressed in GSCs relative to normal neural stem cells (NSCs) and differentiated glioblastoma cells (DGCs) (Fig. 1e–g). Therefore, we selected RBBP6 for further study.
RBBP6 maintained GSCs
To investigate the role of RBBP6 in GSCs, we performed RBBP6 KO with two independent sgRNAs in 3 patient-derived GSCs (GSC468, GSC738, and GSC3565) to confirm the CRISPR screen results. RBBP6 KO inhibited cell proliferation compared to cells transduced with a sgRNA control (Fig. 2a–c). We confirmed this phenotype in orthogonal studies by applying 2 nonoverlapping shRNAs to knock down RBBP6 mRNA expression and confirmed that RBBP6 inhibition impaired GSC proliferation (Fig. 2d, e). A similar loss of growth was detected upon RBBP6 knockdown in two patient-derived primary cells (Supplementary Fig. S2a–d). RBBP6 KO induced apoptosis, as measured by annexin V staining and PARP cleavage in GSCs but not non-stem tumor cells (NSTCs) and iPSC-induced astrocytes and neural progenitor cells (NPCs) (Fig. 2f–h; Supplementary Fig. S2e–h). Extreme limiting dilution assays (ELDAs) demonstrated that depletion of RBBP6 decreased both the frequency and self-renewal of GSCs (Fig. 2i, j). Taken together, these findings indicate that RBBP6 is essential for GSC maintenance.
a–c Cell viability in the GSC468 cell model (a), GSC738 cell model (b), and GSC3565 cell model (c) transduced with two separate sgRNAs targeting RBBP6 or sgNT. Three technical replicates were used for each group (Top). The error bars show the standard deviations (SDs). Western blot results from the Top assays (Bottom). d, e The mRNA level of RBBP6 in GSC468 (d) and GSC3565 (e) cells transduced with two separate shRNAs targeting RBBP6 or shNT (Left). Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. Cell viability in the GSC468 cell model (d) and GSC3565 cell model (e) transduced with two separate shRNAs targeting RBBP6 or shNT (Right). Three technical replicates were used for each group. The error bars show the SDs. f, g Annexin V staining of GSC3565 and GSC468 cells transduced with two separate sgRNAs targeting RBBP6 or sgNT. Quantification of Annexin-V staining using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ***P < 0.001, ****P < 0.0001, n = 3. h Western blot of PARP in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting RBBP6 or sgNT. i, j The tumor sphere formation capacity was measured in vitro by ELDAs in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting RBBP6 or sgNT.
RBBP6 depletion decreased MYC expression and downstream MYC effectors
To elucidate the molecular mechanisms of RBBP6 in GSCs, we performed RNA-seq following knockdown of RBBP6 mRNA expression to explore the signaling pathways modulated by RBBP6 (Fig. 3a). Hallmark enrichment analysis revealed that RBBP6 knockdown downregulated several signaling pathways essential for cancer progression, including MYC, TNFA-NFKB, and estrogen response signaling pathways (Fig. 3b). Gene set enrichment analysis (GSEA) showed that genes downregulated in RBBP6 knockdown were highly enriched in the terms MYC targets and translation elongation, while genes upregulated were enriched in apoptotic process (Fig. 3c). Both hallmark enrichment analysis and GSEA demonstrated that RBBP6 modulated the MYC signaling pathway. We then performed qPCR and western blot analyses to confirm that RBBP6 regulated MYC expression (Fig. 3d–g). Analysis of TCGA and Chinese Glioma Genome Atlas (CGGA) datasets revealed that RBBP6 mRNA levels were positively correlated with MYC mRNA levels (Fig. 3h, i). Collectively, these data demonstrate that RBBP6 controls the expression of MYC.
a Volcano plot showing the fold changes in the normalized RNA read counts (shRBBP6#2 vs shNT) on the x-axis (Log2) and the FDR values on the y-axis (–Log10). The blue dots indicate significantly downregulated genes and the red dots indicate upregulated genes in GSC468 shRBBP6 cells compared with GSC468 shNT cells (FDR ≤ 0.05 and Log2FoldChange difference ≥ 1). b Hallmark enrichment analysis of downregulated genes following RBBP6 knockdown. c GSEA of differentially expressed genes (DEGs) in RBBP6 knockdown and control GSC468 cells. d mRNA levels of RBBP6 and MYC in GSC468 (left) and GSC3565 (right) cell models transduced with two separate shRNAs targeting RBBP6 or (shNT). Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. e The mRNA level of MYC was measured in GSC468 (left) and GSC3565 (right) cells transduced with two separate sgRNAs targeting RBBP6 or sgNT. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. f The protein level of MYC was measured in GSC468 and GSC3565 cells transduced with two separate shRNAs targeting RBBP6 or shNT. g The protein level of MYC was measured in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting RBBP6 or sgNT. h Correlation between RBBP6 and MYC expression in glioma tissues from the TCGA database. i Correlation between RBBP6 and MYC expression in glioma tissues from the CGGA database.
RBBP6 controlled MYC expression by regulating alternative polyadenylation (APA) events
Next, we explored how RBBP6 regulates MYC expression. We considered molecular interactions with other ubiquitin-related targets. MAGE-A11-HUWE1 mediates the polyubiquitination and degradation of PCF11 to regulate APA in cancer, which supports correlation between the ubiquitin‒proteasome pathway and APA14. Almost 70% of genes contain multiple polyA sites, resulting in varying mRNA isoforms. The 3’ end processing complex mediates the usage of different polyA sites15. APA is an essential posttranscriptional regulation mechanism modulating the translation, stability, and cellular localization of mRNAs15, including in glioblastoma16. However, the role of different APA regulators in GBM is still largely unknown. RBBP6 has been reported to be involved in the regulation of APA, although the mechanism is unclear17,18,19. Thus, we hypothesized that RBBP6 may regulate MYC expression by modulating APA events. We used Dynamic analysis of APA from RNA-Seq (DaPars)16 to detect the APA events. We found 2683 3’UTR with APA events in RBBP6 knockdown (shRBBP6) compared with controls. Out of 2683 APA events, 61.68% (1655 of 2683) were shortening and 38.32% (1028 of 2683) were lengthening events (Fig. 4a, b). Overall, there was global 3’ UTR shortening and a tendency to use shorter isoforms in shRBBP6 relative to controls (P < 2.2 × 10−16; Fig. 4b), which suggested that RBBP6 was a potential APA regulator.
a Scatterplot of PDUIs in GSC468 cells transduced with a shRNA targeting RBBP6 (shRBBP6#2) or shNT. The red dots indicate genes with significant 3’US and the blue dots indicate genes with 3’UTR lengthening following RBBP6 knockdown. b Graph showing the counts of genes with 3’UTR lengthening and shortening. c Representative RNA-seq density plots along with ΔPDUI values for MYC. The numbers on the y-axis indicate the RNA-seq read coverage. d The relative expression levels of OGs that are ceRNAs with 3’US (n = 37, right box) are lower than those of control OGs (n = 514, left box). Welch’s t-test, which accounts for different variances in the two groups being compared, was employed to compare means. To verify the normality assumption for the t-test, a Shapiro–Wilk normality test for small sample sizes (n < 50) was conducted. e Bubble plot showing OGs with MAR3UTR scores in the top 1%. f RBBP6 knockdown induces 3’US of a set of genes, allowing miRNAs to repress MYC expression. g Representative RNA-seq density plots of ZNF281 and DNMT3B, whose 3’UTRs are shortened in response to RBBP6 knockdown. RNA-seq read coverage is plotted on the y-axis. h qPCR results of dPAS usage for APA analysis in GSC468 and GSC3565 cells transduced with two separate shRNAs targeting RBBP6 or a shNT. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. i Diagram showing miR-590-3p binding sites in the 3’UTRs of MYC, ZNF281, and DNMT3B. j Luciferase reporter assay showing miR-590-3p binding to the MYC 3’UTR. Renilla luciferase activity was used for luciferase activity normalization. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. k The luciferase reporter assay showed miR-590-3p binding to the ZNF281 3’UTR. Renilla luciferase activity was used for luciferase activity normalization. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. l The luciferase reporter assay showed miR-590-3p binding to the DNMT3B 3’UTR. Renilla luciferase activity was used for luciferase activity normalization. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ***P < 0.001, n = 3. m The luciferase reporter assay showed ZNF281, DNMT3B, and MYC competitively binding to miR-590-3p. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ****P < 0.0001, n = 3. n Level of miR-590-3p or miR-590-3p mutant in GSC468 and GSC3565 cells transduced with vector, hsa-mir-590, and hsa-mir-590 mutant. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, ***P < 0.001; ****P < 0.0001, n = 3. o Western blot of MYC in GSC468 and GSC3565 cells transduced with empty vector, hsa-mir-590, or hsa-mir-590 mutant. p Cell viability in the GSC468 cells transduced with empty vector, hsa-mir-590, or hsa-mir-590 mutant. Three technical replicates were used for each group. The error bars show the SDs. q Cell viability in the GSC3565 cells transduced with empty vector, hsa-mir-590, or hsa-mir-590 mutant. Three technical replicates were used for each group. The error bars show the SDs.
We next asked how RBBP6 induced a decreased expression of MYC. As the 3’UTR length could affect mRNA decay15, we hypothesized that APA-mediated 3’UTR lengthening in MYC could contribute to its decreased expression in cis. However, we found that these 3’UTR changed genes are not enriched in either oncogenes (OGs) or tumor suppressor gene pathways (Supplementary Fig. S3a–c). Besides, our results showed that RBBP6 knockdown did not alter MYC 3’UTR length (ΔPDUI = –0.025, Fig. 4c). The 3’UTR is also engaged in competing-endogenous RNA (ceRNA) regulation in trans, which downregulates ceRNA through miRNA-mediated repression20. We hypothesized that knockdown of RBBP6 induced widespread 3’UTR shortening (3’US) of genes in cancer cells, which in turn resulted in releasing their miRNAs to repress their ceRNA partners in trans, such as MYC. To test this hypothesis, we used model-based analysis of the trans-effect of 3’UTR shortening (MAT3UTR) to predict the trans-effect of 3’US to their ceRNA partners20. We found 37 trans-target OGs of 3’US (top 1% MAT3UTR score). Thirty-seven expressed OGs from 3’US ceRNAs were more likely downregulated than 514 control OGs not in ceRNET (P = 8 × 10−4, Fig. 4d), indicating an association between 3’US and OG repression. Notably, among 37 OGs, MYC ranked at the top (log2FoldChange = –1.23 and adjusted P = 1.94 × 10–179, Fig. 4e) and was predicted to be a ceRNA of five 3’US genes in GSCs (Fig. 4f). 3’US genes would be expected to release miRNA from their 3’UTR, with the released miRNA available to repress MYC expression (Fig. 4f). The RNA-seq density plots and RT-qPCR analysis results confirmed the 3’US of ZNF281 and DNMT3B, which were predicted to be the MYC ceRNAs with the highest confidence scores (Fig. 4g, h). Additionally, we employed the 3’ Rapid Amplification of cDNA Ends (3’ RACE) method to validate that RBBP6 knockdown induces the shortening of the 3’ UTRs of ZNF281 and DNMT3B. This observation aligns with our RT-qPCR results and confirms that RBBP6 knockdown leads to the shortening of the 3’ UTRs of ZNF281 and DNMT3B (Supplementary Fig. S4a–c).
To identify the miRNAs released from the shortened 3’UTRs of MYC ceRNAs to inhibit MYC expression, we selected miR-590-3p, with the highest predicted score, for validation (Fig. 4i). Next, we confirmed that RBBP6 knockdown did not change the expression level of miR-590-3p (Supplementary Fig. S4d). This suggests that the downregulation of long 3’UTR isoforms of ZNF281 and DNMT3B mRNAs is not due to an increase in miR-590-3p. We then leveraged the 3’UTR luciferase reporter system to further confirm the binding of miR-590-3p to MYC and MYC ceRNAs (DNMT3B and ZNF281). miR-590-3p overexpression specifically decreased MYC-, DNMT3B- and ZNF281-3’UTR-driven luciferase activities and miR-590-3p expression without changing the luciferase activities driven by the 3’UTR binding site mutation (Fig. 4j–l). Reciprocally, overexpression of the 3’UTRs of MYC ceRNAs (DNMT3B and ZNF281) reversed the miR-590-3p-mediated reduction in MYC-UTR luciferase activity, supporting the hypothesis that MYC and its ceRNAs competitively bind to miR-590-3p (Fig. 4m). Overexpression of miR-590-3p but not the miR-590-3p mutation (mutant miR-590-3p seed region) in GSCs via a lentiviral vector decreased MYC expression and inhibited GSC proliferation (Fig. 4n–q). Furthermore, we utilized a chemically modified single-strand miRNA inhibitor called miR-590-3p antagomir to inhibit miR-590-3p. Our findings indicate that the suppression of miR-590-3p led to an increase in the mRNA levels of MYC, ZNF281, and DNMT3B in both GSC468 and GSC3565 cells (Supplementary Fig. S4e, f). In addition, we conducted a 3’UTR luciferase reporter assay, which revealed that the inhibition of miR-590-3p by miR-590-3p antagomir enhanced the luciferase activity driven by MYC-, DNMT3B-, and ZNF281-3’UTR. In contrast, no changes in luciferase activity were observed when the miR-590-3p binding sites in the 3’UTR were mutated (Supplementary Fig. S4g–i). These findings indicate that miR-590-3p targets MYC, ZNF281, and DNMT3B mRNAs. Additionally, overexpression of the DNMT3B and ZNF281 3’UTRs but not their respective mutations (mutant miR-590-3p binding sites) partially reversed the RBBP6 knockdown-induced inhibition of cell proliferation (Supplementary Fig. S4j, k). Moreover, we utilized antisense morpholino to obstruct the usage of distal PASs of the ceRNAs to confirm the ceRNA network under physiological conditions. The antisense morpholino targeting DNMT3B or ZNF281 was employed to block the binding of miR-590-3p to DNMT3B or ZNF281. This blockade of miR-590-3p binding site on DNMT3B or ZNF281 consequently inhibited the expression of MYC (Supplementary Fig. S5a, b) and impaired GSC proliferation (Supplementary Fig. S5c–f). In summary, RBBP6 modulated MYC expression by regulating the MYC ceRNA 3’UTR length.
RBBP6 ubiquitinated cleavage and polyadenylation specificity factor 3 (CPSF3)
To elucidate the molecular mechanisms of RBBP6 in APA event regulation, we performed an unbiased analysis of RBBP6-interacting proteins by immunoprecipitation coupled with mass spectrometry (IP-MS). Gene Ontology (GO) analysis showed that RBBP6-interacting proteins were enriched in RNA process and splicing pathways (Fig. 5a). The IP-MS data indicated that RBBP6 interacted with a set of 3’ UTR processing factors, including CPSF3, CPSF2, NUDT21, and CSTF2 (Fig. 5b; Supplementary Table S2). We confirmed these interactions by co-immunoprecipitation (Co-IP) (Fig. 5c). In addition, we analyzed the public knockdown experiment datasets and also observed significant overlap of 3’UTR changed genes between RBBP6 and CPSF3 (Fig. 5d). Recently, RBBP6 was reported to play a vital role in 3’ end processing, and RBBP6 function is dependent on CPSF319. We found that CPSF3 KO led to 3’UTR shortening in MYC ceRNAs, MYC downregulation, and GSC proliferation defects, consistent with the effects of RBBP6 depletion on GSCs (Fig. 5e, f; Supplementary Fig. S6a, b). Furthermore, CPSF3 KO induced apoptosis in GSC but not non-neoplastic cells (iPSC-induced astrocyte and NPC), as measured by annexin V staining (Supplementary Fig. S6c–f). Therefore, we focused on the RBBP6-interacting protein CPSF3 for further investigation. RBBP6 is a ubiquitin ligase that interacts with CPSF3, which prompted us to test whether RBBP6 mediates CPSF3 ubiquitination. The overexpression of wild-type RBBP6, but not its enzymatically inactive form RBBP6-ΔRING, led to an increase in polyubiquitination of CPSF3 (Fig. 5g). To characterize the type of CPSF3 ubiquitination mediated by RBBP6, different ubiquitin mutants were co-transfected with RBBP6 and CPSF3 into HEK293T cells followed by CPSF3 immunoprecipitation (IP). Western blot analysis showed that RBBP6 mediated mainly the K63-linked polyubiquitination of CPSF3 (Fig. 5h, i). To characterize the polyubiquitination sites in CPSF3, we overexpressed HA-tagged RBBP6, Flag-tagged CPSF3, and Myc-tagged UB in HEK293T cells, then performed IP with CPSF3, and analyzed the precipitate by MS. The MS results indicated 6 potential ubiquitin-binding sites in the CPSF3 protein (K306, K410, K549, K604, K381, and K487) (Supplementary Table S3). We then generated point mutations at these 6 sites in CPSF3 to further determine the major ubiquitination site. The ubiquitination assay showed that only the K410R mutation abolished RBBP6-mediated CPSF3 polyubiquitination (Fig. 5j, k). RBBP6 KO decreased CPSF3 protein levels (Fig. 5l). We then tested whether RBBP6 regulates CPSF3 stability through K63-linked polyubiquitination. Overexpression of RBBP6 stabilized CPSF3 (Supplementary Fig. S7a, b), while RBBP6 KO in GSCs accelerated CPSF3 degradation (Fig. 5m, n).
a GO enrichment analysis of the RBBP6-interacting proteins identified by IP-MS. b Heatmap showing that RBBP6 interacted with pre-mRNA 3’ end processing factors identified by IP-MS. c HEK293T cells were transfected with HA-RBBP6 plasmids for 48 h and were collected for IP/IB. IB, immunoblotting. d The number of the pre-mRNA 3’ end processing factors shortening genes that overlap with RBBP6 shortening genes follows a hypergeometric distribution. The y-axis is the signed Log10 P-values of the hypergeometric test P-values. e qPCR results of dPAS usage for APA analysis in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting CPSF3 or sgNT. Statistical significance was assessed using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test, **P < 0.01; *** P < 0.001; **** P < 0.0001, n = 3. f The protein level of MYC was measured in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting CPSF3 or sgNT. g HEK293T cells transfected with Flag-CPSF3, Myc-UB, HA-RBBP6 and HA-RBBP6-ΔRING plasmids were collected for IP/IB. h HEK293T cells transfected with Flag-CPSF3, Myc-RBBP6, and HA-UB mutant plasmids were collected for IP/IB. i HEK293T cells transfected with Flag-CPSF3, Myc-RBBP6, and HA-UB mutant plasmids were collected for IP/IB. j HEK293T cells transfected with HA-RBBP6, Myc-UB, and Flag-CPSF3 mutant plasmids were collected for IP/IB. k MS spectrum of an identified polyubiquitinated CPSF3 peptide. l The protein level of CPSF3 was measured in GSC468 and GSC3565 cells transduced with two separate sgRNAs targeting RBBP6 or sgNT. m The GSC468 cells transduced with a sgRNA targeting RBBP6 or sgNT followed by CHX (50 µg/mL) treatment for the indicated times were collected for IB. n Quantitation of the results shown in m. Two-way ANOVA was used for statistical analysis with Sidak’s multiple comparisons test. *P < 0.05, n = 3.
Next, we conducted CHX experiments to compare half-lives of wild-type and ub-mutant CPSF3 in normal and RBBP6 KO conditions. Our data showed that RBBP6 KO accelerated the degradation of wild-type CPSF3, but not ub-mutant CPSF3 K410R (Supplementary Fig. S7c–f). Moreover, wild-type CPSF3 exhibited a longer half-life than CPSF3 K410R (Supplementary Fig. S7c–f). To confirm the requirement of RBBP6’s E3 ligase activity for the RBBP6-CPSF3-MYC axis, we conducted a rescue experiment in RBBP6 knockdown GSC468 cells by overexpressing either RBBP6-WT or enzyme activity dead form RBBP6 ΔRING. The overexpression of RBBP6-WT successfully restored the downregulation of CPSF3 and MYC caused by RBBP6 knockdown. In contrast, the overexpression of RBBP6-ΔRING failed to rescue the RBBP6 knockdown-induced downregulation of CPSF3 and MYC (Supplementary Fig. S7g). Taken together, these findings showed that RBBP6 interacted with and stabilized CPSF3 through K63-linked polyubiquitination, and the ubiquitination of CPSF3 by RBBP6 plays a crucial role in CPSF3’s function on MYC.
Targeting RBBP6 and CPSF3 inhibited GSC growth in vivo
To explore the potential benefit of therapeutic targeting of RBBP6 in vivo, we intracranially implanted GSCs bearing RBBP6 KO sgRNAs or controlled nontargeting sgRNAs into immunocompromised NSG mice. Consistent with the in vivo screening results, RBBP6 KO inhibited tumor growth and prolonged survival of tumor-bearing mice (Fig. 6a, b; Supplementary Fig. S8a). As CPSF3 appears to be a critical downstream molecule of RBBP6 in GSCs, we investigated the function of CPSF3 in tumor growth in vivo. Concordantly, mice bearing CPSF3-KO GSCs displayed longer survival times and smaller tumors than mice bearing control GSCs (Fig. 6c, d).
a Kaplan‒Meier survival curves of immunocompromised NSG mice bearing intracranial GSC468 cells transduced with sgNT, sgRBBP6#1, or sgRBBP6#7 (Top). Representative images of H&E staining of mouse brains. Brains were isolated after the presentation of the first neurological sign in any cohort (Bottom). b Kaplan‒Meier survival curves of immunocompromised NSG mice bearing intracranial GSC3565 cells transduced with sgNT, sgRBBP6#1, or sgRBBP6#7 (Top). Representative images of H&E staining of mouse brains (Bottom). Brains were isolated after the presentation of the first neurological sign in any cohort. c Kaplan‒Meier survival curves of immunocompromised NSG mice bearing intracranial GSC468 cells transduced with sgNT, sgCPSF3#1, or sgCPSF3#2 (Top). Representative images of H&E staining of mouse brains (Bottom). Brains were isolated after the presentation of the first neurological sign in any cohort. d Kaplan‒Meier survival curves of immunocompromised NSG mice bearing intracranial GSC3565 cells transduced with sgNT, sgCPSF3#1, or sgCPSF3#2 (Top). Representative images of H&E staining of mouse brains (Bottom). Brains were isolated after the presentation of the first neurological sign in any cohort. Scale bars: 5 mm. Log-rank test was used for statistical analysis (a–d). e The bioluminescence image of the vehicle or JTE-607 treated mice. f Kaplan‒Meier curve showing survival of mice following implantation with GSC3565 and treatment with vehicle or JTE-607. Log-rank test was used for statistical analysis. g Immunofluorescent staining of MYC in the vehicle or JTE-607 treated tumors (left). Quantification of MYC positive cells (right, t-test, **P < 0.01, n = 4). h Immunofluorescent staining of Ki-67 in vehicle or JTE-607 treated tumors (left). Quantification of Ki-67 positive cells (right t-test, ***P < 0.001, n = 4). i Immunofluorescent staining of TUNEL in vehicle or JTE-607 treated tumors (left). Quantification of TUNEL positive cells (right, t-test, ***P < 0.001, n = 4). Scale bars: 75 μm (g–i). j Kaplan‒Meier curves showing survival based on RBBP6 mRNA expression in glioma patients from the CGGA dataset. Log-rank test was used for statistical analysis. k Kaplan‒Meier curves showing survival based on CPSF3 mRNA expression in glioma patients from the CGGA dataset. Log-rank test was used for statistical analysis.
Due to the lack of a specific RBBP6 inhibitor, we treated GSCs with the CPSF3 enzyme activity inhibitor JTE-607. Treatment with JTE-607 impaired GSC proliferation in a concentration-dependent manner (Supplementary Fig. S8b). To overcome the challenges of poor blood–brain barrier penetration and the short half-life of JTE-607 in vivo (t1/2 α = 0.1 h in humans)21, we utilized osmotic minipumps to deliver JTE-607 directly into intracranial tumors. This approach allowed us to investigate the anti-tumor efficacy of JTE-607. GSC3565 cells were intracranially transplanted for one week to allow tumor seeding, and then osmotic minipumps were transplanted at the site of injection to deliver JTE-607 locally. JTE-607 administration did not result in a decrease in mouse body weight, indicating an absence of significant toxicity (Supplementary Fig. S8c). JTE-607 treatment suppressed tumor growth and prolonged mouse survival (Fig. 6e, f). Additionally, JTE-607 administration resulted in reduced expression of MYC and Ki-67 in the tumors (Fig. 6g, h). Moreover, treatment with JTE-607 demonstrated its ability to induce tumor cell apoptosis, as measured by the TUNEL assay (Fig. 6i). We then performed in silico analysis to confirm that high RBBP6 and CPSF3 expression correlated with worse survival in glioma patients (Fig. 6j, k). Collectively, these results demonstrated that RBBP6 and CPSF3 are potential therapeutic targets in glioblastoma.
Discussion
High-throughput screening of established cell lines in vitro to discover putative genes that regulate cell proliferation or death is a traditional approach to cancer drug discovery. To date, this approach has led to the successful development of many antitumor drugs, but the overall success rates of these drugs remain low. One of the major reasons is the vast differences between in vitro culture conditions and the in vivo tumor growth microenvironment. To overcome this limitation, we performed parallel in vivo and in vitro CRISPR/Cas9 functional screens to investigate targetable dependencies of GSCs and identified RBBP6 as a consistent regulator of GSC maintenance and tumor formation in vitro and in vivo.
The APA mechanism plays a crucial role in posttranscriptional regulation by modulating mRNA stability, translation, and cellular localization15. The majority of mRNAs are cleaved and polyadenylated in their 3’UTRs, events that can result in the production of different mRNA isoforms with varying 3’UTR lengths to evade or be affected by miRNA-mediated regulation. The ratio of proximal polyA sites to dPASs varies across tissues and diseases22. APA events are regulated by four complexes: CPSF, cleavage factors I and II (CF I and II), and cleavage stimulation factor (CSTF)23. CPSF3 constitutes a central element of the seven-subunit CPSF complex, playing a crucial role in the cleavage and polyadenylation processes of mRNAs. The assembly of CPSF3 with specific proteins forms the processing machinery required to activate its endonuclease activity. Depletion of CPSF3 results in significant alterations in APA and gene expression. Recent studies have suggested that CPSF3 could serve as a prognostic marker and potential therapeutic target in various cancers, including non-small cell lung cancer24, triple-negative breast cancer25, prostate cancer26, colorectal cancer27, acute myeloid leukemia, and Ewing’s Sarcoma28. Ross et al. utilized phenotypic screening in conjunction with chemical genetics to identify CPSF3 as the target of JTE-607. JTE-607-mediated inhibition of CPSF3 led to alterations in the expression of known downstream effectors in both acute myeloid leukemia and Ewing’s Sarcoma. Additionally, treatment with JTE-607 induced apoptosis and impeded tumor growth in mouse xenograft models28. Consequently, the CPSF3-mediated APA processing pathway holds promise as a target for cancer treatment.
Dysregulation of APA contributes to cancer initiation and progression. It was reported that the knockdown of one of the CPSF complex members, NUDT21 (also called CPSF5), led to 3’US of oncogenes and increased glioblastoma cell proliferation16. Here, we found that RBBP6 regulates APA by ubiquitinating CPSF3. Depletion of RBBP6 or CPSF3 resulted in 3’US of the ceRNA of the OG MYC, thereby reducing MYC expression and leading to inhibition of tumor growth. Our finding indicated that the ubiquitination of CPSF3 by RBBP6 plays a crucial role in CPSF3’s function on MYC. Two recent papers19,29 showed that RBBP6 interacts with CPSF3 and promotes its endonuclease activity, independently of its RING domain. In comparison with the substantial quantity of CPSF3 protein utilized in the in vitro assays, the endogenous protein levels of CPSF3 within cells are significantly lower. Consequently, protein stability may play a more critical role in CPSF3’s functionality in cellular contexts. These results indicated that different APA regulators may play distinct roles in glioblastoma development and progression. Our identification of the dependency on the RBBP6/CPSF3-APA-MYC axis in glioblastoma offers novel strategies for lethal cancers.
RBBP6 was originally identified as an Rb-interacting protein and later proven to be a ubiquitin E3 ligase. Recent studies have indicated that RBBP6 may be a prognostic marker for and potential therapeutic target in many cancers, including cervical carcinoma30, colorectal cancer31 and non-small cell lung cancer32. Few RBBP6 ubiquitination substrate proteins have been reported thus far, and these include Zinc Finger And BTB Domain Containing 38 (ZBTB38)33, NFKB Inhibitor Alpha (NFKBIA)31, Y-Box Binding Protein 1 (YBX1)34 and Growth Factor Receptor Bound Protein 2 (GRB2)35. We demonstrated here that CPSF3 is a novel ubiquitination substrate of RBBP6. RBBP6 mediates K63-linked polyubiquitination of CPSF3 and promotes its stability. K48- and K63-linked chains are the two most abundant ubiquitin chain types. Unlike K48-linked ubiquitination, which is a typical signal for proteasomal degradation, K63-linked ubiquitination has many well-studied nondegradative roles. For example, K63-linked ubiquitination of IKK enhances its binding to the activation upstream kinase TAK136, and K63-linked ubiquitination of Akt promotes its cell membrane translocation37. Interestingly, a recent study also reported that the E3 ligase Pellino-1 stabilized the transcription factors Slug and Snail via K63-linked ubiquitination38, which is consistent with our findings, although the exact mechanism requires further investigation.
In summary, by utilizing an in vitro and in vivo CRISPR screening, we identified the dependency on the RBBP6/CPSF3-APA-MYC axis in glioblastoma (Fig. 7). Our data suggest that approaches to modulate APA in GSCs by targeting RBBP6 or CPSF3 showed high therapeutic efficacy, offering novel strategies for glioblastoma treatment.
RBBP6 regulates the maintenance of GSCs. Depletion of RBBP6 leads to the elimination of K63-linked ubiquitination of CPSF3, resulting in the destabilization of CPSF3 and the subsequent shortening of the 3’ UTRs of MYC ceRNAs. This shortening promotes the release of miR-590-3p from the truncated UTRs, thereby reducing MYC expression and hindering the survival and progression of GSCs.
Materials and methods
GSC derivation
The GSC468, GSC738, and GSC3565 cell lines were generated in our laboratory as reported previously39. Patient-derived xenografts were generated and maintained as a reproducible source of GSC cells. The glioblastoma primary cells GBM12388157, and GBM12479390 in this study were obtained from the Department of Neurosurgery, the Second Affiliated Hospital of Zhejiang University School of Medicine. All glioblastoma tissues were obtained from excess surgical resection samples from patients at the Second Affiliated Hospital Zhejiang University School of Medicine with appropriate consent and in accordance with an IRB-approved protocol (IR2022453). All GSC and primary GBM cells were cultured as neurospheres in neurobasal medium (Gibco, Cat# 12349-015) supplemented with 2% B27 supplement (Gibco, Cat# 12587-010), 1% GlutaMax Supplement (Gibco, Cat# 35050-061), 1% sodium pyruvate (Gibco, Cat# 11360-070), 1% penicillin/streptomycin (Invitrogen, Cat# SV30010), 20 ng/mL basic human fibroblast growth factor (R&D systems, Cat# 4114-TC), and 20 ng/mL human epidermal growth factor (R&D systems, Cat# 236-EG). Short Tandem Repeat (STR) analyses were performed to authenticate the identity of each cell line used in this article. Mycoplasma testing was performed by qPCR with cellular supernatants on a yearly basis. Cells were grown for fewer than 20 in vitro passages from xenografts.
Other cell models
The HEK293T cells were purchased from the American Type Culture Collection (ATCC, Cat# CRL-3216). HEK293T cells were cultured in DMEM (Gibco, Cat# C11995500CP) supplemented with 1% GlutaMax Supplement, 1% penicillin/streptomycin and 10% fetal bovine serum (CellMax, Cat# SA211.02). The iPSC cells were purchased from Cellapy (Cellapy, Cat# CA4002106). iPSC-NPCs were differentiated from the iPSC cells using serum-free STEMdiff™ SMADi Neural Induction Kit (StemCell Technologies, Cat# 08581) according to the manufacturer’s instructions. Astrocytic precursors were generated from iPSC-NPCs using STEMdiff™ Astrocyte Differentiation Kit (StemCell Technologies, Cat# 100-0013) according to the manufacturer’s instructions. These astrocytic precursors are then matured further into astrocytes using STEMdiff™ Astrocyte Maturation Kit (StemCell Technologies, Catalog# 100-0016) according to the manufacturer’s instructions. iPSC-astrocytes were cultured in DMEM supplemented with 1% GlutaMax Supplement, 1% penicillin/streptomycin, and 10% fetal bovine serum. iPSC-NPC were cultured in a neurobasal medium supplemented with 2% B27 supplement, 1% GlutaMax Supplement, 1% sodium pyruvate, 1% penicillin/streptomycin, 20 ng/mL basic human fibroblast growth factor, and 20 ng/mL human epidermal growth factor. STR analyses were performed to authenticate the identity of each cell line used in this article. Mycoplasma testing was performed by qPCR with cellular supernatants yearly.
Animal experiments
All mouse experiments were performed under an animal protocol approved by the Institutional Animal Care and Use Committee of Westlake University and in accordance with the relevant guidelines. Intracranial transplantation of GSCs was performed as previously described39. In brief, GSC spheres were dissociated into single cells, and 10,000 cells were intracranially injected into the right cerebral cortex of individual NSG immunocompromised mice. When any mouse was observed to exhibit neurological signs or signs of morbidity, including lethargy, gait changes, hunched posture, and weight loss, we sacrificed all mice in that cohort. To compare tumor growth in vivo, we isolated brains from mice transplanted with GSCs on the day that neurological signs or signs of morbidity were observed. Then, the brains were used for histologic analysis by H&E staining. In parallel survival experiments, mice were observed until the development of neurological signs or signs of morbidity.
Evaluation of JTE-607 in GBM model
GSC spheres were dissociated into single cells, and 10,000 cells were intracranially injected into the right cerebral cortex of individual NSG immunocompromised mice. Osmotic minipumps (ALZET, Cat# 21-3032) were implanted for the direct delivery of the vehicle or JTE-607 (dissolved in water, 7.143 μg per day per mouse) to the tumor site through a brain infusion kit (ALZET, Cat# 0008663) on one week after intracranial GSC injection.
Immunofluorescence
The tissues were fixed overnight at 4 °C using 4% paraformaldehyde, followed by a 48-h incubation in 30% sucrose. Then, the tissues were embedded in an OCT embedding medium. Cryosections with a thickness of 10 µm were prepared for the immunofluorescence experiment. For immunofluorescence staining, the sections were fixed with 4% paraformaldehyde at room temperature for 10 min. Subsequently, the sections were washed three times with 1× PBS. To permeabilize the sections, 0.25% Triton-X100 was applied at room temperature, followed by another three washes with 1× PBS. After the washing steps, the sections were blocked with 5% serum derived from the donkey and then washed with 1× PBS. Staining for MYC (Cell signaling Technology, Cat# D3N8F, 1:200) or Ki-67 (Proteintech, Cat# 27309-1-AP, 1:200) was carried out at room temperature for 1 h. Following the staining, the sections were washed three times and cover-slipped using a fluorescent mounting medium containing DAPI (ZSGB-BIO, Cat# ZLI-9557). To detect apoptosis in the tumors, the sections were processed using a TUNEL kit (Beyotime, Cat# C1086) according to the manufacturer’s instructions.
Chemicals and oligos
JTE-607 was purchased from MedChemExpress (Cat# HY-110133). The antisense morpholino was purchased from Gene Tools. The antisense oligonucleotide sequences employed in this study are provided below: NC-morpholino: 5’-CCTCTTACCTCAGTTACAATTTATA-3’; ZNF281- morpholino: 5’-AATTTTGGATCAGCCCAGATGGAGA-3’; DNMT3B- morpholino: 5’-GGCTCCAGTTACAAAAAAAATTTTA-3’. Hsa-miR-590-3p antagomir was purchased from Ribobio (Cat# miR30004801-4-5).
CRISPR/Cas9 library
The CRISPR/Cas9 library in this study included 100 nontargeting controls and 3704 unique sgRNAs individually targeting 463 ubiquitin E3 ligases and deubiquitinases. A customized single-strand sgRNA oligonucleotide pool was synthesized by Twist Bioscience, amplified by PCR and then inserted into the lentiCRISPR-v2 vector (lentiCRISPR-v2, RRID:Addgene_52961, Addegene) using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB, Cat# E5520). Next-generation sequencing was performed to ensure sgRNA abundance.
Lentivirus production
HEK293T cells were cotransfected with a lentiviral expression vector, the packaging plasmid psPAX2 (psPAX2, RRID:Addgene_12260, Addegene) and the envelope plasmid pMD2.G (pMD2.G, RRID:Addgene_12259, Addegene) using polyethylenimine (PEI) transfection reagent (Polyscience, Cat # 23966-1) following the manufacturer’s instructions. Lentiviral particles were collected 48 h after the medium change and concentrated using the lentivirus concentration kit (Genomeditech, Cat# GM-040801-100).
CRISPR screen of GSCs
GSC spheres were dissociated into single cells with TrypLE (ThermoFisher Scientific, Cat# 12604021) and were transduced with the lentiviral CRISPR library at a multiplicity of infection of 0.3–0.5 before puromycin selection. For the in vitro screens, GSCs were cultured for 1–2 weeks to ensure at least 10-fold cell doubling. For the in vivo screens, GSCs were implanted into the brains of NSG mice (1 million GSCs per mouse), and the brains were harvested when the mice showed neurological signs. Genomic DNA was isolated from GSCs at the experimental start points as well as from GSCs harvested at the experimental endpoints to prepare the library for next-generation sequencing.
Analysis of CRISPR screen data
FASTQ files were trimmed with Trim Galore (Trim Galore, RRID:SCR_011847). The trimmed FASTQ files were then analyzed using MAGeCK software to obtain the read count of each sgRNA. MAGeCK robust rank aggregation (RRA) algorithm was used to identify significantly enriched and depleted sgRNAs and genes by comparing the in vitro cultured samples and in vivo tumor samples with the control samples.
Plasmids
The following sgRNA sequences were chosen from our CRISPR library or designed with CHOP-CHOP (CHOPCHOP, RRID:SCR_015723). SgRNA sequences were inserted into the lentiCRISPR v2 plasmid (lentiCRISPR v2, RRID:Addgene_52961, Addgene). SgRNAs used in this study were sgNT: 5’-CTCTGCTGCGGAAGGATTCG-3’, sgRBBP6#1: 5’-AAGTCGAACTGAACCAGCGA-3’, sgRBBP6#7: 5’-AAGTCGAACTGAACCAGCGA-3’, sgCPSF3#1: 5’-ATGTTCATGATTGAGATCGC-3’, sgCPSF3#2: 5’-ATTCATAGACTAACCACATG-3’. The shRNA sequences were inserted into the pLKO.TRC.1 plasmid (pLKO.TRC.1, RRID: Addgene_10878, Addgene). ShRNAs used in this study were shNT: 5’-CAACAAGATGAAGAGCACCAA-3’, shRBBP6#2: 5’- GACTCTCCTTCTCGGAATAAA-3’, shRBBP6#7: 5’-GATGACTCTTCCGCGTCTATT-3’, shRBBP6#UTR: 5’-GGGTCTCTGGATTATTGTT-3’.
Quantitative RT-PCR
Total cellular RNA was extracted with TRIzol reagent (Invitrogen, Cat# 15596018) according to the manufacturer’s instructions. Then, RNA was reverse transcribed to cDNA with a cDNA reverse transcription kit (Novoprotein, Cat# E047-01B). Additionally, miRNA was reverse transcribed to cDNA with a miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Cat# MR101-01) and specific probes (miR-590-3p: GTCGTATCCAGTGCA-GGGTCCGAGGTATTCGCACTGGATACGACACTAGC, GADPH: GGCTGTTGTCAT-ACTTCTCATGG; RNU6: AGGGGCCATGCTAATCTTCT). qPCR was performed with Novostart SYBR qPCR Supermix Plus (Novoprotein, Cat# E096) in a Bio-Rad CFX instrument. The qPCR primers used in this study were as follows: GAPDH forward primer: 5’-GGAGCGAGATCCCTCCAAAAT-3’, GAPDH reverse primer: 5’-GGCTGTTGTCATACTTCTCATGG-3’; 18 S forward primer: 5’-GGCCCTGTAATTGGAATGAGTC-3’; 18 S reverse primer: 5’-CCAAGATCCAACTACGAGCTT-3’; RBBP6 forward primer: 5’-CATCTCCCTCTGCGACTTAAAG-3’; RBBP6 reverse primer: 5’-TAGTTGCCATCGCTGGTTCAG-3’; MYC forward primer: 5’-GGCTCCTGGCAAAAGGTCA-3’; MYC reverse primer: 5’- CTGCGTAGTTGTGCTGATGT-3’; DNMT3B (coding sequence, CDS) forward primer: 5’-AGGGAAGACTCGATCCTCGTC-3’; DNMT3B (CDS) reverse primer: 5’-GTGTGTAGCTTAGCAGACTGG-3’; DNMT3B (UTR) forward primer-: 5’-TGGAGCCACGACGTAACAAA-3’; DNMT3B (UTR) reverse primer: 5’-GCATCCGTCATCTTTCAGCC-3’; ZNF281 (CDS) forward primer: 5’-AGGACCTCAGTATTCTCCACC-3’; ZNF281 (CDS) reverse primer: 5’- CCATCTCCAACCAAAGAAGGTTT-3’; ZNF281 (UTR) forward primer: 5’- TGCTTTACTCTCAGGAAAGTGT-3’; ZNF281 (UTR) reverse primer: 5’-TGTTACAGTTGAGATCAAGAGAGGG-3’; miR-590-3p forward primer: 5’-GCGCGCGCGCTAATTTTATGTATAA-3’; miR-590-3p reverse primer: 5’-CAGTGCAGGGTCCGAGGTAT-3’.
APA analysis of a single gene was performed as previously described16. In brief, the levels of transcripts amplified with common primers targeting the CDS were used for normalization to the total transcript level. The distal primers targeted sequences just upstream of the dPAS and were used to detect long transcripts that used the dPAS. Values were calculated as previously described16. ΔCT (common or distal) = CTcommon or distal−CTGAPDH. ΔΔCT = ΔCTdistal− ΔCTcommon. Normalized ΔΔΔCT = ΔΔCTaverage of case – ΔΔCTaverage of control.
Apoptosis assay
The apoptosis assay was performed with Annexin V-Alexa Fluor 647 (Yeasen, Cat# 40304ES60). Samples were analyzed using a Beckman Coulter Cytoflex Cytometer.
ELDA
The neurosphere formation capacity was assayed by ELDA. In brief, different numbers of cells per well (100, 50, 20, 10, and 5) were plated into 96-well plates. Seven days later, the number of each well containing neurospheres was recorded. ELDA software was used to analyze the number of wells containing neurospheres as previously described40.
Cell proliferation assay
Twenty-five hundred cells per well were plated in 96-well plates. Cell viability was measured with a CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, Cat# G7572) at the indicated times.
Western blotting analysis
Cells were collected, washed, lysed with RIPA buffer (Beyotime, Cat# P0013C) and incubated on ice for 30 min. Lysates were centrifuged at 4 °C for 15 min at 12,000 rpm, and the supernatants were collected. A Bradford (Beyotime, Cat# P0006C) kit was used to determine protein concentrations. Samples containing equal amounts of protein were mixed with LDS Sample Buffer (Invitrogen, Cat# B0007), boiled for 10 min, separated using SDS-PAGE, and then transferred onto PVDF membranes. The membranes were blocked with 5% nonfat milk for 1 h. Next, the membranes were incubated with a primary antibody at 4 °C overnight. The membranes were washed with TBST buffer and were then incubated with secondary antibodies in 5% nonfat milk for 1 h. For all western blot analyses, membranes were imaged using Bio-Rad Image Lab software (Image Lab, RRID:SCR_003073). The antibodies used in this study wereRBBP6 (Bethyl Laboratories, Cat# A304-975A), CPSF3 (Proteintech, Cat# 11609-1-AP), CPSF2 (Proteintech, Cat# 17739-1-AP), NUDT21 (Proteintech, Cat# 10322-1-AP), CSTF2 (Proteintech, Cat# 26825-1-AP), HA-tag (Cell signaling Technology, Cat# 3724 S), Flag-tag (Sigma, Cat # F1804), MYC-tag (Proteintech, Cat # 16286-1-AP), MYC (Cell signaling Technology, Cat# D3N8F), GAPDH (Proteintech, Cat# 60004-1-Ig).
IP experiments
Cells were collected, washed, lysed with NP-40 lysis buffer (Beyotime, Cat# P0013F) and incubated on ice for 30 min. Lysates were centrifuged at 4 °C for 15 min at 12,000 rpm, and the supernatants were collected. A Bradford (Beyotime, Cat# P0006C) kit was used to determine protein concentrations. For IP experiments, samples containing one milligram of protein were incubated with the indicated anti-Flag-M2 beads (Sigma, Cat# M8823) or anti-HA beads (ThermoFisher Scientific, Cat# 88838) for 1 h at room temperature. After incubation, the beads were washed with lysis buffer five times. After washing, the beads were mixed with LDS Sample Buffer (Invitrogen, Cat# B0007) and boiled for 10 min for western blot analysis.
Luciferase reporter assay
The indicated plasmids were transfected into HEK293T cells (ATCC, Cat# CRL-3216) for 48 h. Then, we used a Dual-Luciferase Reporter Gene Assay Kit (Yeasen, Cat# 11402ES60) to measure firefly and Renilla luciferase activity.
3’ RACE
The full length 3’UTR mRNA were generated with GoScript™ Reverse Transcriptase (Promega, Cat# A5001) kit, following the manufacturer’s protocol with Oligo dT18-N(22) primer. GSP-1 primers and Oligo N(22) primer were used to perform the first-round PCR. GSP-2 primers and Oligo N(22) primer were used to perform the second-round PCR. The 3’RACE primers used in this study were as follows: Oligo dT18-N(22) primer: 5’-CTGATCTAGAGGTACCGGATCCTTTTTTTTTTTTTTTTTT-3’; Oligo N(22) primer: 5’-CTGATCTAGAGGTACCGGATCC-3’; ZNF281 GSP-1 primer: 5’-GGAGTGTGGTTTCGGCCAA-3’; ZNF281 GSP-2 primer: 5’- AGTGTGGTTTCGGCCAATCT-3’; DNMT3B GSP-1 primer: 5’- TCTTTGGCTTTCCTGTGCAC-3’; DNMT3B GSP-2 primer: 5’- TGGCTTTCCTGTGCACTACA-3’. The PCR products were further sequenced.
RNA-seq analysis
Total RNA was extracted with an RNeasy Small RNA Isolation Kit with Spin Columns (Beyotime, Cat# R0028) according to the manufacturer’s instructions. RNA samples, namely, 4 samples of GSC468 cell lines under two different conditions (Control: GSC468_shNT; Case: GSC468_RBBP6 knockdown) with 2 biological replicates per condition, were sequenced on the Illumina platform, and data were obtained in FASTQ format. For each FASTQ file, quality checks were conducted using FastQC (FastQC, RRID:SCR_014583)41. Contaminating data, such as low-quality reads, adaptor sequences, and poor-quality bases, were removed with Trimmomatic software (Trimmomatic, RRID:SCR_011848)42, and 252,765,061 clean reads were generated. The trimmed reads were mapped to the human reference genome (GRCh38) using STAR (STAR, RRID:SCR_004463)43 and were then sorted by SAMtools (SAMtools, RRID:SCR_002105)44. Subsequently, the uniquely mapped reads were quantified by featureCounts (featureCounts, RRID:SCR_002105)45. The transcript per million (TPM) values were calculated using RSEM. The genes with TPM < 1 in more than 80% of samples were removed. The remaining genes were then used to perform downstream analysis. DEG analysis was performed with the DESeq2 package (DESeq, RRID:SCR_000154)46.
APA event analysis
We used DaPars2 to identify the most significant APA events between the shNT and RBBP6 knockdown conditions. To quantify APA events, we first downloaded the human gene annotation file (GRCh38) from UCSC and extracted a 3’UTR annotation for each transcript. Subsequently, .wig files were generated using STAR (STAR, RRID:SCR_004463). The PDUI values were extracted by the DaPars2 algorithm (version 2.0)47 with the .wig files. A PDUI score close to 0 indicates that the gene tends to use a proximal PAS, whereas a PDUI score close to 1 indicates that the gene tends to use a dPAS. Statistical significance was assumed when the adjusted P-value of the PDUI difference was less than 0.05 and the absolute mean PDUI difference was greater than 0.2.
Trans-effect analysis of 3’US
MAT3UTR software20 was used to quantify the trans-effect of 3’US. First, we extracted the 3’UTR information of transcripts from UCSC. Second, we collected the miRNA binding data from TarBase48, miRecords49, miRTarBase50, and TargetScanHuman version 6.251; these data contained the transcript ID, gene name, miRNA family name, chromosome, start and end coordinate of the binding region, and strand information. Third, we assumed coexpression when a pair of transcripts shared at least five miRNA binding sites in their 3’ UTRs. We identified the potential ceRNA partners of the given genes with 3’US. Finally, we used the script ‘MAT3UTR.py’ to estimate the trans-effect of 3’US for each mRNA and miRNA pair. Enrichment analysis was conducted to examine the ceRNA partner genes’ association with OGs. The OGs utilized in this study were determined by the TUSON algorithm, which identified residue-specific activating mutations in over 8200 tumor/normal pairs through genome sequencing. The genes were ranked based on their TUSON prediction P-values, with the top 1000 genes (P < 0.01) considered as the reference oncogenes for the enrichment analysis. To enhance statistical power, we selected 551 highly expressed OGs (TPM > 1) for further analysis. Among these 551 OGs, 37 were identified as 3’UTR ceRNAs in our study, while 514 were not in the ceRNA networks (ceRNET). Welch’s t-test, which accounts for different variances in the two groups being compared, was employed to compare means. To verify the normality assumption for the t-test, a Shapiro–Wilk normality test for small sample sizes (n < 50) was conducted. All statistical analyses were performed using R (version 3.6).
APA regulatory analysis
The 3’ end processing factors datasets were retrieved from NCBI GEO (GSE151919 and GSE149204) and ENCODE (ENCSR594DNW vs ENCSR067GHD, ENCSR815JDY vs ENCSR856ZRV, ENCSR895BTE vs ENCSR913CAE). We then used DaPars2 to identify the most significant APA events. We applied a hypergeometric test to test for significant overlap between the pre-mRNA 3’ end processing factors shortening genes and the RBBP6 shortening genes.
Data availability
All the RNA-seq raw sequencing data have been deposited into the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA914900. The expression data of RBBP6 in both glioblastoma (GBM) and non-tumor samples was obtained from the Gliovis data portal52. The data of RBBP6 expression in GSCs and NSCs can be found on the Gene Expression Omnibus (GEO) under the accession number GSE119834. H3K27ac ChIP-seq data can be accessed on the GEO with the accession numbers GSE54792 and GSE119755. For survival and gene correlation analyses, data were obtained from the CGGA and TCGA from the Gliovis data portal.
References
Stupp, R. et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA 318, 2306–2316 (2017).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Gimple, R. C., Bhargava, S., Dixit, D. & Rich, J. N. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 33, 591–609 (2019).
Sturm, D. et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 14, 92–107 (2014).
Valor, L. M. & Hervas-Corpion, I. The epigenetics of glioma stem cells: a brief overview. Front. Oncol. 10, 602378 (2020).
Humphreys, L. M., Smith, P., Chen, Z., Fouad, S. & D’Angiolella, V. The role of E3 ubiquitin ligases in the development and progression of glioblastoma. Cell Death Differ. 28, 522–537 (2021).
Jin, W. L., Mao, X. Y. & Qiu, G. Z. Targeting deubiquitinating enzymes in glioblastoma multiforme: expectations and challenges. Med. Res. Rev. 37, 627–661 (2017).
Glickman, M. H. & Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 (2002).
Eichhorn, P. J. A. et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 18, 429–435 (2012).
Chen, Z. et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Invest. 129, 2043–2055 (2019).
Fang, X. et al. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J. Exp. Med. 214, 245–267 (2017).
Suva, M. L. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014).
Mack, S. C. et al. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216, 1071–1090 (2019).
Yang, S. W. et al. A Cancer-specific bbiquitin ligase Drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3’ End processing complex. Mol. Cell 77, 1206–1221.e7 (2020).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Masamha, C. P. et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416 (2014).
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3’ processing complex. Mol. Cell 33, 365–376 (2009).
Di Giammartino, D. C. et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3’ UTRs. Genes Dev. 28, 2248–2260 (2014).
Schmidt, M. et al. Reconstitution of 3’ end processing of mammalian pre-mRNA reveals a central role of RBBP6. Genes Dev. 36, 195–209 (2022).
Park, H. J. et al. 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat. Genet. 50, 783–789 (2018).
Uesato, N., Fukui, K., Maruhashi, J., Tojo, A. & Tajima, N. JTE-607, a multiple cytokine production inhibitor, ameliorates disease in a SCID mouse xenograft acute myeloid leukemia model. Exp. Hematol. 34, 1385–1392 (2006).
MacDonald, C. C. Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond (2018 update). Wiley Interdiscip. Rev. RNA 10, e1526 (2019).
Gruber, A. J. & Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 20, 599–614 (2019).
Ning, Y., Liu, W., Guan, X., Xie, X. & Zhang, Y. CPSF3 is a promising prognostic biomarker and predicts recurrence of non-small cell lung cancer. Oncol. Lett. 18, 2835–2844 (2019).
Mitra, M. et al. Alternative polyadenylation factors link cell cycle to migration. Genome Biol. 19, 176 (2018).
Zhu, Z. H., Yu, Y. P., Shi, Y. K., Nelson, J. B. & Luo, J. H. CSR1 induces cell death through inactivation of CPSF3. Oncogene 28, 41–51 (2009).
Luo, K. et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-beta signalling in colorectal cancer. J. Exp. Clin. Cancer Res. 38, 249 (2019).
Ross, N. T. et al. CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing’s sarcoma. Nat. Chem. Biol. 16, 50–59 (2020).
Boreikaite, V., Elliott, T. S., Chin, J. W. & Passmore, L. A. RBBP6 activates the pre-mRNA 3’ end processing machinery in humans. Genes Dev. 36, 210–224 (2022).
Teng, F. et al. RBBP6 promotes human cervical carcinoma malignancy via JNK signaling pathway. Biomed. Pharmacother. 101, 399–405 (2018).
Xiao, C. et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial-mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 10, 833 (2019).
Wang, Q. S., Wei, S. R. & Xiao, H. L. RBBP6 induces non-small cell lung cancer cell proliferation and high expression is associated with poor prognosis. Oncol. Lett. 19, 2895–2901 (2020).
Miotto, B. et al. The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability. Cell Rep. 7, 575–587 (2014).
Chibi, M. et al. RBBP6 interacts with multifunctional protein YB-1 through its RING finger domain, leading to ubiquitination and proteosomal degradation of YB-1. J. Mol. Biol . 384, 908–916 (2008).
Ye, Z. et al. GRB2 enforces homology-directed repair initiation by MRE11. Sci. Adv. 7, eabe9254 (2021).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Xu, Y. et al. RNF8-mediated regulation of Akt promotes lung cancer cell survival and resistance to DNA damage. Cell Rep. 37, 109854 (2021).
Jeon, Y. K. et al. Pellino-1 promotes lung carcinogenesis via the stabilization of Slug and Snail through K63-mediated polyubiquitination. Cell Death Differ. 24, 469–480 (2017).
Xie, Q. et al. N(6)-methyladenine DNA Modification in Glioblastoma. Cell 175, 1228–1243.e20 (2018).
Gimple, R. C. et al. Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 9, 1248–1267 (2019).
de Sena Brandine, G. & Smith, A. D. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 8, 1874 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Feng, X., Li, L., Wagner, E. J. & Li, W. TC3A: the cancer 3’ UTR atlas. Nucleic Acids Res. 46, D1027–D1030 (2018).
Sethupathy, P., Corda, B. & Hatzigeorgiou, A. G. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 12, 192–197 (2006).
Xiao, F. et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, D105–D110 (2009).
Huang, H. Y. et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48, D148–D154 (2020).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005 (2015).
Bowman, R. L., Wang, Q., Carro, A., Verhaak, R. G. & Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro. Oncol. 19, 139–141 (2017).
Acknowledgements
We thank Dr. Yan Luo (Zhejiang University) for providing the ubiquitin expression plasmids. We thank the MS & Metabolomics Core Facility at the Center for Biomedical Research Core Facilities of Westlake University for sample analysis. We thank Dr. Catherine C.L. Wong (Peking Union Medical College Hospital) for her helpful suggestions regarding the design of proteomic experiments. This work was supported by grants from the National Natural Science Foundation of China (82073268) and Westlake Education Foundation to Q.X.;the National Natural Science Foundation of China (32100533) and Open grant funds from Shenzhen Bay Laboratory. (SZBL2021080601001) to L.L;NIH grants (CA197718, CA238662, and NS103434) to J.N.R.
Author information
Authors and Affiliations
Contributions
P.L.: Data curation, formal analysis, validation, investigation, visualization, methodology, and manuscript writing and editing. W.C.: Data curation, software, formal analysis, methodology, and manuscript writing and editing. Z.L.: Software, formal analysis, and methodology. J. Yu: Data curation and methodology. J. Yang: Data curation and methodology. Z.X.: Data curation and methodology. Q.W.: methodology. X.M.: Data curation and methodology. J.T.: Software, formal analysis, and methodology. Y.C.: Software, formal analysis, and methodology. F.L.: Methodology and resources. C.W.: Methodology and resources. J.Z.: Methodology and resources. W.L.: Conceptualization, methodology, and manuscript editing. J.N.R.: Conceptualization, resources, data curation, supervision, manuscript writing and editing, funding acquisition, and project administration. L.L.: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, manuscript editing, funding acquisition, and project administration. Q.X.: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, manuscript writing and editing, funding acquisition, and project administration.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, P., Chen, W., Long, Z. et al. RBBP6 maintains glioblastoma stem cells through CPSF3-dependent alternative polyadenylation. Cell Discov 10, 32 (2024). https://doi.org/10.1038/s41421-024-00654-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-024-00654-3