Abstract
Unlike conventional αβT cells, invariant natural killer T (iNKT) cells complete their terminal differentiation to functional iNKT1/2/17 cells in the thymus. However, underlying molecular programs that guide iNKT subset differentiation remain unclear. Here, we profiled the transcriptomes of over 17,000 iNKT cells and the chromatin accessibility states of over 39,000 iNKT cells across four thymic iNKT developmental stages using single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) to define their developmental trajectories. Our study discovered novel features for iNKT precursors and different iNKT subsets and indicated that iNKT2 and iNKT17 lineage commitment may occur as early as stage 0 (ST0) by two distinct programs, while iNKT1 commitments may occur post ST0. Both iNKT1 and iNKT2 cells exhibit extensive phenotypic and functional heterogeneity, while iNKT17 cells are relatively homogenous. Furthermore, we identified that a novel transcription factor, Cbfβ, was highly expressed in iNKT progenitor commitment checkpoint, which showed a similar expression trajectory with other known transcription factors for iNKT cells development, Zbtb16 and Egr2, and could direct iNKT cells fate and drive their effector phenotype differentiation. Conditional deletion of Cbfβ blocked early iNKT cell development and led to severe impairment of iNKT1/2/17 cell differentiation. Overall, our findings uncovered distinct iNKT developmental programs as well as their cellular heterogeneity, and identified a novel transcription factor Cbfβ as a key regulator for early iNKT cell commitment.
Similar content being viewed by others
Introduction
Invariant natural killer T (iNKT) cells are innate-like T cells that share the characteristics of T cells and NK cells1,2 and modulate a broad spectrum of immune responses and diseases3. In the mouse thymic cortex, rare CD4+CD8+ double positive (DP) thymocytes expressing a Vα14-Jα18 T-cell receptor (TCR) chain, preferentially paired with a diverse set of TCR Vβ chains (Vβ8s, Vβ7 or Vβ2 chains), are positively selected by CD1d on DP thymocytes and highly express CD24 (CD24+, defined as stage 0 (ST0)). These newly selected iNKT cells enter the medulla directed by the expression of Ccr74. In the thymic medulla, they sharply downregulate CD24 (CD24−CD44−NK1.1−, defined as stage 1 (ST1)), then upregulate the adhesion molecule CD44 and acquire a memory or activate phenotypes (CD44hiNK1.1−, defined as stage 2 (ST2)). ST2 iNKT cells either emigrate to the peripheral organs or remain as long-lived resident cells and mature after acquiring NK1.1 and other NK lineage markers (CD44hi NK1.1+, defined as stage 3 (ST3)) in the thymus1.
Unlike conventional Th1/2/17 T cells, which differentiate in the peripheral lymphoid tissues upon antigen encounter or specific cytokine treatment, iNKT cells acquire their effector function and differentiate into iNKT1 (PLZFloT-bethi), iNKT2 (PLZFhiRORγt−), and iNKT17 (PLZFintRORγt+) cells prior to thymic export. These iNKT subsets have cytokine profiles similar to their Th1/2/17 counterparts, but are less strict. For example, iNKT1 cells also produce IL-4, although they mainly produce IFN-γ. Most ST3 iNKT cells are either CD4+ single positive (CD4SP) or CD4−CD8− double negative (DN) cells, while ST2 cells are more diverse, including CD4SP iNKT2 cells, DN iNKT17 cells, and immature iNKT1 cells5.
Although advanced studies have been conducted in iNKT cells recently, several central questions remain to be answered including: (1) What is the early biological event post iNKT positive selection? (2) How do the specific transcription factors coordinated with their chromatin background guide iNKT cell sub-lineage commitment and differentiation in the thymus? (3) What is the potential checkpoint for different iNKT subset differentiation? (4) Are iNKT1, iNKT2 and iNKT17 cells phenotypically/functionally heterogeneous or homogenous? Recent advances in single-cell assays provide an avenue to explore the transcriptomic and epigenetic heterogeneity of cells at single-cell resolution. Single-cell RNA sequencing (scRNA-seq) can be utilized to assess cell-to-cell variation and has been used to discover rare populations and to infer lineage relationships6,7, which offers an unbiased approach to study iNKT cell developmental trajectory and heterogeneity. Single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) offers a similar resolution and provides additional information about gene regulatory processes. Here, we profiled the transcriptomes of over 17,000 iNKT cells and the chromatin accessibility states of over 39,000 iNKT cells across four thymic iNKT developmental stages. By integrating transcriptome and chromatin accessibility profiles, we identified two developmental programs in ST0 that contribute to iNKT2 and iNKT17 differentiation, while iNKT1 commitment occurs in ST1. Both iNKT2 and iNKT1 cells exhibit extensive heterogeneity, while iNKT17 cells are relatively homogenous. We identified a co-transcription factor Cbfβ highly expressed in the iNKT commitment checkpoint, and conditional deletion of Cbfβ in the thymocytes almost totally blocked early iNKT cell development and severely impaired iNKT1/2/17 cell differentiation. Overall, our study captured iNKT cell developmental trajectories, revealed their cellular heterogeneity, and identified Cbfβ as a key regulator for early iNKT cell commitment.
Results
Clustering thymic iNKT cells across successive developmental stages by scRNA-seq and scATAC-seq
To unveil the iNKT cell developmental landscape, thymic iNKT cells across successive developmental stages were harvested by fluorescence-activated cell sorting (FACS) for scRNA-seq and scATAC-seq assays (Fig. 1a, b). Given the rarity of thymic ST0 (CD24+) iNKT cells in C57BL/6 mice, we utilized Vα14-Jα18-transgenic mice (also called rec-Vα14Tg) for ST0 iNKT cell analysis, which closely mimic the endogenous TCR locus but have abundant ST0 iNKT cells8,9 (Supplementary Fig. S1a, b). As expected, we found a high similarity in scATAC-seq profile in ST0 iNKT cells between the rec-Vα14Tg and C57BL/6 mice (Supplementary Fig. S1c, d). Therefore, we further performed scRNA-seq analyses using rec-Vα14Tg mice for ST0 iNKT cells and C57BL/6 mice for ST1 (CD24−CD44loNK1.1−), ST2 (CD24−CD44hiNK1.1−) and ST3 (CD24−CD44hiNK1.1+) iNKT cells (Fig. 1a–c and Supplementary Fig. S1b).
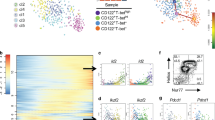
a iNKT cells collected for scATAC-seq and scRNA-seq analysis. b Sorting strategy of ST0 (CD24+), ST1 (CD24−CD44loNK1.1−), ST2 (CD24−CD44hiNK1.1−) and ST3 (CD24−CD44hiNK1.1+) iNKT cells. c Overview of study design. d t-SNE plots from 10X genomics scRNA-seq dataset. Cells from sorted thymic ST0, ST1, ST2 and ST3 iNKT cells were pooled. Displaying relationships between individual cells with color-coded on ST0/1/2/3 iNKT cells (left); t-SNE plots of data identical to those in the left but color-coded on the different clusters (right). Bar graph represents cell numbers in each cluster (top), n = 2. e The fractions of sixteen clusters defined in aggregated iNKT cells across ST0–3. f Heatmap of the top ten DEGs from each cluster derived from (d). Each column represents gene expression for an individual cell with color-coded on gene expression profiles. Yellow is upregulated, and purple is downregulated. g Violin plots of cluster-defining genes in each cluster derived from (d).
The scRNA-seq and scATAC-seq libraries were generated using the 10X Genomics platform. After the quality control filtering and excluding cell outliers, a total of 17,944 high-quality single thymic iNKT cells with a total of 13,578 expressed genes were retained for the subsequent scRNA-seq analysis. A total of sixteen clusters were identified using the R Seurat package10,11 in the aggregated iNKT cells (ST0–ST3), from as few as 255 cells to as many as 2817 cells per cluster (Fig. 1d, e). The most differentially expressed genes (DEGs) in each cluster were shown in the heatmap (Fig. 1f) and violin plots (Fig. 1g). Among these clusters, four clusters (C2, C4, C9, and C14) were from ST0, eight clusters were from ST1 (C3, C5, C6, C7, C10, C11, C15, and C16), nine clusters were from ST2 (C1, C3, C5, C6, C7, C10, C11, C13, and C15) and four clusters were from ST3 (C1, C5, C8, and C12) (Fig. 1d and Supplementary Fig. S2a). ST0 iNKT cells were clearly separated from the rest of the iNKT stages, and the clusters from ST1 and ST2 iNKT cells moderately overlapped, whereas iNKT clusters from ST3 were closely adjacent to those from ST2 iNKT cells (Fig. 1d and Supplementary Fig. S2b). Moreover, the correlation analysis indicated that distinct clusters within the same stage exhibit a relatively similar transcriptomic pattern. For example, the correlation between clusters in ST0 ranged from 0.37 (C2 vs C14) to 0.79 (C2 vs C4), and the correlation between clusters in ST3 ranged from 0.39 (C5 vs C12) to 0.87 (C1 vs C8). As expected, the clusters in different stages did not exhibit a significant correlation, indicating that transcriptomic patterns are greatly distinct in the clusters belonging to different stages (Supplementary Fig. S2c).
Cellular differentiation is accompanied by the expression of genes controlled by cis-regulatory elements, which must be in an open state in order to function properly. We therefore performed scATAC-seq analyses on thymic iNKT cells from different developmental stages as those in scRNA-seq analysis and mapped the chromatin accessibility landscape of individual iNKT cells using the R Seurat and Signac packages. A total of 39,428 cells were analyzed, with a median of 11,398 fragments per cell mapped to the nuclear genome (Supplementary Fig. S2d). As expected, those sixteen iNKT cell clusters were well identifiable after the integration with scRNA-seq data (Fig. 2a). Since scRNA-seq allows us to identify the current cell state as implicated by the transcriptome, we then assigned each of the clusters (C1–C16) into defined iNKT1, iNKT2, and iNKT17 functional subsets based on their signature transcriptomes as previously published12 and cytokine transcript expression. We found that C3, C6, C7, C10, C11, C15, and C16 clusters from ST1 and ST2 were categorized into iNKT2 subset; C1, C5, C8, and C12 clusters, majorly from ST3 were categorized into iNKT1 (Fig. 2b and Supplementary Fig. S2a, e); while a unique iNKT C13 from ST2 was assigned into iNKT17 (Fig. 2b and Supplementary Fig. S2a, d). As expected, ST0 clusters (C2, C4, C9, and C14) did not stand out in any iNKT subsets (Fig. 2b) since they did not exhibit strong effector signatures. Overall, by integrating transcriptomic and epigenetic profiles, we mapped the dynamic transcriptome and chromatin landscapes of thymic iNKT cells with sixteen clusters and uncovered the heterogeneity of iNKT1 and iNKT2 cells and the relative homogeneity of iNKT17 cells.
a UMAP plots showing integrated analysis of scRNA-seq and scATAC-seq from sorted ST0–3 iNKT cells. b Bubble plots showing gene expression in individual clusters (C1–C16) from aggregated iNKT cells. Gene names labeled in blue are iNKT17 signature genes, in red are iNKT1 signature genes and in green are iNKT2 signature genes. y axis shows different clusters identified in (a).
iNKT cells proliferate briskly during development, especially at ST1 and ST2, or iNKT2/17 subsets (Supplementary Fig. S3a, b), we wondered whether the cell cycle genes may mask other functionally key genes for iNKT cells. To eliminate this confounding factor and unmask the underlying iNKT cell heterogeneity, we regressed out cell effects followed by re-clustering of these iNKT cells. As shown in Supplementary Fig. S3c–e after removing the effects of the cell cycle on the transcriptome, the clusters observed in ST0 and ST3 are very consistent with our primary data (Fig. 1d). Given that cell proliferation is the nature of ST1 and ST2, and those cells have a very strong capability to expand iNKT1, iNKT2, and iNKT17 subsets, we included those cells for our following analysis.
Two developmental trajectories in ST0
iNKT cell development in the thymus relies on the pool of around 1000 ST0 iNKT precursors (iNKTp) located in the thymic cortex13. Here, we identified four clusters (C2, C4, C9, and C14) in ST0 (Figs. 1d and 3a) and highlighted the specific signatures for each cluster (Fig. 3b). To further test the robustness of ST0 cells from rec-Vα14Tg mice, we then compared ST0 from rec-Vα14Tg mice with selected CD24+ ST0 cells from C57BL/6 mice recently reported by Krovi et al.14. After correction of batch effect, merging, and aligning of data from two libraries, ST0 cells from C57BL/6 mice essentially mirrored the ones from rec-Vα14Tg mice with similar clusters (Supplementary Fig. S4a, b). Furthermore, the co-expression of these genes in individual clusters were verified in both rec-Vα14Tg and C57BL/6 mice by flow cytometry (Supplementary Fig. S4c, d). Alternatively, based on CD4 and CD8 expression, these ST0 clusters can be classified into three groups: C14 are CD4+CD8+ (DP) iNKTp; C9 are CD4+CD8− (CD4SP) iNKTp, enriched with the regulators of T lymphocyte survival, including Id2 and Il7r; C2 and C4 are CD4−CD8− (DN) iNKTp, with C4 highly expressing Cd5 and Cd6, while C2 iNKTp abundantly expresses Egr2 and Slamf6, which are essential for iNKT cell development via regulation of PLZF expression and TCR signaling strength, respectively15,16 (Fig. 3b–d).
a t-SNE plots from scRNA-seq dataset from sorted ST0 iNKT precursors (iNKTp). b Feature plots depicting specific gene expression in each cluster in ST0 iNKTp. c Feature plot depicting single-cell gene expression of Cd4, Cd8a and their co-expression (top). Bar graph represents the average (Ave) expression of Cd4 and Cd8a in the clusters (bottom). d Representative flow plots of CD4 vs CD8 expression. The bar graph represents means ± SD, n = 19. Data represent seven independent experiments (bottom). DP CD4+CD8+ double positive, DN CD4−CD8− double negative, CD4SP CD4+ single positive. e Violin plots of Cd8 expression in each cluster-derived ST0 iNKT cells from C57BL/6 mouse (red) and rec-Vα14Tg mouse (green). f The ordering of ST0 iNKTp along pseudotime in a state-space defined by Monocle 3. Each color represents a cluster. g The same pseudotime plot as in (f), feature plots depicting single-cell gene expression trajectory of Ccr7 in ST0 iNKTp development (left). The bar graph represents Ave expression of Ccr7 (right). h Representative flow plots of Ccr7 expression in ST0 iNKTp (left). Bar graph presents mean CCR7+ iNKT ± SD, n = 3 (right). i, l Aggregate scATAC-seq browser tracks for Zbtb16 (i) and Rorc (l) in ST0 iNKTp clusters. The bar graph represents Zbtb16 (i) and Rorc (l) Ave expression (right). j, m The same pseudotime plot as in f; feature plots depicting single-cell gene expression of Zbtb16 (j) and Rorc (m) in the ST0 iNKTp development trajectory. k, n Representative flow plots of PLZF (k) and RORγt (n) expression patterns in ST0 iNKTp (bottom). Bar graphs represent mean PLZF+ iNKT ± SD (k) and mean RORγt+ iNKT ± SD (n) (top). n = 5, data represent three independent experiments.
Although previous studies claimed that iNKT cells are either DN or CD4SP 17, a small DP iNKTp cluster (C14) was indeed uncovered in ST0 (Fig. 3c–e and Supplementary Fig. S4a–d). To rule out the possibility that C14 DP iNKTp might be the contaminated un-signaled DP thymocytes, we analyzed the usage of TCR Vβ8s/β7 in both rec-Vα14Tg and C57BL/6 mice, and found that the Vβ repertoire of DP iNKTp was similar to non-DP ST0 iNKTp and mature iNKT cells. More importantly, the expression of TCR Vβ chains in DP iNKTp was significantly higher than that in un-signaled DP thymocytes18 (Supplementary Fig. S5a, b). In addition, this cluster also highly expressed recombinase subunits (e.g., Rag1, Rag2, and Dntt) (Supplementary Fig. S6) and early T-cell decision molecule Ly6d19 (Fig. 3b), indicating that C14 DP iNKTp had recently been TCR signaled. Therefore, we assumed that C14 ST0 iNKT cells could be the transient iNKT precursors from DP thymocytes, which were recently positively selected to iNKT cell lineage.
To explore the developmental programs after selection, we applied the Monocle toolkit20 in R to organize ST0 iNKTp into a pseudotime trajectory. Two potential developmental branches, C14-C2-C4 branch (termed as DP-DN) and C14-C9 branch (DP-CD4SP) were identified (Fig. 3f). Both branches eventually meet at the developmental ends with increased expression of chemokine receptor Ccr7 (Fig. 3g), which is required for ST0 iNKTp migration from the thymic cortex to medulla21. Ccr7 expression patterns in DP, DN, and CD4SP iNKTp were further confirmed at the protein level by flow cytometry (Fig. 3h). Thus, after selection, DP iNKTp downregulate CD8 or both CD4 and CD8 expression to initiate DP-CD4SP or DP-DN developmental programs in the thymic cortex, which eventually migrate into the thymic medulla.
Functional iNKT subset-lineage commitments in ST0
We next asked how these two developmental programs in ST0 contribute to iNKT subset-lineage commitments. Although PLZF is annotated as a key iNKT2 signature, it is also critical for overall iNKT cell development, including iNKT1 and iNKT17 cells. Thus, we first assessed the expression pattern and chromatin accessibility of Zbtb16 (encodes PLZF) in ST0 iNKTp. We found that the region +40 kb and TSS to Zbtb16 was not accessible in the DP iNKTp (C14) but was accessible in CD4SP (C9) and DN (C2 and C4) iNKTp with much higher levels of openness in C2 over C4, which closely resembled the Zbtb16 expression pattern (Fig. 3i, j). The pseudotime trajectory and flow cytometry further indicated that Zbtb16 was enriched in both DN and CD4SP iNKTp (Fig. 3j, k). Interestingly, PLZFhi iNKT cells in CD4SP exhibit stronger TCR signaling strength compared to that in DN iNKT cells, indicated by the increased expression levels of PLZF, TCR, Nur77, and Vβ722,23 (Supplementary Fig. S7a). Given that iNKT2 differentiation requires the strongest TCR signaling compared to iNKT1 and iNKT1724,25, it is likely that PLZFhi iNKTp in CD4SP (C9) prefer to commit into the iNKT2 cell lineage.
RORγt is a key transcription factor to regulate iNKT17 differentiation but is also highly expressed in un-signaled DP thymocytes and promotes iNKT cell selection26. Unlike Zbtb16, Rorc (encode RORγt) at the +4 kb region was accessible in both DP (C14) and DN (C2 and C4) iNKTp, but not in the CD4SP (C9) cluster, while +13 kb to Rorc was only accessible in DP iNKTp (C14). The Rorc expression pattern is consistent with its chromatin accessibility status in each cluster (Fig. 3l). Rorc was significantly highly expressed in DP iNKTp and was gradually downregulated in the DN branch (C4 and C2) but was barely detected in CD4SP (C4) (Fig. 3m), which was further validated by flow cytometry (Fig. 3n). Interestingly, the distinct PLZFintRORγthi iNKT population in DN iNKTp exhibits a similar phenotype to the mature iNKT17 cells (Supplementary Fig. S7b). Thus, it is likely that iNKT17 commitment underwent the DP-DN developmental program in ST0 (C14-C4-C2). T-bet (encoded by Tbx21) is a key transcription factor to regulate iNKT1 cell differentiation27. Interestingly, we did not observe obvious Tbx21 expression in any clusters or open regions near Tbx21 in ST0 iNKTp (data not shown). Overall, our pseudotime-based analysis of developmental trajectories revealed that there might be two potential development programs in ST0, at which iNKT cells may initiate their commitment to iNKT2 and iNKT17 cells that occur as early as ST0 and may initiate iNKT1 cells post ST0. However, this hypothesis is still under further investigation.
iNKT cell developmental trajectory
We understand that iNKT cells that are presorted from ST0, ST1, ST2, and ST3 may not perfectly present the actual developmental path of their development. To test the robustness of our approach and analyze whether these cells can unveil the model of iNKT cell development, we projected cells from different developmental stages on Uniform Manifold Approximation and Projection (UMAP) of a recently published study from unbiased iNKT cells thymic population14. We found a similar distribution of developmental stages and clusters along the iNKT cell development (Supplementary Fig. S8a–c). Furthermore, the gene expressions of cells in each stage for those two datasets are highly correlated (Pearson’s r ≥ 0.95; Supplementary Fig. S8d). Thus, we thereafter focused our stage-based iNKT cells, which also provided another clue (e.g., stage related) for iNKT cell development. We found that iNKT cells, especially for iNKT1 cells, were following linear “stage” of development; however, iNKT2/17 cells were terminated their differentiation at ST2.
To understand the iNKT subset development post ST0, we mapped aggregated thymic iNKT cells (ST0–3) into a pseudotime trajectory (Fig. 4a and Supplementary Fig. S9a, b). Two branches stemming from ST0 were merged into a narrow window-C16 at ST1, where Ccr7 chromatin was much more accessible compared with that of other ST1 clusters (Supplementary Fig. S9c). The dynamic expression pattern of Zbtb16, Tbx21, and Rorc described three iNKT subset developmental trajectories, respectively (Supplementary Fig. S10a). Zbtb16hi iNKT2 (C3, C6, C7, C10, C11, C15, and C16) may initiate their development from both DP-CD4SP (predominately) and DP-DN branches in ST0 and continued throughout ST1 and ST2. Interestingly, C6 cells were terminally ended at ST1 as DN iNKT cells (Supplementary Fig. S10a, b); Tbx21+ iNKT1 cells start from CD4SP in ST1 and undergo brisk proliferation transitioned through iNKT2 clusters prior to the terminal iNKT1 differentiation in ST3. Among the iNKT1 pool, C12 as DN iNKT cells reach the end of the developmental journey (Supplementary Fig. S10a, b). Rorc+ iNKT17 cells (C13) rooted from the DP-DN branch in ST0 became terminally differentiated at ST2 as DN iNKT cells (Supplementary Fig. S10a, b). Collectively, our pseudotime-based analysis of developmental trajectories revealed that both iNKT1 and iNKT17 cells were at the ends of the trajectory, indicating that they were well differentiated. However, the majority of iNKT2 clusters (except C6) were centrally positioned along the iNKT cell differentiation axis suggesting a high plasticity in iNKT2 cells.
a The ordering of iNKT cells along pseudotime in a state-space defined by Monocle 3. Each color represents an iNKT cluster (left) and stage (right). b The same pseudotime plot as in (a); feature plots depicting single-cell gene expression trajectory of Zbtb16 in iNKT cell development. c Aggregate scATAC-seq browser tracks for Zbtb16 for iNKT cell clusters (left). Bar graph represents Zbtb16 average (Ave) expression in iNKT cell clusters (right). d UMAP projection colored by the activity of GATA3-binding motifs. e Heatmap showing pseudotime ordering of most DEGs selected from iNKT2 clusters C3, C6, C10, and C16 of scRNA-seq data. f Feature plots depicting single-cell gene expression trajectory of G1, G2/M, and S phages in aggregated iNKT cell development (left). Bar graph represents fraction of G1, G2/M and S cells in iNKT cells clusters (right). g DEGs in cluster C3, C6, C10, and C16. Pathway enrichment is expressed as the –log10 (P value) adjusted for multiple comparison.
Furthermore, an alternative computational approach, URD28, was performed, whereby cluster C14 from ST0 was used as the root point. Consistently, C13 (iNKT17) was the first to branch off from the trunk, followed by iNKT2 clusters (C10, C6, and C3), and subsequently, the iNKT1 clusters (C1, C5, C8, and C12) emerged. Notably, cluster C5 was branched out earlier than C1, C8, and C12 (Supplementary Fig. S10c). Overall, using URD approach, we further validated iNKT cell thymic development trajectory.
Cellular diversity in iNKT2 cells
iNKT2 cells showed an extensive diversity, including C3, C6, C7, C10, C11, C15, and C16 clusters (Figs. 2b and 4a, b). We first assessed Zbtb16 chromatin accessibility in integrated iNKT cell clusters (C1–C16). In 181 kb of the Zbtb16 locus, we found highly accessible regions in iNKT2, iNKT1 and iNKT17 clusters, but much weaker regions in ST0 clusters (C2, C4, C9, and C14). Zbtb16 expression was closely matched to its chromatin accessibility (Fig. 4c). iNKT2 clusters and C2 and C9 in ST0 displayed high activity of GATA3 motif-binding activity, which is critical for iNKT2 differentiation5 (Fig. 4d). As expected, most signature genes for iNKT2 cells gradually increased until reaching their peaks at an intermediate stage, followed by a down-regulation during the terminal differentiation. A few genes, however, including Btg1, Cebpb, and Osgin1, only appeared near the iNKT developmental end, which may be related to iNKT2 cell terminal events29,30,31 (Fig. 4e).
Previous studies indicated that iNKT cells in ST1 and ST2 undergo high proliferation. Cell-cycle pathway enrichment analysis suggested that iNKT2 clusters (C7, C11, and C15) exhibited highly proliferative characteristics, which were either in S phase (C7) or G2/M phase (C11 and C15) (Fig. 4f). Ingenuity Pathway Analysis (IPA) indicated that clusters C3, C6, C10, and C16 are functionally different. C16 is a transient phase of iNKT cells from ST0 to ST1, and these cells sharply elevated expression of genes associated with both glycolysis and oxidative phosphorylation (Fig. 4g), suggesting their increased energy demands32. C6 terminated at ST1, and these cells enriched with genes in TCR signals, co-stimulation signaling, cytoskeleton, TNFR, and cell death and exhaustion pathways (Fig. 4g). This cluster highly expresses Cd74 (Supplementary Fig. S11a), a gene associated with class II major histocompatibility complex and related to T cell–T cell interaction33. Consistently, flow cytometry analysis further confirmed that the majority of CD74+ iNKT cells were in ST1 (Supplementary Fig. S11b). However, the functional profiles of C6 are still under investigation. Taken together, iNKT2 exhibit a great cellular diversity.
Heterogeneity of iNKT1 cells
iNKT1 cell differentiation was controlled by transcription factor Tbx21 and assigned to four clusters (C1, C5, C8, and C12) (Figs. 2b and 5a). Within 17.2 kb of the Tbx21 locus, the promoter regions –3 kb and –4 kb were highly accessible in C1, C5, and C12, but were less accessible in C8 (Fig. 5b). The same regions were also accessible in hyper-proliferative C7, C11, and C15 iNKT2 clusters in ST1 and ST2. Importantly, Tbx21-binding motif activity also occurred in these clusters, but not in ST0 clusters (Fig. 5c). Flow cytometry analysis further confirmed that a small fraction of PLZFhiiNKT2 cells express T-bet in ST1 and ST2, with considerable proliferation ability as measured by Ki-67 (Fig. 5d). Therefore, it is possible that iNKT1 progenitors might hide in these so-called proliferative PLZFhiT-bethi iNKT2 cells.
a The same pseudotime plot as in Fig. 4a; feature plots depicting single-cell gene expression trajectory of Tbx21 in iNKT cell development. b Aggregate scATAC-seq browser tracks for Tbx21 for iNKT cell clusters (left). Bar graph represents Tbx21 average (Ave) expression in iNKT cell clusters (right). c UMAP projection colored by the activity of Tbx21-binding motif. d Representative flow plots of PLZF vs T-bet gated on iNKT cells, iNKT2 (PLZFhiT-bet−) in green and different iNKT1 (T-bethi) cells, marked by PLZF high (PLZFhiT-bethi) in blue, PLZF medium (PLZFintT-bethi) in purple, and PLZF low (PLZFloT-bethi) in red (top left). Histogram showing Ki-67 expression in the indicated iNKT subsets derived from the right. Representative flow plots of CD44 vs NK1.1 expression in the indicated iNKT sub-population derived from top (bottom). e The same pseudotime plot as in Fig. 4a; feature plots depicting single-cell gene expression trajectory of Slamf7 in iNKT cell development. f Representative flow plots of SLAMF7 vs NK1.1 expression in iNKT cells. PLZFhiT-betlo in orange, PLZFhiT-bethi in blue, PLZFint/loT-bethi in red. g Bar graph represents Slamf4 Ave expression in iNKT cell clusters. h Representative flow plots of SLAMF4 expression in iNKT2 (PLZFhiT-bet−) in green and different iNKT1 (T-bethi) cells, marked by PLZF high (PLZFhiT-bethi) in blue, PLZF medium (PLZFintT-bethi) in purple, and PLZF low (PLZFloT-bethi) in red. i Dot graph represents mean SLAMF4+ ± SD. n = 9, Data represent three independent experiments, and were analyzed by a two-sided paired t test, ****P < 0.0001. j Representative flow plots of IL-4 vs IFN-γ production in SLAMF4− (left) and SLAMF4+ (right) iNKT1 cells post PMA/Ionomycin stimulation for 4 h, n = 5, Data represent three independent experiments. k DEGs in clusters C1, C5, C8, and C12. Pathway enrichment is expressed as the –log10 (P value) adjusted for multiple comparison.
Pseudotime analysis showed that most iNKT1 signatures were not highly expressed until reaching the end of the iNKT development continuum (Supplementary Fig. S12a). We found that a novel signature, signaling lymphocytic activation molecule family member 7 (Slamf7), is enriched in iNKT1 cell clusters (Fig. 5e). Flow cytometry further confirmed this based on the strong correlation between SLAM7 and NK1.1 expression in PLZFloT-bethi iNKT1 cells (Fig. 5f). NK cell-related signature Slamf4 further distinguished terminal ended C12 from other iNKT1 cell clusters. SLAMF4+ iNKT cells (C12) were mainly DN, and gradually sprouted out from PLZFloT-bethi iNKT1 cells (Fig. 5g–i and Supplementary Fig. S12b, c). SLAMF4+ iNKT1 cells mainly secreted IFN-γ with less IL-4, like “classical iNKT1 cells”, while the majority of SLAMF4− iNKT1 cells secreted both IL-4 and IFN-γ upon stimulation (Fig. 5j). Furthermore, a small population of SLAMF4+PLZFloT-bethi iNKT1 cells also expressed soluble cytotoxic mediator Gzma, identified as a cytotoxic iNKT1 population (Supplementary Fig. S12d–f). However, these cytotoxic GZMA+SLAMF4+ iNKT1 cells were barely detected in the peripheral organs (Supplementary Fig. S12g). C5 cells were enriched with Ifit1 and Ifit3, which are involved in the interferon signaling pathway34 (Fig. 5k and Supplementary Fig. S12h). Overall, iNKT1 cells start their journey from as currently defined “iNKT2 cells” at ST1, and gradually complete their differentiation and turn into SLAMF4+ iNKT1 cells at the end, as classical IFN-γ-secreting iNKT1 cells.
iNKT17 cells exhibit limited heterogeneity
C13 was assigned as iNKT17 cells, which was segregated distinctively from other clusters (Figs. 2b and 6a). We observed that +4 kb and +14 kb regions of Rorc were accessible in ST0 clusters (C2, C4, and C14) and mature iNKT17 cluster (C13) (Fig. 6b), and binding motifs of RORC were activated in iNKT17 cluster as well as in ST0 iNKTp (Fig. 6c). These data suggested that iNKT cells may start their iNKT17 commitment at ST0 and complete their differentiation at C13. At mRNA level, Rorc was highly expressed in C14 in ST0, which was consistent with the chromatin accessibility at Rorc, but was gradually downregulated in C4 and C2, and re-upregulated in C13, indicating that other transcription factor(s) may target the open Rocr sites and manipulate Rorc expression during iNKT17 differentiation.
a The same pseudotime plot as in Fig. 4a; feature plots depicting single-cell gene expression trajectory of Rorc in iNKT cell development. b Aggregate scATAC-seq browser tracks for Rorc for iNKT cell clusters (left). Bar graph represents Rorc average (Ave) expression in iNKT cell clusters (right). c UMAP projection colored by the activity of RORC-binding motif. d Heatmap showing pseudotime ordering of top 30 genes in cluster 13 of scRNA-seq data. e The same pseudotime plot as in Fig. 4a; feature plots depicting single-cell gene expression trajectory of Aqp3 in iNKT cell development. f Representative flow plots of Aqp3 expression in iNKT1 (PLZF−RORγt−, gray), iNKT2 (PLZFhi RORγt−, orange) and iNKT17 (PLZFintRORγt+, purple). Bar graph represents mean Aqp3+ iNKT ± SD, n = 8. Data represent three independent experiments, and were analyzed by a two-sided paired t test, ****P < 0.0001.
To trace the developmental trajectory of iNKT17 cells, we further checked iNKT17 signature genes in an ordered iNKT cell trajectory and found that few iNKT17 signatures including Rorc were initially expressed in the early iNKTp, before they reached the mature iNKT17. However, the majority of iNKT17-related signatures were barely expressed in iNKTp (Fig. 6d). We further found that a novel signature aquaporin-3 (Aqp3) was specifically expressed in thymic iNKT17 (PLZFintRORγt+) cells (Fig. 6e, f). Overall, iNKT cells likely initiate iNKT17 commitment at ST0 and these iNKT17 cells exhibits limited heterogeneity.
Cbfβ regulates iNKT cell early commitment
Egr2 and Slamf6 were reported to control early iNKT cell development by modulating Zbtb16 expression and TCR in ST015,16,35. Here, we identified a novel co-transcription factor, Cbfβ, which showed a similar expression pattern with Egr2 and Slamf6, and exhibited a great enrichment at ST0 of iNKT cells, specifically in the DP-DN branch C2 (Fig. 7a, b).
a iNKT ST0 pseudotime plot; feature plots depicting single-cell gene expression trajectory of Cbfβ, Egr2, and Slamf6 at ST0 iNKT cells development (Fig. 3f). b Violin plots of Cbfβ expression in different stages of iNKT cells, and Cbfβ expression in different clusters (C2, C9, C4, and C14) from ST0 iNKT cells. c Representative flow plots of iNKT cells from Cbfβ KO and WT mice (left). Bar graphs represent mean ± SD of iNKT cell frequency and iNKT cell number (right), n = 5 for Cbfβ KO and WT controls. Data represent three independent experiments. d Representative flow plots of different stages of iNKT (after anti-CD1d-tetramer enrichment) from Cbfβ KO and WT mice (left). Bar graphs represent mean ± SD of iNKT cell frequency and iNKT cell number in Cbfβ KO and WT controls (right). e Representative flow plots of CD8 and CD4 expression in ST0 iNKT cells from Cbfβ KO and WT controls (left). Bar graph represents means ± SD of DP, DN, and CD4 SP ± SD (right). WT controls, n = 6; Cbfβ KO, n = 7. Data represent three independent experiments, data were analyzed by a two-sided unpaired t test, *P < 0.05. f, g Histogram showing PLZF expression (f) and Ki-67 expression (g) in ST1 iNKT cells from Cbfβ KO and WT mice. Bar graph represents means ± SD of indicated iNKT population. WT, n = 4. Cbfβ KO, n = 6. Data represent two independent experiments, and were analyzed by a two-sided unpaired t test, *P < 0.05, ***P < 0.001. h Representative flow plots of PLZF vs T-bet expression in ST1 iNKT cells from Cbfβ KO and WT mice. i Representative flow plots of PLZF vs RORγt expression in ST2 iNKT cells from Cbfβ KO and WT mice. j The speculated schematic model of mouse iNKT cell developmental trajectory (left) and the role of Cbfβ in iNKT cell development (right).
Cbfβ-encoded CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX (RUNX1, RUNX2, and RUNX3), and therefore modulates the transcription of their target genes. Here we observed that Runx1 shows a similar expression trajectory as Cbfβ in thymic iNKT cells (Supplementary Fig. S13a) even though the expression level is relative lower. More interestingly, consistent with the high expression of Cbfb in C2, we observed a greater RUNX1-binding activity in C2, compared with in other clusters in ST0 (Supplementary Fig. S13b). In mammals, two RNA splice variants, Cbfβ1 and Cbfβ2, are generated from a single Cbfβ gene, and each variant has distinct amino acid sequences at the C terminus36. To determine if Cbfβ regulates iNKT development, we examined iNKT cell development in thymic-specific Cbfβ knockout mice (CD4CreCbfβ f/f, Cbfβ KO), in which both Cbfβ1 and Cbfβ2 were deleted. Cbfβ deletion was further confirmed in protein level and mRNA level in different T-cell subsets (Supplementary Fig. S14a, b). We observed that conventional αβT cells with Cbfβ deletion phenotypically mimicked those in RUNX3 KO mice37, as Cbfβ KO mice displayed abnormalities in CD4 expression, impairment of CD8 T cells maturation in thymus, and a large proportion of DP cells in peripheral (Supplementary Fig. S14c, d). Interestingly, deletion of Cbfβ led to a severe reduction in the frequency and absolute number of thymic iNKT cells (Fig. 7c), as well as in the peripheral lymphoid organs (Supplementary Fig. S14e), which phenotypically mimicked iNKT cells in Runx1 deletion mice38, but not Runx3 KO mice. This phenomenal explained a similar expression pattern of Cbfβ and Runx during iNKT cell development and higher binding activity of RUNX1 at C2 ST0 (Supplementary Fig. S13a, b). A more comprehensive analysis revealed a selective and significant reduction in the frequency and absolute number of ST2 and ST3 iNKT cells in Cbfβ KO mice. The frequencies of ST0 and ST1 iNKT cells were significantly increased in Cbfβ KO mice, but the absolute numbers were comparable between Cbfβ KO and WT controls (Fig. 7d). Of interest, DP iNKTp were increased in ST0 iNKT cells in Cbfβ KO mice (Fig. 7e), suggesting that the deletion of Cbfβ partially blocks DP iNKTp conversion to either CD4SP or DN lineages. We assume that Cbfβ may affect iNKT cells selection at DP. Egr2 showed a similar expression pattern as Cbfβ in ST0 (Fig. 7a), and is critical for iNKT lineage commitment at DP35, we therefore detected Egr2 expression at DP stage. As shown in Supplementary Fig. S14f, Egr2 expression was significantly reduced in DP thymocytes from Cbfβ KO mice, indicating that Egr2 may involve in Cbfβ-mediated iNKT cell development.
ST1 iNKT cell undergoes briskly proliferation and contains the progenitors of iNKT subsets with high Zbtb16-encoded PLZF expression. Here we observed that PLZF expression at ST1 was significantly downregulated in Cbfβ KO iNKT cells (Fig. 7f). Similar phenomenon was also observed on a proliferating marker, Ki-67 (Fig. 7g). These data suggested that Cbfβ-deficient iNKT cells entered a relative quiescent status and were unable to normally upregulate PLZF expression at ST1. Furthermore, the remnant PLZF+ iNKT cells in ST1 fail to co-express T-bet to initiate iNKT1 differentiation and to co-express RORγt to initiate iNKT17 differentiation (Fig. 7h, i and Supplementary Fig. S14g). Bone marrow chimera transfer experiments showed that there was a sever defect on iNKT cell development from the Cbfβ-deficient donors (Supplementary Fig. S15a, b), suggesting that the defective iNKT cell development in Cbfβ KO mice was cell-intrinsic. Overall, our study suggests that Cbfβ serves as a key regulator to control early iNKT cell development at ST0, iNKT cell differentiation at ST1/2, and final maturation at ST3 (Fig. 7j).
Discussion
iNKT cell development was previously considered as a linear model with four successive stages (ST0–ST3). However, recent studies have indicated that thymic iNKT cell development is a complex cellular differentiation process and the linear developmental model does not apply to all iNKT subsets13. A new functional classification of three terminally differentiated subsets, iNKT1/2/17, was proposed based on transcription factor expression and cytokine production patterns12,26. Very recently, Thomas Baranek et al. performed scRNA-seq on thymic whole iNKT cells pool and proposed a model for iNKT cell effector differentiation in which iNKT1 and iNKT17 subsets derive from iNKT2. Moreover, iNKT1 subset arises linearly and sequentially from iNKT2 cells39. This study yielded strong evidence for iNKT cell development and differentiation models. However, are iNKT2 cells the earliest cells pool for iNKT1/17 lineages commitment? If no, how early can their progenitor be identified? To this end, we applied both scRNA-seq and scATAC-seq analyses of thymic iNKT cells and mapped the developmental landscape of terminal iNKT1/2/17 subsets on iNKT development. We did single-cell analysis based on “stages” of development, given the following reasons: (1) There might be some key features of iNKT cells at the very early stage, which only take up about 0.5% of total iNKT cells in mouse thymus, and sorting whole iNKT cells as a pool may hide key features in this rare population. Thus, sorting iNKT cells based on the stages would make sure that enough iNKT cells at earlier stages were included for further clustering analysis; in this case, we included a total of 7591 ST0 cells for scRNA-seq and scATAC-seq analysis; (2) By pooling different stages of iNKT cells, we were also able to assign iNKT1/2/17 into different clusters from different developmental stages based on published signature markers. And this could allow us to trace iNKT1/2/17 differentiation trajectory in different stages and identify their progenitors. With the integration of single-cell transcriptome and chromatin accessibility analysis, we found that iNKT2 and iNKT17 lineage commitment may occur in ST0 by two distinct programs, and iNKT1 lineage commitment may occur post ST0. Finally, we identified that transcription factor Cbfβ plays a key role in iNKT cell commitment.
Previous studies suggested that iNKT cells arise from a common progenitor designated as PLZFhiCD24+ in ST0, which further differentiate into iNKT1/2/17 subsets5,40. However, it is still unclear how and at which specific time window their effector programs unfold during their development. In our current study, we found that ST0 iNKTp exhibit extreme heterogeneity, suggesting that iNKTp may be destined to the specific subset lineages in ST0. A rare DP iNKT cell population that has been ignored previously and hidden in ST0 is likely the earliest iNKTp post-positive selection. These DP iNKTp then differentiate to either CD4SP or DN iNKTp in ST0, and form two distinct developmental programs, DP-CD4SP and DP-DN. The two developmental branches differed in many aspects including distinct transcriptomes, diversity of TCR Vβ usage, and TCR signaling strength. Interestingly, Zbtb16 upregulation only occurred at the end of DP-CD4SP and DP-DN developmental branches. Zbtb16+ iNKTp in the CD4SP path showed a stronger TCR signaling strength and increased Vβ7 usage compared to that in DP-DN iNKTp, suggesting that the iNKTp in the DP-CD4SP development program prefer to differentiate into iNKT2 cells. However, Rorc chromatin accessibility and gene expression are highly enriched in DP iNKTp and the DP-DN developmental branch, supporting the notion that iNKTp in the DP-DN branch prefer to be differentiated into iNKT17 cells. We did not observe any open-chromatin regions near Tbx21 and its gene expression in ST0 iNKTp, but Tbx21 chromatin accessibility and its gene expression occurred as early as ST1. These findings highly suggest that iNKT2 and iNKT17 are pre-determined at ST0; however, iNKT1 are progressively determined at ST1.
Although three subsets of iNKT cells were defined post ST0 based on transcription factor and cytokine secretion profiles, iNKT1 and iNKT2 cells exhibit extensive phenotypic and functional heterogeneity, while iNKT17 cells are relatively homogenous. The trajectory of iNKT development showed that iNKT17 and iNKT1 branches were both sprouted from the developmental tree. However, the iNKT2 cells, which were highly proliferative and heterogeneous, were located at the center of developmental trunk. Interestingly, we found that both Tbx21 and Rorc chromatins were also accessible in these proliferative iNKT2 clusters, indicating that these proliferative clusters hidden in so-called “iNKT2 cells” might contribute to early iNKT1/2/17 expansion and differentiation. These results further supported a recent notion that currently defined iNKT2 cells may contain mature iNKT2, transitioning iNKT17, and transitioning iNKT141. Aqp3 belongs to a family of highly conserved transmembrane channels that transport water and, in some cases, small solutes such as glycerol. Recent studies indicated that Aqp3 is expressed on T cells and regulates their trafficking in skin and lung immune reactions42,43. T-cell migration toward chemokines is dependent on Aqp3-mediated hydrogen peroxide (H2O2) uptake. Here, our fate-mapping identified that Aqp3, as a new biomarker, is specifically expressed in iNKT17 cells. It will be very interesting to further investigate Aqp3’s functions in iNKT17 cells, especially for their trafficking to peripheral organs.
iNKT cells express multiple Slam family receptors, but only Slamf1, Slamf5 and Slamf6 are highly expressed in ST0 iNKTp (data not shown), which have been reported to be required for iNKT cell development15. Here, we found that Slamf7, as a new marker, is specifically expressed in iNKT1 cells. Most importantly, we found that Slamf4 was enriched in terminal DN iNKT1cluster (C12). Given the dynamic transcription factor expression and cytokine production profiles, the Slamf4+ iNKT1 cells are likely the terminal differentiated iNKT1 cells as “classical iNKT1 cells” that highly secrete IFN-γ. Interestingly, the few Slamf4+ iNKT1 cells might receive attention as novel cancer therapeutic targets.
iNKT10 cells are a novel iNKT subset identified upon stimulation of the strong agonist αGalcer, which are characterized by transcription factor Nfil3 (also term E4bp4) and IL-10 production44. Previous study suggested that the appearance of iNKT10 after αGalcer stimulation could be the result of selective expansion of a rare population of pre-existing iNKT10 cells or some non-iNKT10 cells converting into the iNKT10 phenotype. It is, however, important to point out that Nfil3 was surprisingly found to be enriched in thymic iNKT17 cluster and a small fraction of iNKT1 cells. However, IL-10 expression was undetectable in thymic iNKT cells (data not shown). It is still unknown whether these Nfil3+ iNKT cells can be converted into iNKT10 cells following αGalcer stimulation. More comprehensive study is still ongoing.
Collectively, our study generated a comprehensive atlas of thymic iNKT cells and their developmental trajectory, providing a valuable resource for future studies of iNKT cell biology. We have also uncovered Cbfβ as a novel regulator of early iNKT cell development (Fig. 7j).
Materials and methods
Mice
C57BL/6 were purchased from Jackson Laboratory (Bar Harbor, ME). Rec-Vα14Tg TCR transgenic mice were generated in Dr. Derek Sant’Angelo laboratory9, which require Rag-mediated recombination to produce a functional TCR (Supplementary Fig. S1a). These mice have increased numbers of iNKT cells, as compared with C57BL/6 mouse. Mice carrying a conditional floxed allele of Cbfβ (Cbfβfl/fl) were previously described45 and provided by Dan R. Littman (New York University, New York, NK). Mice were backcrossed to the C57BL/6 background for 7 generations and then mated to C57BL/6 mice carrying the Cd4 enhancer/promoter/silence Cre allele (obtained from The Jackson Laboratory), to generate CD4CreCbfβ fl/fl conditional knockout mice (Cbfβ KO). The full list of mouse strains used can be found in Supplementary Table S1. 5-week-old, sex-matched mice were utilized in this study. All studies, protocol, and mouse handling were approved by the Institutional Animal Care and Use Committee.
Flow cytometry gating strategy and antibodies
Single-cell suspensions were washed twice with FACS staining buffer (1× PBS, 2% FBS) and incubated with Fc block (clone 2.4G2). Cells were stained with anti-mouse PBS57-loaded and -unloaded CD1d-tetramer (provided by the NIH Tetramer Core Facility), the following fluorescence conjugated antibodies were used: anti-TCRβ (H57-597), anti-CD24 (M1/69), anti-CD44 (IM7), anti-NK1.1 (PK136), anti-TCR Vβ8.1/8.2 (KJ16-133.18), anti-TCR Vβ8.3 (1B3.3), anti-TCR Vβ7 (TR310), anti-Nur77 (12.14), anti-Ly6d (49-H4), anti-CD5 (53-7.3), anti-CD6 (IM348), anti-Egr2 (erongr2), anti-ID2 (ILCID2), anti-IL7R (A7R34), anti-Cbfβ, anti-Aqp3, anti-CD8 (53-6.7), anti-CD4 (GK1.5), anti-RORγt (B2D), anti-PLZF (Mags.21F7), anti-T-bet (eBio4B10 (4B10)), anti-Slamf7 (520914), anti-Slamf4 (eBio244F4), anti-CD45.1(A20), and anti-CD45.2 (104). Cell surface staining was performed with staining buffer; intranuclear staining for anti-Aqp3, anti-PLZF, anti-T-bet, and anti-RORγt were performed with eBioscience Fixation/permeabilization buffer. The flow cytometry assay was performed through BD FACSCelesta and data were analyzed using FlowJo V10.2 software. Gating strategy: after gating on lymphocyte, doublets were excluded by using forward scatter (FSC), and side scatter (SSC); mouse iNKT cells were further identified as TCRβ+CD1d-tetramer+. The full list of antibodies, reagent, and software used can be found in Supplementary Table S1.
Bone marrow chimeras transfer experiment
To generate bone marrow chimeras, 7- to 8-week-old C57BL/6. SJL (B6.SJL) recipient mice were lethally irradiated initially with 9.5 Gy with a dose rate of 2.5 Gy per min. Quality assurance of the radiation exposure was performed using multiple dosimetry endpoints including an electrometer, micro-TLDs, and Gafchromic film. Donor bone marrows were harvested from age- and sex-matched SJL (CD45.1+) and Cbfβ KO (CD45.2+) or WT control mice (CD45.2+). After erythrocyte lysis, mature T cells (CD3+) were depleted by biotin-conjugated anti-mouse CD3 (BD Biosciences) mAbs and anti-biotin magnetic beads (BD Biosciences) from bone marrows of each donor, using Magni SortTM. Over 90% of mature T-cell depletion was confirmed by flow cytometry. CD45.1+ SJL and CD45.2+ Cbfβ KO or CD45.2+ WT littermate control bone marrows were mixed at a 1:1 ratio, and 1 × 107 cells per mouse (in a volume of 100 μL) were then injected into the irradiated recipients via tail vein. The chimeras were analyzed 8 weeks after reconstitution.
Mouse iNKT cell enrichment and sorting
Mouse thymi were harvested from 5-week-old C57BL/6 mice and rec-Vα14Tg mice. For the enrichment of iNKT cells, thymocytes were stained with APC-conjugated CD1d-tetramer, bound to anti-APC MicroBeads (Miltenyi Biotec), and enriched with an autoMACS Pro Separator (Miltenyi Biotec) using POSSEL_S program. ST1 (CD24−CD44loNK1.1−), ST2 (CD24-CD44hiNK1.1−), and ST3 (CD24−CD44hiNK1.1+) iNKT cells were further sorted from C57BL/6 mouse thymus, and ST0 iNKT cells (CD24+) were sorted from both C57BL/6 and rec-Vα14Tg mice using FACSAria II Usage.
Quantitative RT-PCR analysis
Total RNA was isolated by Sigma-Aldrich kit and was quantified by a NanoDrop ND-1000 spectrophotometer. The A260/A280 ratio was > 1.9 for all the samples. cDNA was prepared by using a cDNA synthesis kit (Sigma-Aldrich) following the manufacturer’s instruction. Primer pairs for Cbfβ are: forward (F): 5′-GGTTAGGAGTCATTGTGATCA-3′; reverse (R): 5′-CCTCCTCATTCTAACAGGAATC-3′. The PCR amplification was carried out on the Applied Biosystem 7900 Real-time PCR system; relative quantification using ΔCT values in the cells from Cbfβ KO vs WT control was carried out, and fold changes were calculated.
Western blot analysis
The T-cell protein lysates were prepared using a RIPA buffer containing protease inhibitors. Next, the T-cell protein lysates were subjected to 12% SDS-PAGE analysis with 10 µg of lysate loaded per lane. The anti-Cbfβ rabbit polyclonal antibody was obtained from ThermoFisher Scientific. The anti-GAPDH monoclonal antibody was purchased from Sigma-Aldrich.
ScRNA-seq library generation
Two biological repeats for each stage of scRNA-seq libraries were generated using the 10X Genomics Chromium Single Cell 3′ Reagent Kit (v2 Chemistry) and Chromium Single Cell Controller as previously described46. Briefly, ~5000 cells were loaded into each reaction for gel bead-in-emulsion (GEM) generation and cell barcoding. Reverse transcription of the GEM (GEM-RT) was performed in a Thermocycler (Veriti™ 96-Well Fast Thermal Cycler, Applied Biosynthesis; 53 °C 45 min, 85 °C 5 min, 4 °C hold). cDNA amplification was performed after GEM-RT cleanup with Dynabeads MyOne Silane (ThermoFisher Scientific) with the same Thermocycler (98 °C 3 min; 98 °C 15 s, 67 °C 20 s, 72 °C 1 min, repeat 12 cycles; 72 °C 1 min, 4 °C hold). Amplified cDNA was cleaned up with SPRIselect Reagent Kit (Beckman Coulter) followed by library construction procedure, including fragmentation, end repaired, adaptor ligation, and library amplification. A Bioanalyzer (Agilent) was used for library quality control. cDNA libraries were sequenced on an Illumina HiSeq 4000 using paired-end flow cells (Read 1, 26 cycles, i7 index 8 cycles; Read 2: 110 cycles) by the University of Michigan DNA Sequencing Core facility following the manufacturer’s protocol.
ScRNA-seq data analysis
Sequenced reads from scRNA-seq libraries were demultiplexed, aligned to the mm10 mouse reference, barcode processed, and Unique Molecular Identifier (UMI) counted using the 10X Genomics Cell Ranger (v2.0.1) pipeline46. Estimated number of cells captured per sample were between 1296–3290 with 34,258–95,264 mean reads per cell, 764–1892 median genes per cell, and 1266–6412 median UMI counts per cell. A total of 19,044 cells with 4238 UMI counts/cell in average were selected via Cell Ranger for further analysis for all of six samples. Datasets were subsequently analyzed using the R Seurat package10,11. Principle Component Analysis (PCA) was employed to analyze combined samples. Quality control metrics employed are as follows. We employed two strategies to identify potential doublets. Firstly, cells expressing both Xist and Y chromosome genes (Kdm5d, Eif2s3y, Gm29650, Uty, and Ddx3y) were excluded from the dataset. Secondly, cells expressing uncharacteristically high numbers of genes (> 4000) were excluded. Low-quality cells were excluded based on a low number of genes detected (< 300) and/or having high mitochondrial genetic content (> 5%). Additionally, uninteresting sources of variation within the data were removed. Genes removed include ribosomal structural proteins (as identified by gene ontology term GO:0003735 and the Ribosomal Protein Gene (RPG) database47), non-coding rRNAs, Hbb, and genes not expressed in ≥ 3 cells. A total of 13,578 genes in 17,944 cells passed these quality control measures.
A global-scaling normalization method “LogNormalize” in Seurat was employed to normalize gene expression measurements of each cell by the total expression, multiplied by a factor of 10,000, followed by log-transformation. Highly variable genes in each data analysis were identified, and the intersecting top 1000 genes in each dataset were used for clustering and downstream analyses. Datasets underwent scaling and regressing on the number of detected molecules per cell and the percentage mitochondrial gene content (pct.mito). The number of principal components (PCs) used to cluster cells was determined by manual inspection of the scree plot. After identifying the number of PCs to be included for downstream analyses (25 PCs), a graph-based clustering approach implemented in Seurat was used to iteratively cluster cells into groups, based on similarities of those components among cells. The t-Distributed Stochastic Neighbor Embedding (t-SNE) method was utilized to visualize resulting clusters. To assess the effects of cell cycle heterogeneity, cell cycle phase scores (G2/M and S phases) were calculated based on canonical markers and used to regress out the data48. The FindAllMarkers function in Seurat was then implemented to identify DEGs between clusters with a fold-change of > 2 and a Bonferroni adjustment of P value < 0.05 as a statistical significance threshold. To determine if DEGs belong to identifiable groups, pathway analysis was carried out IPA (Qiagen Bioinformatics, Redwood City, CA).
Comparison analysis between our iNKT scRNA-seq dataset with the one from ref. 14
To compare our scRNA-seq dataset with recently published scRNA-seq data from unbiased iNKT cell thymic populations by Krovi et al.14 (GEO accession: GSE152786), an integration analysis implemented in the R Seurat package was employed49. After performing a normalization using regularized negative binomial regression (the SCTransform function) for each dataset, a set of cell pairs or “anchors” between datasets that are assumed to have a similar biological state was identified (the SelectIntegrationFeatures, PrepSCTIntegration and FindIntegrationAnchors functions) using a canonical correlation analysis (CCA) and mutual nearest neighbors (MNNs). These ‘anchors’ are utilized as the reference to merge the datasets using the IntegrateData function. The iNKT stages and cluster labels from our scRNA-seq dataset are then projected onto the Krovi et al.’s dataset via the FindTransferAnchors and TransferData functions.
ScATAC-seq library generation
FACS-sorted stages 0, 1, 2, and 3 iNKT cells from C57BL/6 and rec-Vα14Tg (ST0) mouse thymi were used to generate Chromium Single Cell ATAC Libraries following the recommended protocol (10X Genomics, Pleasanton, CA) as previously described50. Briefly, after isolating, washing, and counting of nuclei suspensions, the recovered nuclei were combined with the ATAC buffer and enzyme to create a transposition mix in a tube and incubated at 37 °C for 60 min. A master mix that includes the barcoding reagent, reducing agent, and barcoding enzyme was then added to the same tube. This solution was loaded into the 10X Genomics Chromium Single Cell Controller instrument along with the vortexed scATAC gel beads and partition oil. The resulting GEMs were amplified in a Thermocycler (Veriti™ 96-Well Fast Thermal Cycler, Applied Biosynthesis; 72 °C for 5 min, 98 °C for 30 s, cycled 12×: 98 °C for 10 s, 59 °C for 30 s, and 72 °C for 1 min). After cleaning up using the Recovery Agent, Dynabeads MyOne Silane, and SPRIselect Reagent, the resulting accessible DNA fragments underwent the barcoded and indexed sequencing library construction. These scATAC-seq libraries were sequenced on the Illumina NovaSeq 6000 using paired-end flow cells (50 bp Read 1, 8 bp i7 index, 16 bp i5 index, and 49 bp Read 2) by the University of Michigan DNA Sequencing Core facility following the manufacturer’s protocol.
ScATAC-seq data processing
Sequenced reads from scATAC-seq libraries were processed using the 10X Genomics Cell Ranger ATAC (v1.0.1). Specifically, raw sequencing reads were demultiplexed and converted into FASTQ files using the cellranger-atac mkfastq pipeline. Then, the sequencing reads underwent the cellranger-atac count pipeline consisting of read filtering, alignment to the reference genome using STAR, barcode counting, identification of transposase cut sites and accessible chromatin peaks, cell calling, generation of count matrices for chromatin peaks and transcription factors. Estimated number of cells captured per sample were between 2536–11,015 with 11,401–16,343 median fragments per cell and 55,349–87,603 total peaks detected. Then, the output files from multiple samples were combined using the cellranger aggr command with read depth normalization to generate a single feature-barcode matrix for all the data. A total of 39,428 cells with a median of 11,398 fragments per cell were selected for further analysis for all samples using the R Seurat and Signac packages (https://satijalab.org/signac/). Cells with sufficient numbers of fragments in peaks (> 3000 and < 100,000), a high percentage (> 20%) of fragments in peaks, a blacklist ratio (a ratio of reads in the regions associated with artificial signals based on the ENCODE Project) of < 0.02 and a nucleosome signal strength of < 10 were kept. A frequency-inverse document frequency (TF-IDF) normalization was performed to normalize sequencing depth differences across cells and provide higher weights to rare peaks using the RunTFIDF function. For dimensionality reduction, a singular value decomposition (SVD) was performed using the RunSVD function after selecting the top 95% of peaks via the FindTopFeatures(object, min.cutoff = “q5”) function. Non-linear dimensionality reductions using t-SNE (the RunTSNE function on the first 30 dimensions) and UMAP51, the RunUMAP function on the first 30 dimensions) methods for visualization, constructing a Shared Nearest Neighbor (SNN) graph (the FindNeighbors function), cell clustering (the FindClusters function with a resolution of 0.6), and cluster differential accessibility (the FindAllMarkers function) were conducted subsequently. To quantify the chromatin accessibility at the gene levels, the fragments for each cell that intersect the gene and promoter (2000 bp upstream) regions were counted. The gene coordinates were obtained from the Ensembl Mus musculus v79 (EnsDb.Mmusculus. v79 R package). To perform motif analysis, the presence of known transcription factor-binding motifs based on the JASPAR 2018 database was scanned within the DNA sequence of each peak. Hence, over-representative motifs in a specific cluster can be identified using the FindMotifs function.
Integrated analysis of scRNA-seq and scATAC-Seq
To perform an integrative analysis of scATAC-seq and scRNA-seq data, an anchoring approach as implemented in the R Seurat and Signac packages was employed49. Briefly, a set of cell pairs or “anchors” between datasets assumed to share a similar biological state was first identified (the FindTransferAnchors function) using a CCA and MNNs. These “anchors” are then used as the reference to integrate the datasets and the cluster labels from scRNA-seq data can be projected onto the scATAC-seq data via the TransferData function. Hence, single-cell measurements across different modalities can be evaluated.
Pseudotime analysis
Pseudotime analysis of scRNA-seq data was performed using Monocle52 (v. 2.99.3) to infer the trajectories of cells during their development. Monocle utilizes the reversed graph embedding technique to construct the trajectories based on the cell similarity in gene expression profiles. Specifically, the size factors for each cell and dispersion function for the genes in the data were first calculated. Next, the data were projected onto the top principal components using the preprocessCDS function with parameters as follows: (method = “PCA"', norm_method = “log”, num_dim = 30) and the UMAP dimensionality reduction using the reduceDimension function (method = ‘UMAP’, metric = “cosine”, min_dist = 0.75, n_neighbors = 100, num_dim = 30) were applied based on the top 2000 of highly variable genes. After partitioning the cells into groups using the partitionCells function, Monocle organizes cells into trajectories based on the concept of reversed graph embedding called SimplePPT, which learns a tree-like trajectory, using the learnGraph function (RGE_method = ‘SimplePPT’, prune_graph = F, close_loop = T).
IPA
To infer the functionality of subset-specific gene signatures and open-chromatin regions, pathway analysis using the IPA (Qiagen Bioinformatics, Redwood City, CA) was performed. This analysis provides information on the potential functions of novel cell subsets that can be validated in mechanistic studies.
Statistical analysis
For comparison between groups, statistical analysis was performed by unpaired t test with GraphPad Prism 8.0.
Data availability
ScRNA-seq and scATAC-seq data that support the findings of this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with the accession numbers GSE130184 and GSE141825, respectively (Supplementary Table S1). All relevant data are available from the authors upon reasonable request.
References
Bendelac, A., Savage, P. B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Godfrey, D. I. & Berzins, S. P. Control points in NKT-cell development. Nat. Rev. Immunol. 7, 505–518 (2007).
McEwen-Smith, R. M., Salio, M. & Cerundolo, V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol. Res. 3, 425–435 (2015).
White, A. J. et al. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J. Immunol. 192, 2659–2666 (2014).
Lee, Y. J., Holzapfel, K. L., Zhu, J., Jameson, S. C. & Hogquist, K. A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154 (2013).
Grun, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015).
Nguyen, A., Khoo, W. H., Moran, I., Croucher, P. I. & Phan, T. G. Single cell RNA sequencing of rare immune cell populations. Front. Immunol. 9, 1553 (2018).
Kovalovsky, D., Pezzano, M., Ortiz, B. D. & Sant’Angelo, D. B. A novel TCR transgenic model reveals that negative selection involves an immediate, Bim-dependent pathway and a delayed, Bim-independent pathway. PLoS ONE 5, e8675 (2010).
Thapa, P. et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat. Commun. 4, 1582 (2013).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Engel, I. et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17, 728–739 (2016).
Das, R., Sant’Angelo, D. B. & Nichols, K. E. Transcriptional control of invariant NKT cell development. Immunol. Rev. 238, 195–215 (2010).
Harsha Krovi, S. et al. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun. 11, 6238 (2020).
Lu, Y. et al. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat. Immunol. 20, 447–457 (2019).
Lazarevic, V. et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009).
Coquet, J. M. et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc. Natl. Acad. Sci. USA 105, 11287–11292 (2008).
Cameron, G. et al. Antigen specificity of type I NKT cells is governed by TCR beta-chain diversity. J. Immunol. 195, 4604–4614 (2015).
Inlay, M. A. et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23, 2376–2381 (2009).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Wang, H. & Hogquist, K. A. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. eLife 7, e34793 (2018).
Schumann, J., Mycko, M. P., Dellabona, P., Casorati, G. & MacDonald, H. R. Cutting edge: influence of the TCR V beta domain on the selection of semi-invariant NKT cells by endogenous ligands. J. Immunol. 176, 2064–2068 (2006).
Wei, D. G. et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J. Exp. Med. 202, 239–248 (2005).
Zhao, M. et al. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat. Commun. 9, 2627 (2018).
Tuttle, K. D. et al. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 9, 2650 (2018).
Constantinides, M. G. & Bendelac, A. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25, 161–167 (2013).
Townsend, M. J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494 (2004).
Farrell, J. A. et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018).
Winkler, G. S. The mammalian anti-proliferative BTG/Tob protein family. J. Cell Physiol. 222, 66–72 (2010).
Gade, P., Roy, S. K., Li, H., Nallar, S. C. & Kalvakolanu, D. V. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol. Cell. Biol. 28, 2528–2548 (2008).
Brennan, M. S., Matos, M. F., Richter, K. E., Li, B. & Scannevin, R. H. The NRF2 transcriptional target, OSGIN1, contributes to monomethyl fumarate-mediated cytoprotection in human astrocytes. Sci. Rep. 7, 42054 (2017).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019).
Holling, T. M., Schooten, E. & van Den Elsen, P. J. Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum. Immunol. 65, 282–290 (2004).
Fensterl, V. & Sen, G. C. Interferon-induced Ifit proteins: their role in viral pathogenesis. J. Virol. 89, 2462–2468 (2015).
Seiler, M. P. et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13, 264–271 (2012).
Ogawa, E. et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194, 314–331 (1993).
Woolf, E. et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA 100, 7731–7736 (2003).
Mao, A. P., Ishizuka, I. E., Kasal, D. N., Mandal, M. & Bendelac, A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 8, 863 (2017).
Baranek, T. et al. High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. 32, 108116 (2020).
Georgiev, H., Ravens, I., Benarafa, C., Forster, R. & Bernhardt, G. Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat. Commun. 7, 13116 (2016).
Verykokakis, M. & Kee, B. L. Transcriptional and epigenetic regulation of innate-like T lymphocyte development. Curr. Opin. Immunol. 51, 39–45 (2018).
Hara-Chikuma, M. et al. Chemokine-dependent T cell migration requires aquaporin-3-mediated hydrogen peroxide uptake. J. Exp. Med. 209, 1743–1752 (2012).
Ikezoe, K. et al. Aquaporin-3 potentiates allergic airway inflammation in ovalbumin-induced murine asthma. Sci. Rep. 6, 25781 (2016).
Sag, D., Krause, P., Hedrick, C. C., Kronenberg, M. & Wingender, G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J. Clin. Invest. 124, 3725–3740 (2014).
Egawa, T. & Littman, D. R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9, 1131–1139 (2008).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Nakao, A., Yoshihama, M. & Kenmochi, N. RPG: the ribosomal protein gene database. Nucleic Acids Res. 32, D168–D170 (2004).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Diaz-Papkovich, A., Anderson-Trocme, L., Ben-Eghan, C. & Gravel, S. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet. 15, e1008432 (2019).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Acknowledgements
We thank the National Institutes of Health Tetramer Core Facility for supplying CD1d tetramers for mouse iNKT cell flow cytometry analysis and for cell sorting. We thank Dr. Karla Passalacqua for editorial assistance and Dr. Stephen Brown for assisting irradiation for bone marrow charisma transfer experiments. We thank all laboratory members for their support and encouragement. This work was partially supported by Henry Ford Immunology Program Grants T71017 (to L.Z.) and T71016 (to Q.-S.M.) and National Institutes of Health Grants 1R01 AI119041-01A1 (to Q.-S.M.), 1R61AR076803-01, R33AR076803, and R21AR079089 (to Q.-S.M. and I.A.), 1R56AI119041-01 (to Q.-S.M.), and AI71922 (to M.K.).
Author information
Authors and Affiliations
Contributions
Q.-S.M. and L.Z. designed the study; J.W., T.L., K.S., Q.Y., and X.W. performed the experiments; X.W. and C.Y prepared scRNA-seq and scATAC-seq libraries; I.A. and J.W. analyzed and interpreted the data, I.D. and I.L. performed single-cell sequence processing with 10X cell Ranger, Seurat, Signac, and IPA; D.B.S. generated rec-Vα14Tg mice; M.K. provided critical comments and contribution to manuscript writing; J.W., I.A., L.Z., and Q.-S.M. wrote the manuscript, which was reviewed and approved by all the authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, J., Adrianto, I., Subedi, K. et al. Integrative scATAC-seq and scRNA-seq analyses map thymic iNKT cell development and identify Cbfβ for its commitment. Cell Discov 9, 61 (2023). https://doi.org/10.1038/s41421-023-00547-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-023-00547-x