Abstract
The prokaryotic CRISPR-Cas adaptive immune systems provide valuable resources to develop genome editing tools, such as CRISPR-Cas9 and CRISPR-Cas12a/Cpf1. Recently, CRISPR-Cas12b/C2c1, a distinct type V-B system, has been characterized as a dual-RNA-guided DNA endonuclease system. Though being active in vitro, its cleavage activity at endogenous genome remains to be explored. Furthermore, the optimal cleavage temperature of the reported Cas12b orthologs is higher than 40 °C, which is unsuitable for mammalian applications. Here, we report the identification of a Cas12b system from the Alicyclobacillus acidiphilus (AaCas12b), which maintains optimal nuclease activity over a wide temperature range (31 °C–59 °C). AaCas12b can be repurposed to engineer mammalian genomes for versatile applications, including single and multiplex genome editing, gene activation, and generation of gene mutant mouse models. Moreover, whole-genome sequencing reveals high specificity and minimal off-target effects of AaCas12b-meditated genome editing. Our findings establish CRISPR-Cas12b as a versatile tool for mammalian genome engineering.
Similar content being viewed by others
Introduction
CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated protein) systems provide adaptive immunity in archaea and bacteria by employing a combination of Cas effector proteins and CRISPR RNAs (crRNAs)1,2,3,4. To date, two classes (class 1 and 2) including six types (type I–VI) CRISPR-Cas systems are characterized based on pronounced functional and evolutionary modularity5. Among class 2 CRISPR-Cas systems, type II Cas9 systems and type V-A Cas12a/Cpf1 systems have been harnessed for genome editing and hold tremendous promise for biomedical research6,7,8,9.
Recently, the type V-B CRISPR-Cas12b/C2c1 system has been identified as a dual-RNA-guided DNA endonuclease system with distinct features from Cas9 and Cas12a10. First, Cas12b was reported to generate staggered ends distal to the protospacer adjacent motif (PAM) site in vitro when reconstituted with the crRNA/tracrRNA duplex10,11. Second, although the RuvC domain of Cas12b is similar to that of Cas9 and Cas12a, its putative Nuc domain shares no sequence or structural similarity to the HNH domain of Cas9 and the Nuc domain of Cas12a11,12. Moreover, Cas12b possesses a smaller size than the most widely-used SpCas9 and Cas12a (AacCas12b: 1,129 amino acids (aa), SpCas9: 1,369 aa, AsCas12a: 1,353aa, LbCas12a: 1,274 aa)9,10,13, making it suitable for adeno-associated virus (AAV)-mediated in vivo delivery for gene therapy. Compared with the small-sized Cas9, such as the SaCas9 and CjCas9, Cas12b recognizes simpler PAM sequences (AacCas12b: 5′-TTN-3′, SaCas9: 5′-NNGRRT-3′, CjCas9: 5′-NNNNRYAC-3′)8,10,14, which can significantly increase the targeting range of the genome. Most importantly, Cas12b has minimal off-target effects12 and thus may serve as a safer choice for therapeutic and clinical applications. However, the previously identified Cas12b nuclease (AacCas12b) maintains optimal cleavage activity in vitro at the temperature ranging between 40 °C and 55 °C10, which is not suitable for mammalian genome editing. Given the broad applications of Cas9 and Cas12a as genome engineering tools15,16,17,18,19, and the diversity of CRISPR-Cas systems5, we sought to explore the potential applications of Cas12b systems in mammalian genome engineering.
Here, we report the identification and engineering of Cas12b from Alicyclobacillus acidiphilus (AaCas12b), which enables robust genome editing in mammalian cells and mice using a chimeric single-guide RNA (sgRNA). Meanwhile, AaCas12b has features, including relatively small size, increased stability in human plasma, and high specificity, which makes it suitable for therapeutic genome editing.
Results
In vitro characterization of AaCas12b
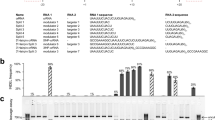
To seek Cas12b orthologs capable of mammalian genome editing, we synthesized the human codon-optimized Cas12b sequences, and their cognate crRNA and tracrRNA sequences from Alicyclobacillus acidiphilus (NBRC 100859, AaCas12b), Alicyclobacillus kakegawensis (NBRC 103104, AkCas12b), Alicyclobacillus macrosporangiidus (DSM 17980, AmCas12b), and Bacillus sp. (NSP2.1, BsCas12b) (Fig. 1a, Supplementary Fig. S1a and Supplementary Table S1 and Supplementary Sequences). Then we expressed and purified these Cas12b proteins from E. coli, and applied them to reconstitute Cas12b ribonucleoproteins (RNPs) with in vitro-transcribed crRNAs and tracrRNAs for in vitro DNA cleavage assay (Supplementary Fig. S1b and Supplementary Table S2 and Supplementary Sequences). Given that Cas12b mediates DNA interference by recognition of the T-rich PAM at the 5′-end of the protospacer sequence10, we used PCR-generated double-stranded DNAs (dsDNAs) bearing the 5′-NTTN PAM as templates (Supplementary Table S2). The in vitro DNA cleavage assay results showed that at 37 °C, only AaCas12b and AkCas12b RNPs could cleave the target DNAs, with AaCas12b being more potent than AkCas12b (Supplementary Fig. S1c).
a Schematic illustration of the genomic architecture of CRISPR-Cas12b from Alicyclobacillus acidiphilus (NBRC 100859) (left) and the crRNA/tracrRNA duplex (right). b In vitro cleavage activity of AaCas12b at various temperatures. The cleavage rate is shown under the cleaved lanes. c In vitro validation of the PAM requirements of AaCas12b showing that PAMs matching the 5′-TTN sequence can be efficiently cleaved. The cleavage rate is shown under the cleaved lanes. d Cleavage site determination of AaCas12b by sequencing the cleavage products. The cleavage sites are indicated by red triangles in the left panel. TS target strand, NTS non-target strand
Intriguingly, we found that as a thermoacidophilic bacterium-derived protein, AaCas12b could maintain the nuclease activity over a wide range of temperature and pH values (Fig. 1b and Supplementary Fig. S1d, e). AaCas12b could cleave target DNAs between 4 °C and 100 °C in vitro, and maintained the maximal cleavage activity between 31 °C and 59 °C (Fig. 1b and Supplementary Fig. S1d). For pH value, AaCas12b adapted to a wide range from pH 1.0 to pH 12.0, with highest cleavage activity between pH 1.0 and pH 8.0 (Supplementary Fig. S1e). Since the nuclease activity of Cas12b is metal-dependent, we further determined the metal ion dependence of AaCas12b. The results showed that Mg2+ as well as Ca2+, Mn2+, Sr2+, Ni2+, Fe2+, and Co2+, but not Zn2+ or Cu2+, enabled AaCas12b to cleave target DNAs (Supplementary Fig. S1f, g).
Next, we performed in vitro cleavage assay using DNA templates bearing various PAMs, and identified that AaCas12b utilized the 5′-TTN PAM for target DNA recognition (Fig. 1c and Supplementary Table S2), while AkCas12b recognized the 5′-TTTN PAM (Supplementary Fig. S1h and Supplementary Table S2). We further employed Sanger sequencing of the cleaved dsDNAs to map the cleavage sites of AaCas12b. The results showed that AaCas12b cleaved the complementary DNA strand at 23-base pair (bp) upstream of the PAM sequence, and the non-complementary DNA strand at 17-bp upstream of the PAM sequence, which generated a six-nucleotide (nt) 5′-overhang (Fig. 1d and Supplementary Fig. S1i). This cleavage pattern is distinct from the blunt ends generated by Cas913 but similar to the Cas12a-mediated target cleavage9.
AaCas12b can robustly edit the mammalian genomes in human and mouse cells
Next, we explored the capacity of the Cas12b systems to cleave endogenous genomic loci in mammalian cells. We cloned the four Cas12b sequences into eukaryotic expression vectors under the CAG promoter, and crRNA/tracrRNA within one polycistron gene hijacked by pre-tRNAGly20 under human U6 polymerase III promoter (Fig. 2a and Supplementary Sequences). Two nuclear localization signals (NLSs) were attached to each end of the Cas12b proteins to ensure their nuclear compartmentalization in mammalian cells (Supplementary Fig. S2a). Then the vectors were transfected into human 293FT cells and mouse embryonic stem cells (ESCs) to target two sites of the human RNF2 gene and one site of the mouse Nrl gene, respectively (Fig. 2b, Supplementary Fig. S2b, c and Supplementary Table S3). T7 endonuclease I (T7EI) assay showed that both AaCas12b and AkCas12b induced targeted mutations at site 1 of RNF2 gene (Fig. 2c), but only AaCas12b induced targeted mutations at site 2 of RNF2 gene and in Nrl gene (Supplementary Fig. S2b, c), suggesting that AaCas12b was more robust than AkCas12b. Consistently, sequence insertions and deletions (indels) at the target sites were detected by Sanger sequencing (Fig. 2d and Supplementary Fig. S2d, e), demonstrating that the engineered Cas12b systems could generate site-specific mutations in mammalian cells. The thermostable nuclease activity of AaCas12b suggested that it might be more stable in human blood. Indeed, although SpCas9 lost its activity even after incubation with low concentrations of human plasma, similarly as previously reported21, AaCas12b maintained a robust nuclease activity to cleave target DNAs even after 12 h-incubation with human plasma at 37 °C (Fig. 2e), suggesting that AaCas12b might be suitable for in vivo RNP delivery to achieve genome editing. Collectively, these results showed that AaCas12b could serve as a genome editing tool to engineer human and mouse cells.
a Schematic illustration of the eukaryotic expression strategy of the Cas12b and its cognate guide RNA (gRNA). The pre-tRNAGly, an endogenous RNA processing system for cleavage of transcripts, was hijacked to simultaneously express the tracrRNA and crRNA using a single human U6 promoter. b Schematic illustration of the human RNF2 target site 1 and crRNA/tracrRNA duplex. Red letters indicate the PAM sequences. c T7EI analysis of the indels produced by Cas12b orthologs (AaCas12b, AkCas12b, AmCas12b, and BsCas12b) at the human RNF2 target site 1. The indel rate is shown under the lanes with mutation. mock, an U6 empty vector without crRNA/tracrRNA expression. GFP, an empty backbone vector without Cas12b protein expression. d Sanger sequencing results showing the indels in human RNF2 target site 1 produced by AaCas12b and AkCas12b. Blue dashes, deleted bases; purple lowercases, insertions or mutations; red uppercases, PAM. e Effects of human plasma incubation on the nuclease activity of SpCas9 and AaCas12b. After incubation in human plasma at indicated concentrations for 12 h at 37 °C, in vitro DNA cleavage assay was conducted. The cleavage rate is shown under the cleaved lanes
Multiplex genome engineering in mammalian cells using a chimeric single-guide RNA
Next, we characterized the guide RNA requirements for the AaCas12b-mediated genome editing. To simplify the AaCas12b-mediated genome editing, a chimeric single-guide RNA was designed by fusion of crRNA/tracrRNA transcripts (Fig. 3a), which contained three stem-loop structures according to previous reports10. T7EI assay showed that the sgRNA could direct AaCas12b to target sites for cleavage in human cells as efficiently as the dual-RNA guide (Supplementary Fig. S4a). To characterize the sequence requirements of the sgRNA, we truncated the sequences of each stem-loop (Fig. 3a and Supplementary Fig. S4b). We found that truncations in stem-loop 1 and stem-loop 2 completely abolished both the in vitro and in vivo endonuclease activity of AaCas12b (Fig. 3b, c and Supplementary Fig. S4c, d), suggesting that they were indispensable. However, truncations in stem-loop 3, which was formed by the crRNA/tracrRNA duplex, did not affect the AaCas12b nuclease activity (Fig. 3c and Supplementary Fig. S4c, d). We further explored the programmability of sgRNA modules by attaching a functional MS2 RNA hairpin to the sgRNA scaffold22,23 (Supplementary Fig. S5a-c). We showed that insertion of MS2 sequences into the 5′-end of the sgRNA or the stem-loop 3 had no effect on the target recognition and cleavage activity of the AaCas12b systems in human cells (Supplementary Fig. S5d), indicating that the sgRNA could be repurposed for other functions.
a Schematic illustration of the sequences and structure of chimeric single guide (sgRNA) (top), and depiction of the truncation strategy of the sgRNA scaffold (bottom). b Effects of truncation of stem-loop 1 within the sgRNA scaffold on the nuclease activity of AaCas12b in human cells. The indel rate is shown under the lanes with mutation. c Effects of truncation of stem-loop 2 and stem-loop 3 within the sgRNA scaffold on the nuclease activity of AaCas12b in human cells. The indel rate is shown under the lanes with mutation. d AaCas12b facilitates multiplex genome editing by simultaneously targeting CCR5, CD34, DNMT1, and RNF2, using four individual sgRNAs containing spacers. (Top) Schematic illustration of the target sites in the human genome. (Bottom) All the four sgRNAs mediate efficient protospacer cleavage. The indel rate is shown under the lanes with mutation
As a type of RNA-guided nucleases, we showed that simultaneous transfection of AaCas12b and paired sgRNAs targeting human RNF2 gene resulted in large fragment deletions (Supplementary Fig. S6a, b). Moreover, by simultaneously transfecting human 293FT cells with four sgRNAs targeting the human CCR5, CD34, DNMT1, and RNF2 genes, all the four genes were mutated as detected by the T7EI assay (Fig. 3d). Simultaneous mutations could also be generated in mouse ESCs by the CRISPR-AaCas12b system (Supplementary Fig. S6c). These results demonstrated that AaCas12b could enable robust multiplex genome editing in mammalian cells.
To further compare the frequencies of target mutations generated by AaCas12b, AsCas12a, and SpCas9, we conducted endogenous gene editing experiments at eight genomic sites. Each site contained three PAM sequences that can be recognized by AaCas12b (5′-TTN-3′), SpCas9 (5′-NGG-3′)13, and AsCas12a (5′-TTTN-3′)9, respectively (Supplementary Fig. S3 and Supplementary Table S3). T7EI assay results showed that each nuclease had a wide range of mutation frequencies at these sites tested, and the overall efficiency of AaCas12b was similar to or higher than AsCas12a, but was lower than SpCas9 (Supplementary Fig. S3). Moreover, AaCas12b recognized 5′-TTN PAM (Fig. 1c), instead of the 5′-TTTN PAM utilized by AsCas12a9, indicating that AaCas12b has wider genome coverage and may edit some genomic sites that AsCas12a couldn’t target (Supplementary Fig. S3).
Gene activation based on deactivated AaCas12b (dAaCas12b)
We next intended to abolish the nuclease activity of AaCas12b, as the CRISPR effectors with abolished nuclease activity, can be repurposed for broad applications beyond genome editing. Using protein alignment (Supplementary Fig. S7), we identified six potential catalytic residues of AaCas12b in either the REC lobe or the Nuc lobe according to the structural annotations11,12. Then we constructed AaCas12b variants containing point mutations of these residues and tested their cleavage activity in mammalian cells (Fig. 4a). Three AaCas12b variants with catalytic residue mutation (R785A, R911A, and D977A) lost the ability to cleave dsDNAs in vitro, whereas three variants (R122A, D570A, and D806A) still partially retained the catalytic activities (Fig. 4b and Supplementary Fig. S8a). Subsequently, the T7EI assay showed that the three AaCas12b variants (R785A, R911A, and D977A) lost nuclease activities in human cells (Fig. 4c and Supplementary Fig. S8b). Furthermore, in vitro cleavage assays showed that AaCas12b(R785A) could not cleave the Nb.BtsI- and Nt.BstBNI-nicked double-stranded DNA templates, suggesting that the R785A mutation converted AaCas12b nuclease into a deactivated form (dAaCas12b) rather than a nickase (Supplementary Fig. S8c). As a proof-of-concept, we explored the potential of applying the dAaCas12b to targeted gene activation. After co-transfection of dAaCas12b, sgRNAs with MS2 fusion, and the MCP-VP64 or MCP-VPR (VP64-p65-Rta fusion) effectors in 293FT cells22,23,24,25 (Fig. 4d), the expression levels of two target genes, the IL1B and HBG1 (Supplementary Table S3), were substantially elevated (Fig. 4e), proving the feasibility of dAaCas12b-mediated target gene activation. However, the gene activation level induced by dAaCas12b was still lower that by dSpCas9, suggesting that further improvement of the dAaCas12b-based gene activation platform is required (Fig. 4e).
a Schematic illustration of AaCas12b domain structure showing the positions of catalytic residue mutations. The catalytic residues were identified based on sequence homology of AaCas12b and A. acidoterrestris Cas12b (AacCas12b) (PDB: 5WQE). b In vitro DNA cleavage analysis of mutation of indicated nuclease mutant or wild-type (WT) AaCas12b proteins. The cleavage rate is shown under the cleaved lanes. c T7EI analysis showing effects of mutation of AaCas12b catalytic residues on DNA targeting in 293FT cells. The indel rate is shown under the lanes with mutation. GFP, an empty backbone vector without Cas12b protein expression. d Schematic illustration of sgRNA scaffold-based recruitment enabling simultaneous activation of independent target genes. The sgRNA construct with MS2 RNA hairpin recruits MCP-VP64 or MCP-VP64-p65-Rta (VPR) to activate endogenous gene expression in human 293FT cells. e The sgRNA scaffold recruits MCP-VPR or MCP-VP64 to simultaneously activate endogenous expression of IL1B and HBG1 in human 293FT cells combined with dAaCas12b or dSpCas9 expression, respectively. Cells transfected with only empty vectors were used as control (Ctrl)
Targeted mutagenesis in mice by microinjection of AaCas12b RNPs
We next explored the potential of Cas12b in generation of targeted mutations in a whole organism. To generate mutated mice with the AaCas12b system, we conducted embryo microinjection of recombinant AaCas12b RNPs. First, we assessed the activity of preassembled AaCas12b RNPs in mouse embryos at target site 1 in the mouse Nrl gene (Fig. 5a). We microinjected the AaCas12b RNPs into 16 one-cell-stage mouse embryos (Fig. 5b), cultured them in vitro and obtained eight blastocysts (Supplementary Fig. S9a). T7EI assay and subsequent Sanger sequencing results showed that six out of eight (75%) blastocysts carried AaCas12b-medaited mutations in the Nrl gene (Supplementary Fig. S9b, c). To generate knockout mice, we further transplanted one-cell-stage embryos microinjected with the AaCas12b RNPs into surrogate mothers, and obtained 12 Nrl mutant mice and 2 Prmt7 mutant mice in total as determined by T7EI assay (Fig. 5c, d and Supplementary Fig. S10a, b). Subsequently, Sanger sequencing analysis validated that most mutations were indels with the frequency at up to 66.7% (Fig. 5e, and Supplementary Fig. S10c, d).
a Schematic illustration of AaCas12b sgRNA targeting exon 3 of the mouse Nrl gene. The PAM and target sequences are colored in red and blue, respectively. The cleavage sites are indicated with red triangles. b Schematic illustration of AaCas12b RNP delivery into 1-cell stage mouse embryos by microinjection. c Summary of the mutants generated via AaCas12b RNP microinjection with sgRNA targeting the Nrl and Prmt7 genes, respectively. d T7EI-based genotyping assay identifying founder mice derived from embryos injected with AaCas12b RNPs targeting the mouse Nrl gene. The numbers in red denote newborn mice with induced indel mutations. The indel rate is shown under the lanes with mutation. e Mutated Nrl alleles observed in the founder mice in Fig. 5d. Blue dashes, deleted bases; purple lowercases, insertions or mutations; red uppercases, PAM. Indel frequencies are indicated
Successful germline transmission is a trademark for establishing genetically-modified animal models, particularly for CRISPR-based methods. To assess the transmission of genetic mutations in Nrl knockout founders, we analyzed the genotypes of offspring produced from Nrl mutants crossed with wild-type (WT) mice. T7EI and Sanger sequencing results demonstrated that the gene mutations in the founders could be successfully transmitted to their offspring (Supplementary Fig. S11). Collectively, these results demonstrate that Cas12b-based genome editing is a useful tool for generating knockout mouse models.
Minimal off-target effects of AaCas12b in cells and mice
Off-target effects pose a major challenge to the applications of genome editing tools. Next, we assessed the off-target effects of AaCas12b-mediated genome editing in mammalian cells. We found that at least 18 nt of the guide sequence was required to produce detectable indels, and a minimum of 20 nt of the guide sequence was required to achieve efficient mutagenesis in human and mouse genomes (Supplementary Fig. S12a). To further test the sensitivity of AaCas12b to Watson–Crick mismatches at the sgRNA-DNA interface, we used sgRNAs containing single substitutions or adjacent double substitutions at the position within 19 nt proximal to the PAM sequence to target the human RNF2 and mouse Nrl genes (Supplementary Table S3). No off-target mutation by any mismatched sgRNAs was detected in these genome loci (Fig. 6a and Supplementary Fig. S12b), suggesting that AaCas12b could hardly tolerate any mismatches between sgRNA and target DNA sequences. To directly assess the off-target mutagenesis in the genome induced by AaCas12b, the potential off-target sites containing one, two, or three mismatches to the guide RNAs were computationally identified from the whole human genome (Supplementary Table S4). No off-target mutation in the 58 predicted genomic loci was detected in the AaCas12b group as shown by T7EI assay and targeted deep sequencing results (Fig. 6b and Supplementary Table S4). We next assessed the degree of genome-wide off-target mutagenesis in 293FT cells targeted with AaCas12b. By performing whole-genome sequencing (WGS) of two mutant cell lines targeted by AaCas12b complexed with sgRNAs, we detected no off-target effects induced by AaCas12b/sgRNA targeting the RNF2 sites in the human genome (Fig. 6c). These results indicated that AaCas12b had a minimal off-target effect, consistent with the notion of a previous report12.
a Analysis of cleavage specificity of AaCas12b/sgRNA in human and mouse cells using sgRNAs carrying single base-pair mismatches in the guide sequence. Error bars indicate standard errors of the mean (s.e.m.), n = 3. b (Top) Schematic illustration of AaCas12b target sites in the human CCR5 and RNF2 loci, respectively. (Bottom) Indel frequencies induced by AaCas12b directed by sgRNAs targeting endogenous CCR5 and RNF2 sites and their corresponding off-target sites in human 293FT cells. Mutation frequencies were assessed by T7EI assay. Error bars indicate s.e.m., n = 2. c WGS analysis of genomic DNAs of RNF2-targeted 293FT cells. None of 2598 (RNF2-site 1) and 3394 (RNF2-site 2) sites identified with Cas-OFFinder in the reference genome (hg38) that differed from the on-target site by up to five mismatches harbored indels in the mutated genome. d Whole-genome sequencing (WGS) analysis of genomic DNAs of Nrl-mutated mouse gonads. None of the 2657 sites identified using Cas-OFFinder in the reference genome (mm10) that differed from the on-target site by up to five mismatches harbored indels in the mutated genome
To further determine the specificity of AaCas12b RNPs in gene targeting in vivo, we assessed the potential off-target mutations in the Nrl-mutated founder mice. Potential off-targets were predicted in the mouse whole genome with similarity to the target sequence in Nrl gene (Supplementary Table S4). Five mutated founders and two WT siblings were analyzed by the T7EI assay and deep sequencing (Supplementary Fig. S12c, d). We found no detectable off-target effects at these 30 potential off-target sites in the five mutants (Supplementary Fig. S12c, d and Supplementary Table S4). WGS data further validated that no off-target effects were induced by AaCas12b RNPs in mice (Fig. 6d and Supplementary Fig. S13). These results demonstrated AaCas12b owned a high cleavage specificity and had minimal off-target effects in both mammalian cells and in mice.
Fewer off-target effects of AaCas12b compared with SpCas9
Next, we sought to directly compare the off-target effects of AaCas12b, SpCas9, and AsCas12a. We used guide RNAs (gRNAs) containing single mismatches within 19 nt from the PAMs to target the same genomic loci (Supplementary Table S3). Both SpCas9 and AsCas12a generated substantial mutations with mismatched gRNAs in mouse and human cells (Fig. 7a and Supplementary Fig. S14). However, AaCas12b did not generate any mutations with mismatched gRNAs (Fig. 7a and Supplementary Fig. S14), suggesting the requirement of a perfect match between the sgRNA and target sequence for the AaCas12b-mediated mammalian genome editing. To directly assess the off-target mutagenesis in the genome induced by AaCas12b and SpCas9, the potential off-target sites, containing one, two, or three mismatches to the gRNAs, were computationally identified from the whole human genome26 (Supplementary Table S5). No off-target mutation in the 88 predicted genomic loci was detected in the AaCas12b group as T7EI results showed (Fig. 7b, Supplementary Fig. S15 and Supplementary Table S5). However, 3 out of 82 predicted off-target sites in the SpCas9 group were mutated (Fig. 7b, Supplementary Fig. S15 and Supplementary Table S5). We also performed whole-genome sequencing (WGS) analysis. Two mutated off-targets were induced by SpCas9 when targeting CCR5 gene in the human genome, however, no off-target mutation was detected by AaCas12b (Fig. 7c). These results indicated that AaCas12b possessed fewer off-target effects than SpCas9.
a (Left) Schematic showing the targeting sequences of AaCas12b, AsCas12a and SpCas9 in mouse Nrl locus. (Right) Activities of AaCas12b, AsCas12a and SpCas9 targeted to mouse Nrl locus using respective guide RNAs with single mismatches in mouse ES cells. Mutation frequencies were assessed by T7EI assay. Error bars indicate standard errors of the mean (s.e.m.), n = 3. b (Upper) Schematic showing AaCas12b and SpCas9 targeting sites in the human RNF2 locus. (Lower) Frequencies of induced indels induced at on- and off-target sites by AaCas12b and SpCas9 in human 293FT cells. Mutation frequencies were assessed by T7EI assay. Error bars indicate s.e.m., n = 2. c Whole-genome sequencing (WGS) analysis potential off-target sites with one to five mismatches to gRNAs induced by AaCas12b or SpCas9 in the human genome and the amount of mutated sites observed by WGS
Discussion
In this work, we demonstrated that type V-B CRISPR-Cas12b systems can enable robust engineering of mammalian genomes. Similar to Cas9, Cas12b is a dual-RNA-guided endonuclease10, in contrast to the single-RNA-guided Cas12a9. We showed that the crRNA/tracrRNA duplex can be engineered into a shorter chimeric RNA, which works as efficiently as the dual-RNA duplex. Also, we demonstrated that the 5′-end and the stem-loop 3 can be programmed as a scaffold to recruit effector proteins, together with the dAaCas12b, suggesting that the AaCas12b system can be repurposed for versatile applications, such as regulation of endogenous gene expression. Additionally, Cas12b generates a staggered DSB with a 5′ overhang, which is similar to Cas12a9. This structure of the cleavage products might be advantageous for facilitating non-homologous end joining (NHEJ)-based gene modifications27, particularly in nondividing cells28.
Importantly, we showed that AaCas12b RNPs can efficiently introduce targeted indel mutations in mouse embryos, which can be successfully transmitted into the next generation via germline. Our results also indicated that AaCas12b possess minimal off-target effects in cell lines compared with SpCas9 and AsCas12a. Through thorough assays by targeted deep sequencing and WGS, we identified no off-target effects in both cell lines and mice. This high specificity indicates that AaCas12b will be useful for therapeutic applications, though more detailed investigation is required.
Another potential promising feature of CRISPR-Cas12b is that AaCas12b possesses its nuclease activity over a wide range of temperatures and pH values, which will enhance the utility of CRISPR-Cas12b technology under both thermophilic and acidophilic conditions. Moreover, we have shown that AaCas12b possesses longer lifetime and improved stability as RNPs in human plasma, which will benefit the future clinical applications requiring delivery of Cas12b into the bloodstream.
Materials and methods
DNA manipulations
DNA manipulations, including DNA preparation, digestion, ligation, amplification, purification, agarose gel electrophoresis, etc. were conducted according to Molecular Cloning: A Laboratory Manual with some modifications. Briefly, PAM determination plasmids were constructed by ligating annealed oligonucleotides (oligos) (Supplementary Table S1) into the p11-LacY-wtx1 vector29 digested by EcoRI and SphI, and corresponding dsDNA fragments were PCR-generated (Supplementary Table S6). Targeting crRNA/tracrRNA duplexes or sgRNAs (thereafter guide RNAs or gRNAs) for cell transfection assay were constructed by ligating annealed oligos (Supplementary Table S3) into BasI-digested pUC19-U6-gRNA vectors (Supplementary Sequences). Templates for in vitro transcription of tracrRNAs and sgRNAs were PCR-amplified using primers containing a T7 promoter sequence (Supplementary Table S2 and S6). The crRNAs were transcribed from annealed oligos bearing a T7 promoter (Supplementary Table S1).
De novo gene synthesis and plasmid construction
PSI-BLAST program30 were adopted to identify new type V-B CRISPR-Cas12b proteins. And their coding sequences were humanized31, and oligos for the synthetic Cas12b genes and their cognate gRNAs were designed by GeneDesign program32. For Cas12b coding genes which are > 3 kb were split into four “chunks” of ~800 bp (Supplementary Table S1 and Supplementary Sequences). For gRNAs which are < 300 bp were left as is (Supplementary Table S1). All oligos for gene synthesis were commercially purchased (Taihe Biotechnology Co., LTD). Oligos within one chunk were mixed into the final concentration at 300 nM and applied for fragment assembly without primers in a 25 μL PCR amplification system. Then a 2.5-μL product from the first-round PCR was used as a template with 2.5 μL, 3 μM primers added to amplify the designed fragments in a 25-μL system. The route of 1st and 2nd PCR-based gene assembly reaction: 95 °C 15 min; 95 °C 30 s, 59 °C 30 s, 72 °C 30 s/500 bp, 25 cycles; 72 °C 10 min33. Purified products were assembled into expression vectors via homologous recombination in vitro using NEBuilder® HiFi DNA Assembly Master Mix (NEB). The pCAG-2AeGFP vector (Supplementary Sequences) was applied for mammalian cell expression of Cas12b proteins. gRNAs were constructed in the pUC19-U6 vector (Supplementary Sequences) for mammalian cell expression.
Protein purification
The synthetic Cas12b coding sequences were constructed into a BPK2014-ccdB expression vector (Supplementary Sequences) using ligation-dependent cloning. The resulting fusion construct containing a C-terminal fused His10 tag. The proteins were expressed in E. coli strain BL21 (λDE3), grown in CmR + LB medium at 37 °C to OD600 ~0.4, following induction with 0.5 mM IPTG at 16 °C for 16 h. In total, 300mL of induced cells were harvested for protein purification, and all subsequent steps were conducted at 4 °C. Cell pellets were lysed in 30 mL Lysis Buffer (NPI-10: 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 5% glycerol, pH 8.0) supplemented with 1x protease inhibitors (Roche cOmplete, EDTA-free) before lysis by sonication. Lysates were clarified by centrifugation at 8000 rpm, 4 °C for 10 min, and the supernatants were incubated with His60 Ni Superflow Resin (Takara) in batch at 4 °C for 2 h. After the Resin was washed with each 20 mL Wash Buffer 1 (NPI-20: 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 5% glycerol, pH 8.0), Wash Buffer 2 (NPI-40: 50 mM NaH2PO4, 300 mM NaCl, 40 mM imidazole, 5% glycerol, pH 8.0) and Wash Buffer 3 (NPI-100: 50 mM NaH2PO4, 300 mM NaCl, 100 mM imidazole, 5% glycerol, pH 8.0), expressed proteins were eluted with 5 mL Elution Buffer (NPI-500: 50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, 5% glycerol, pH 8.0). Purified Cas12b proteins were dialyzed using 100 kDa dialyzer overnight with storage buffer (50 mM Tris-HCl, 200 mM KCl, 0.1 mM EDTA, 1 mM DTT, 20% glycerol, pH 8.0). Fractions were pooled and concentrated with 100 kDa Centrifugal Filter Unit (Millipore). The purity of enriched proteins was analyzed by SDS-PAGE and Coomassie staining, and the concentration was quantitated using BCA Protein Assay Kit (Thermo Fisher).
In vitro RNA transcription
RNAs were in vitro-transcribed using HiSribeTM T7 Quick High Yield RNA Synthesis Kit (NEB) and PCR-amplified DNA templates carrying a T7 promoter sequence. Transcribed RNAs were purified using Oligo Clean & ConcentratorTM (ZYMO Research) and quantitated on NanoDropTM 2000 (Thermo Fisher). Primers used for template preparation are listed in Supplementary Table S6.
In vitro analysis of PAM sequences
To determine the PAM sequence of Cas12b, 100 nM Cas12b proteins, 400 ng in vitro-transcribed sgRNAs and 200 ng PCR-generated dsDNAs bearing different PAM sequences (Supplementary Table S2) were incubated at 37 °C for 1 h in the cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, pH 8.0). The reactions were stopped by adding RNase A to digest gRNAs at 37 °C for 20 min, and resolved by ~3% agarose gel electrophoresis and ethidium bromide staining.
dsDNA cleavage assay
For dsDNA cleavage assay, 100 nM Cas12b proteins, 400 ng in vitro-transcribed gRNAs and 200 ng PCR-generated dsDNAs containing a 5′-TTN PAM sequence was conducted at 37 °C for 1 h in cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, pH 8.0), if not specified. To determine the thermostability of AaCas12b, the cleavage was reacted at a large-range temperature (4 °C – 100 °C) for 1 h in cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, pH 8.0). For pH tolerance assay, the cleavage reactions were performed at 37 °C for 1 h in cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2) with pH value ranging from 1.0 to 13.0. In Mg2+-dependent assay, cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, pH 8.0) was supplemented with EDTA (0 mM, 1 mM, 5 mM, 10 mM, 20 mN, and 40 mM) or Mg2+ (0 mM, 1 mM, 5 mM, 10 mM, 20 mN, and 40 mM), and the mixture was incubated at 37 °C for 1 h. Further metal-dependent cleavage reactions were conducted at 37 °C for 1 h in cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, pH 8.0) supplemented with 1 or 5 mM of CaCl2, MnCl2, SrCl2, NiCl2, FeCl2, CoCl2, ZnCl2, or CuCl2. The reactions were stopped by adding RNase A to digest sgRNAs at 37 °C for 20 min, and resolved by ~3% agarose gel electrophoresis and ethidium bromide staining.
Thermostability assay
SpCas9 and AaCas12b nucleases were incubated in diluted concentrations of human plasma at 37 °C for 12 h. After incubation, DNA cleavage reactions were conducted as described above. For SpCas9, its target bears a 3′-NGG PAM. The reactions were stopped by adding RNase A to digest sgRNAs at 37 °C for 20 min, and resolved by ~3% agarose gel electrophoresis and ethidium bromide staining.
Cell culture, transfection, and fluorescence-activated cell sorting (FACS)
Human embryonic kidney cell line 293FT and HeLa cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotic–antimycotic (Gibco) at 37℃ with 5% CO2 incubation. Mouse embryonic stem (mES) cell line was maintained in N2B27 medium plus 1 μM PD0325901 (Stemgent), 3 μM CHIR99021 (Stemgent), and 1000 U/mL mLIF (Millipore). The N2B27 medium consists of DMEM/F12 (Gibco) and Neurobasal (Gibco) at a ratio of 1:1 and was supplemented with 1% N-2 supplement (Gibco), 0.5% B-27 supplement (Gibco), 20 ng/ml BSA (Sigma), 10 μg/ml insulin (Roche Applied Science), 1% GlutaMAX (Gibco), 5% knockout serum replacement (KOSR, Gibco), 0.1% 2-mercaptoethanol (Gibco) and 1% antibiotic–antimycotic (Gibco). 293FT cells were transfected using Lipofectamine LTX (Invitrogen) following the manufacturer’s recommended protocol. mES cells were transfected via electroporation using NeonTM transfection system (Invitrogen) following the manufacturer’s recommended protocol. For each well of a 24-well-plate, a total of 750 ng plasmids (Cas12b: gRNA = 2: 1) were used. Then 48 h following transfection, GFP-positive cells were sorted using the MoFlo XDP (Beckman Coulter).
T7 endonuclease I (T7EI) assay and Sanger sequencing analysis for genomic modifications
Harvested or FACS-sorted GFP-positive 293FT or mES cells post transfection were lysed for genomic DNA extraction. Briefly, cells were directly lysed with Buffer L and incubated at 55 °C for 3 h and 95 °C for 10 min. Genomic region surrounding the CRISPR-Cas12b target site for each gene was PCR-amplified (Supplementary Table S6). 200– 400 ng PCR products were mixed with ddH2O to a final volume of 10 μL, and subjected to re-annealing process to enable heteroduplex formation according to previous methods6. After re-annealing, products were treated with 1/10 volume of NEBufferTM 2.1 and 0.2 μL T7EI (NEB) at 37 °C for 30 min, and analyzed on 3% agarose gels. Indels were quantitated based on relative band intensities6. T7EI assay-identified mutated products were subjected to be cloned into TA-cloning vector and transformed into competent E. coli strain. After overnight culture, colonies were randomly picked out and sequenced.
Site-directed Cas12b gene mutagenesis
Two pairs of primers containing the desired site-directed mutations and 5′-end overlaps were used for gene amplification (Supplementary Table S6). The two agarose gel-purified gene fragments were seamlessly assembled into XmaI and NheI double-digested mammalian expression vector (Supplementary Sequences) using NEBuilder® HiFi DNA Assembly Master Mix (NEB) following the manufacture’s recommended protocol. And E. coli expression vectors were reconstructed using digestion- and ligation-dependent methods.
Endogenous gene transcription activation
For dAaCas12b-based transcription activation of endogenous genes, dAaCas12b, targeting sgRNAs bearing MS2 RNA hairpin, and MCP-VP64 or MCP-VPR (VP64-p65-Rta fusion) were co-transfected into human 293FT cells. For each gene, eight target sgRNAs were constructed and mixed into a pool (Supplementary Table S3). All target sites were located within the ~ 500 bp upstream of the transcription start site (TSS).
Animals
Mice were housed in the animal care facility of the Institute of Zoology, Chinese Academy of Sciences, according to the institutional guidelines for the care and use of laboratory animals. Specific pathogen-free (SPF) grade ICR mice (Stock No. 201) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. All animal experiments were conducted according to the guidelines for the care and use of laboratory animals established by the Beijing Association for Laboratory Animal Science and approved under the Animal Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences (1 Beichen West Road, Chaoyang District, Beijing, P.R. China).
Microinjection of AaCas12b RNPs
ICR female mice at 8 weeks of age were superovulated by intraperitoneal injection of PMSG and hCG hormones (Sigma) at a 48 -h interval. These mice were mated with ICR male mice at 10 to 16 weeks of age, and fertilized one-cell embryos were collected from the oviduct. Cumulus cells were removed from embryos by exposure to 0.1% hyaluronidase (Sigma) in PBS buffer. For microinjection, preassembled AaCas12b RNPs were injected into cytoplasm using a Nikon ECLIPSE Ti micromanipulator and a FemtoJet 4i microinjector (Eppendorf). Embryos were cultured in microdrops of KSOM + AA containing D-glucose and phenol red (Millipore) under mineral oil at 37 °C for 3.5 days in a humidified atmosphere consisting of 5% CO2 in air. Two-cell stage embryos were transferred on the following day into the oviducts of 0.5 dpc pseudopregnant foster mothers to obtained mutated founders.
Off-target prediction and detection
The potential off-target sites for CRISPR-Cas12b system in the human or mouse genome with individual on-target sequences were predicted using Cas-OFFinder26. The complementarity region bearing one, two, or three mismatches with requisite PAMs were assessed as potential off-targets by T7EI assay or targeted deep sequencing.
Targeted deep sequencing
Target sites and potential off-target sites were amplified by barcoded PCR and pooled libraries were subjected to paired-end sequencing using HiSeq (Illumina). A reference genome was built using Picard Tools (http://broadinstitute.github.io/picard) and samtools34 from DNA sequences of the considered on-/off-target regions. Raw sequencing data (FASTQ files) were mapped against the created reference genone using BWA35 with standard parameters and resulting alignment files were sorted using smatools. Samples with fewer than 20 reads were excluded.
Whole-genome sequencing (WGS)
Genomic DNA from cultured cells or whole gonad tissues were extracted using MicroElute Genomic DNA Kit (OMEGA) and subjected to quality assessment. The extracted DNA was sequenced using an Illumina NovaSeq sequencer at a sequencing depth of 30x diploid coverage. The pair-ends reads were aligned onto the hg19 (GRCh38) human or mm10 (GRCm38) mouse reference genome using Bowtie 236 and BWA35, following manipulated using Picard Tools (http://broadinstitute.github.io/picard), respectively.
The WGS analysis was performed according to previous report37. Briefly, the Genome Analysis ToolKit (GATK4)38 or pysamstats (https://github.com/alimanfoo/pysamstats) were used for local realignment around indels, base score recalibration, variant calling across the human or mouse samples, and variant score recalibration. Candidate indels were filtered on several criteria using Python, PyVCF, and PyFasta packages. First, we removed indels near low-complexity regions as defined by RepeatMasker (http://repeatmasker.org) and annotated by softmasking in hg19 or mm10. Second, we removed indels that caused expansions or compressions of long (one with > 6 bp or two with > = 5 bp) homopolymers.
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Sorek, R., Kunin, V. & Hugenholtz, P. CRISPR - a widespread system that provides acquired resistance against phage in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186 (2008).
Makarova, K. S. et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 9, 467–477 (2011).
Mohanraju, P. et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016).
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Yang, H., Gao, P., Rajashankar, K. R. & Patel, D. J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167, 1814–1828 e1812 (2016).
Liu, L. et al. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell 65, 310–322 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Sternberg, S. H. & Doudna, J. A. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell 58, 568–574 (2015).
Wang, H., La Russa, M. & Qi, L. S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264 (2016).
Wright, A. V., Nunez, J. K. & Doudna, J. A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016).
Xie, K., Minkenberg, B. & Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112, 3570–3575 (2015).
Harrington, L. B. et al. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 8, 1424 (2017).
Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Maresca, M., Lin, V. G., Guo, N. & Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 (2013).
Chan, F., Hauswirth, W. W., Wensel, T. G. & Wilson, J. H. Efficient mutagenesis of the rhodopsin gene in rod photoreceptor neurons in mice. Nucleic Acids Res. 39, 5955–5966 (2011).
Chen, Z. & Zhao, H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 33, e154 (2005).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Grote, A. et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, W526–W531 (2005).
Richardson, S. M., Wheelan, S. J., Yarrington, R. M. & Boeke, J. D. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16, 550–556 (2006).
Li, G., Dong, B. X., Liu, Y. H., Li, C. J. & Zhang, L. P. Gene synthesis method based on overlap extension PCR and DNAWorks program. Methods Mol. Biol. 1073, 9–17 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Veres, A. et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell. Stem. Cell. 15, 27–30 (2014).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Acknowledgements
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA16030400), the National Natural Science Foundation of China (Grant No. 31621004, 31422038), the National Key Research and Development Program (Grant No. 2017YFA0103803), the National Basic Research Program of China (Grant No. 2014CB964800), CAS Key Projects (Grant No. QYZDY-SSW-SMC022, QYZDB-SSW-SMC002). We thank Shi-Wen Li, Xi-Li Zhu, Qing Meng and Xia Yang for their help with fluorescence-activated cell sorting.
Author information
Authors and Affiliations
Contributions
W.L. and Q.Z. conceived this project, supervised the experiments. W.L., Q.Z., F.T., and T.C. wrote the paper with the help from all authors. W.L., Q.Z., F.T., G.F., and T. C. analyzed the data. F.T., T.C., G.F., L.G., K.X., T.L., and J.L. performed the experiments.
Corresponding author
Ethics declarations
Conflict of interest
A patent application has been filed relating to this work. The authors declare that there is no conflict of financial interests, and they plan to deposit the reagents in Addgene to freely share with the academic community.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Teng, F., Cui, T., Feng, G. et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4, 63 (2018). https://doi.org/10.1038/s41421-018-0069-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-018-0069-3
This article is cited by
-
An Update on the Application of CRISPR Technology in Clinical Practice
Molecular Biotechnology (2024)
-
The applications of CRISPR/Cas-mediated genome editing in genetic hearing loss
Cell & Bioscience (2023)
-
CRISPR/Cas9 therapeutics: progress and prospects
Signal Transduction and Targeted Therapy (2023)
-
VarLOCK: sequencing-independent, rapid detection of SARS-CoV-2 variants of concern for point-of-care testing, qPCR pipelines and national wastewater surveillance
Scientific Reports (2023)
-
CRISPR/Cas systems for the detection of nucleic acid and non-nucleic acid targets
Nano Research (2023)