Abstract
The functional alterations of proteins and nucleic acids mainly rely on their modifications. ADP-ribosylation is a NAD+-dependent modification of proteins and, in some cases, of nucleic acids. This modification is broadly categorized as Mono(ADP-ribosyl)ation (MARylation) or poly(ADP-ribosyl)ation (PARylation). MARylation catalyzed by mono(ADP-ribosyl) transferases (MARTs) is more common in cells and the number of MARTs is much larger than poly(ADP-ribosyl) transferases. Unlike PARylation is well-characterized, research on MARylation is at the starting stage. However, growing evidence demonstrate the cellular functions of MARylation, supporting its potential roles in human health and diseases. In this review, we outlined MARylation-associated proteins including MARTs, the ADP-ribosyl hydrolyses and ADP-ribose binding domains. We summarized up-to-date findings about MARylation onto newly identified substrates including protein, DNA and RNA, and focused on the functions of these reactions in pathophysiological conditions as well as speculated the potential mechanisms. Furthermore, new strategies of MARylation detection and the current state of MARTs inhibitors were discussed. We also provided an outlook for future study, aiming to revealing the unknown biological properties of MARylation and its relevant mechanisms, and establish a novel therapeutic perspective in human diseases.
Similar content being viewed by others
Facts
-
MARTs catalyze MARylation, the hydrolases reverse MARylation and ADP-ribose binding domains recognize MARylation.
-
The newly identified MARylated substrates includes protein, DNA and RNA, and possess physiological and pathological roles in mammal via diverse mechanisms.
-
Targeting MARylation and MARylation-associated enzymes are promising therapeutic perspectives in human diseases.
Open questions
-
What are the biofunctions of nucleic acids MARylation, especially RNA MARylation.
-
Is there any undiscovered enzymes regulating MARylation reactions.
-
What is the potential connection between MARylation and PARylation.
Introduction
ADP-ribosylation is a dynamic covalent modification that highly conserved throughout almost all domains of life [1]. The processes of this modification include transferring the ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to its specific substrate meanwhile releasing nicotinamide. ADP-ribosylation was firstly defined as a post transcriptional modification (PTM) of protein in the sixties [2], followed by described crucial roles in the DNA damage responses [3]. In the following decades, major cellular processes involving cell growth and differentiation, transcription, stress responses, metabolisms and immunity were sequentially proved to be modulated by ADP-ribosylation as a PTM [4]. Of note, recent studies demonstrated ADP-ribosylation of nucleotides, besides protein targets [5]. In this enzymatic reaction, according to their roles, the enzymes can be categorized as ADP-ribosyl transferases (ARTs) which add ADP-ribose and ADP-ribosyl hydrolases which remove the ADP-ribose (Fig. 1). ADP-ribosylation is grouped into two patterns that MARylation identified by a single ADP-ribose unit linked to targets while PARylation containing polymers of ADP-ribose unit modification. Compared to the roles of PARylation and poly-ADP-ribosyl transferases, most notable PARP1, in cellular pathways are well-studied, the biofunctions of MARylation and MARTs are not fully elucidated, and MARylation-targeted therapeutics is still in the initial stage. However, knowledge of MARylation is rapidly developing and expanding. A growing number of evidence support MARylation is not only solely regarded as a type of PTM but also serving as a reversible modification targeting DNA and RNA. Furthermore, with the involvement of MARylation in essential cellular processes such as cancers, DNA repair, viral infection and cell cycle has been continuously reported, novel insights about MARylation attracts increasing attentions.
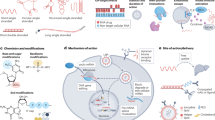
A MARylation of proteins. PARP3, 4, 6, 7, 8, 10, 11, 12, 14, 15 and 16 MARylates acceptor amino acid residues including arginine, cysteine, tyrosine and histidine that cleaved by TARG1, ARH3, MacroD1/2 and NUDT16. B MARylation of dsDNA termini. PARP3, 14 MARylates phosphorylated groups of dsDNA ends that removed by TARG1, PARG, ARH3, MacroD1/2 and NUDT16. C MARylation of ssDNA termini. PARP3, 14 MARylates phosphorylated groups of ssDNA ends that hydrolyzed by PARG. D MARylation of ssRNA termini. PARP10, 11,14, 15 and TRPT1 MARylates 5’ phosphorylated ssRNA which reversed by TARG1, PARG, ARH3, MacroD1/2 and NUDT16.
In this review, firstly, we provided an overview of MARTs catalyzing this novel modification, the hydrolases reversing it and ADP-ribose binding domains recognizing it. Secondly, we summarized the latest information about newly identified MARylated substrates including protein, DNA and RNA. And we emphasized on the physiological and pathological roles of these reactions in mammal as well as revealed the underlying mechanisms. Thirdly, we discussed the current methods for ADP-ribosylation detections and the state of MARTs inhibitors. Finally, we provided an outlook for future study, aiming to further analyzing the unclear biological functions of MARylation and their relevant mechanisms, and providing novel therapeutic perspectives in human diseases via targeting MARylation-related enzymes.
ADP-ribosyl transferases
The ARTs are defined by their catalytic domain, a polypeptide fold binding NAD+ and transferring ADP-ribose onto substrates covalently. There are 22 known human ARTs which grouped into two classes based on key amino acids in their catalytic domain. The ARTs containing a histidine-tyrosine-glutamate triad (H-Y-E motif) are grouped into ADP-ribosyl transferase diphtheria-toxin like (ARTDs) [6]. While the ARTs containing an arginine-serine-glutamate sequence (R-S-E motif) are referred to ADP-ribosyl transferase cholera-toxin like (ARTCs), also called ecto-ARTs [7]. In addition to these two ARTs families, two Sirtuins members are also confirmed to possess ART-like catalytic activity. Based on the number of ADP-ribose units, ADP-ribosylation could be in the form of a single ADP-ribose unit catalyzed by MARTs or polymers of ADP-ribose units modified by poly(ADP-ribosyl) transferases (Table 1).
ARTDs
ARTD family include 17 poly (ADP-ribose) polymerases (PARPs) and a divergent PARP homolog tRNA2 -phosphotransferase (TRPT1) [8,9,10]. Some members generate PAR and others are limited to MARylation. Actually, there are only four ARTD members including PARP1, PARP2, and Tankyrases (PARP5a and PARP5b) are able to synthetize long PAR chains that leading to PARylation. In contrast, majority ARTDs are MARTs which transfer only a single ADP-ribose unit resulting in targets MARylation. These enzymes are PARP3, PARP4, PARP6-12 and PARP14-16 which sharing a common characterization that no glutamate catalytic activity [11]. PARP9 and PARP13 are both regarded as no catalytic activity. During MARylating reaction, cleaving the covalent linkage between a ribose and a nicotinamide molecule is the rate-limiting process, and subsequently, the single ribose is to be connected to targets. The ARTDs possess diverse cellular distribution [12] and modify various substrates. Protein was first identified as an acceptor of ADP-ribosylation and most enzymes are known to be protein-modifying. However, some of others are described to modify DNA, RNA or chemical groups for example phosphate.
ARTCs
There are four ARTCs members in human [7]. These four ARTCs consist of two active members (ARTC1, ARTC5) and two inactive members (ARTC3, ARTC4). Since lacking the R-S-E motif, ARTC3 and ARTC4 cannot interact with NAD+. In despite of different binding mechanisms, ARTCs possess the similar configuration of NAD+ within the binding pocket as ARTDs. To date, protein is the only identified target of ARTCs-mediated MARylation and all active ARTCs conduct MARylation specifically onto arginine residues. ARTC1, 3 and 4 are localized in cellular membrane while ARTC4 is a secreted protein.
Sirtuins
Sirtuins is a class of deacetylases group with seven enzymes in human. Sirt4 and Sirt6 possess MARyation activity in addition to two above ART superfamilies [13]. Sirtuins cleave acyl groups in NAD+-dependent manner and generate O-acyl-ADP-ribose (OAADPr) followed by transferred to target residues such as Lysine [14, 15]. Sirt4 is localized in cytoplasm, mainly mitochondria, while Sirt6 is in nucleus.
Substantially, although current evidence presented the knowledge about ARTs, some of them are yet poorly understood and their structures and modification sites are required to identify. In addition, the modification and conjunction sites of ARTs, in particular, MARTs are not well established that remain to be solved.
ADP-ribosyl hydrolases
As a reversible modification, ADP-ribosylation is tightly regulated by three enzyme families including the DraG (dinitrogenase reductase activating glycohydrolase)-like ADP-ribosyl hydrolase family (ARHs), macrodomain-containing family and the nucleoside diphosphate linked to a variable moiety X (Nudix) family (Table 2). Additionally, an ADP-ribosyl hydrolase named NADAR was very recently found in bacteria and most of the eurkaryotes [16]. Due to no obvious homologues of the NADAR enzyme in humans, we do not further discuss NADAR in this article.
ARHs family
The ARH family contains three enzymes (ARH1, ARH2, and ARH3) with structurally similar catalytic domains including 290-360 residues, especially, ARH1 and ARH2 sharing a higher degree of sequence homology [17, 18]. ARH1, the first described ARH enzyme, specifically reverses MARylated arginine [19]. ARH1 is able to hydrolyze the N-glycosidic bond between a single ADP-ribose and arginine [20], while has weak capacity against PAR and OAADPr. Given that O-glycosidic is the main linkage between ADP-ribose and nucleic acids, ARH1 is not able to remove ADP-ribose from DNA or RNA [5]. The function of ARH1 is poorly understood despite a study showed that ARH1 deficiency promoted persistence of ADP-ribosylation and resulted in more sensitivity to Vibrio cholera infections in mice [21]. Due to lack of an aspartate in the catalytic center, ARH2 appears to be catalytically inactive [5]. ARH3 plays important roles in neurodegenerative disease [22] and efficiently uses PAR chains, MARylated serine and OAADPr as a substrate. ARH3 is the only hydrolase which able to cleave serine-linked mono(ADP-ribose) [23]. According to its specificity, ARH3 removes ADP-ribose from protein, DNA and RNA [23,24,25]. ARH3 is localized in both nucleus and mitochondria [18]. Nucleus-localization is responsible for its role in DNA damage [26] and regulation of parthanatos, a special type of cell death [27]. Mitochondria-localization of ARH3 may participate in the degradation of OAADPr as a metabolite in Sirtuins-mediated deacetylation reaction [28].
Macrodomain-containing family
Macrodomain-containing enzymes are widely distributed in all domains of life, and share a highly conserved ADP-ribose binding domain, known as macrodomain [29]. Macrodomains consist of 150–210 amino acids with the core motif harboring a three-layer sandwich architecture and are able to bind some OAADPr, MARylated, and PARylated proteins [30]. The poly-ADP-ribosyl glycohydrolase (PARG) is the only known members with PAR-hydrolyzing property but unable to remove the terminal ADP-ribose linked to substrates [31]. Notably, PARG is also able to reverse MARylation onto the 5’ and 3’ phosphorylated ssDNA and ssRNA [24, 25]. The functions of PARG are involved in DNA repair, replication forks and recovery from persistent replication stress [32]. Therefore, targeting PARG is regard as a novel approach for modern chemotherapy [33]. Additionally, The mutation of PARG is clearly associated with neurodegeneration and accumulation of PAR in the CNS [34]. There are also three other human macrodomain-containing family members which have hydrolytic activity towards MARylated substrates: MacroD1, MacroD2 and TARG1. These enzymes were demonstrated to remove the single ADP-ribose unit from modified protein substrates instead of PAR chain. TARG1 also cleaves the ester linkage between glutamate-linked PAR although its activity is much lower compared with PARG [35]. Importantly, MarcoD1, MacroD2 and TARG1 can cleave the single ADP-ribose from the 5’ or 3’ terminal phosphates of dsDNA and ssRNA to reverse nucleic acids modification [24, 25]. The roles of MacroD1, MacroD2, and TARG1 in normal cell biological processes are not well understood. A few studies have revealed the biofunctions of these macrodomain-containing enzymes. Deficiency of TARG1 causes neurodegenerative disorder in an autosomal recessive inheritance pattern [35]. TARG1 could also be related to insulin sensitivity. MACROD1, a mitochondrial enzyme, overexpressed in various tumor tissues is proposed to participate in growth and invasion of cancer cells [36, 37]. As, so far, only be detected in the brain, the MACROD2 has been reported to correlated with autism-spectrum disorders [38, 39], which is supported by behavioral phenotypes observed in MACROD2 gene knockout mice model [40].
Nudix family
The Nudix family, a class of pyrophosphatases, has been found to participate in metabolism of ADP-ribose. Instead of completely removing the ADP-ribose moiety, the Nudix digest the phosphodiester bond which links the adenosine to the ribose moiety in ADP-ribose, liberating the AMP or phosphoribose AMP. In Nudix family, only NUDT9 and NUDT16 have been shown to hydrolyze protein linked ADP-ribose [41, 42]. NUDT9 possesses phosphodiesterase activity against PARylated substrates and OAADPr while its hydrolytic capacity is much lower than that of NUDT16 [41, 43]. NUDT16 is the only Nudix member which digest both MARylated and PARylated substrates including PARylated protein and DNA, and MARylated protein, DNA and RNA, as well as OAADPr [44]. Notably, Ectonucleotide Pyrophosphatase/Phosphodiesterase 1 (ENPP1), a type II transmembrane glycoprotein, possesses equivalent activity to NUDT16 on ADP-ribosylated proteins. As reported, ENPP1 is able to convert protein-linked MAR and PAR into ribose-5′-phosphate [45].
In general, ADP-ribosyl hydrolases inversing the ADP-ribose modification functions as key regulators of this signaling. But yet, little is known that how these hydrolyses are modulated for example coordination with undefined cofactors. Furthermore, the binding sites’ specificity and the linkage selectivity are needed further detailed elaboration. Given that, better clarify molecular interactions between ADP-ribosylation hydrolyses and ARTs will contribute to revealing the mechanisms in charge of ADP-ribosylation reversal. The physiological and pathological functions associated with these enzymes remains largely unclear and further studies are required to clarify these issues.
ADP-ribose binding domains
In order to mediating downstream events, it is essential for ADP-ribosylation tags to be recognized by ADP-ribose binding proteins to read this modification. A number of studies have discovered different motifs, domains, or modules in proteins interacting with various forms of ADP-ribosylation.
PAR-binding motifs
The PAR-binding motif (PBM), a short 20 hydrophobic amino acid motif, is the most common PAR-reader domain. PBM’s directly bind to ADP-ribose likely based on electrostatic interactions between the positively charged amino acids in the PBM and the negatively charged PAR chains [46]. Although the exact nature of its interaction with PAR remains unclear, PBM is found in hundreds of proteins which involved in DNA-related processes, RNA regulation, cell-cycle regulation as well as apoptosis and parthanatos [47,48,49]. For instance, hexokinase, the rate-limiting enzyme of glycolysis, contains a strong PBM [50]. PARylation of hexokinase inhibits its catalytic activity and induces bioenergetic collapse in parthanatos [50]. Interestingly, multiple PBMs can simultaneously appear in a same protein and may cooperate in their interaction with PAR chains by increasing affinity and specificity [46, 47].
Macrodomains
The macrodomain, a sizable and globular module containing 130-190 amino acids, has been investigated the most extensively among the widespread ADP-ribose-binding domains. Macrodomains can interact with mono-ADP-ribose or the terminal ADP-ribose unit in poly-ADP-ribose chains and is responsible for reading variety of MARylated and/or PARylated substrates including proteins, free ADP-ribose, nucleic acids and ADPr metabolites [51]. Macrodomains participate in diverse biological events such as DNA repair, cellular signal transduction, transcription, genomic stability, immune response, different types of cell death [52]. Macrodomains are found in diverse proteins. Macrodomains act as readers of ADP-ribosylation in some macrodomain-containing proteins including macroPARPs (PARP9, PARP14 and PARP15), macroH2A1.1 and the chromatin remodeler ALC1 [53,54,55,56]. In contrast, macrodomains act as erasers of ADP-ribosylation in other macrodomain-containing proteins including ADP-ribose hydrolases (PARG, TARG1, MacroD1 and MacroD2) [29, 35, 51]. Interestingly, PARP14 is the only PARP with both transferase and hydrolase activities in one polypeptide due to it possessing both hydrolytic and binding macrodomains [57]. PARP14 exhibit ADP-ribosylhydrolase activity on protein and nucleic acid substrates [57].
WWE domains
The WWE domain is named after its three most conserved amino acids (tryptophan-tryptophan-glutamate). Unlike macrodomains, the WWE domains commonly prefer to recognize PARylated targets due to its pocket binds to the iso-ADP-ribose (the base-ribose linker between two ADP-ribose units) rather than an ADP-ribose unit [58,59,60]. Four residues including Y107A, Y144A, R163A, and Q153A in WWE domains are crucial for the recognition capacity [61]. Interestingly, this domain is also present in some E3 ubiquitin ligases [61, 62], highlighting the crosstalk between ADP-ribosylation and ubiquitylation, and supported by recent reviews [63, 64]. The best-characterized PAR-targeted E3 ligase is RNF146 (also named Iduna) which catalyzing the ubiquitylation and degradation of PARyated protein substrate [65,66,67,68,69], and influences on diverse biological processes [70]. It is very interesting that in ART family, the members who carry WWE domains appear to be MARTs including PARP7 and PARP11-14.
The PAR-binding zinc finger (PBZ)
PAR binding zinc fingers (PBZ) is a Cys2-His2 type zinc finger motif that binds PAR molecules contained in some DNA damage response proteins. The PBZ domain consists of a conserved motif with less than 30 amino acids and recognizes two adjacent ADPr units by hinging on adenine bases [71]. Similar to other zinc fingers, PBZ requires zinc for nanomolar affinity to its binding partner. Notably, compare to a single PBZ, the two zinc finger motifs cooperate to achieve high affinity for ADPr binding [72, 73]. Aprataxin polynucleotide kinase (PNK)-like factor (APLF) is a key regulator in DNA repair. Deletion of APLF in human cells leads to reduction of DNA repair rates following ionizing radiation [74]. APLF contains two tandem PBZ domains which are required for APLF recruitment to DNA damage sites [72].
RNA and DNA binding motifs
One of the most intriguing recent fundings is that ADP-ribose can be recognized by protein motifs that are able to bind to RNA or DNA. Similar to nucleic acids, the ADP-ribose carries a negative charge because of its phosphate backbone. Because these macromolecules tend to bind positively charged amino acid residue, some competition exists among DNA, RNA or PAR for the same binding site.
The RGG/RG motifs
The arginine/glycine-rich (RGG/RG) domain, also called glycine-arginine-rich (GAR) domains, is one of the most common RNA-binding domain throughout eukaryotes [75]. The RGG/RG motifs include RGG and RG repeats of varied lengths interspersed with spacers of different amino acids. Thousands of human proteins are reported to contain the RGG/RG motifs and involved in numerous cellular functions, such as DNA damage pathway, transcription, RNA processing, apoptosis and more [75, 76]. The RGG/RG domain binds to RNA in a non-specific sequence manner based on the interaction between its positively charged arginine residues and the negatively charged RNA chain [77]. Interestingly, The RGG/RG domain can also recognize PAR chain mediate downstream events. A number of studies demonstrated that some RNA-binding proteins containing the RGG/RG domain assembled in PAR-dependent way in response to genotoxic stress [78,79,80,81].
The RRM
The RNA recognition motif (RRM) is an abundant RNA binding domains which targeting multiple RNA sequences and structures. Although with lower affinity than for their preferred substrate, RRMs also recognized PAR molecule as a nucleic acid-like sequence. Some RRM-containing proteins assemble at sites of PAR formation and promote genome stability [82,83,84]. Notably, there is a dynamical competition between RNA and PAR, and various RNA-binding proteins to be assembled at PARylated sites.
The OB-fold
The oligonucleotiode/oligosaccharide binding fold (OB-fold), a 70–150 amino acid containing, is responsible for the interaction with single-stranded DNA (ssDNA) or oligosaccharides [85]. The exposed ssDNA is unstable and vulnerable to chemical attack and nucleolytic degradation, and needs to be properly protected to avoid mutations. Therefore, the OB-fold-containing proteins participate in multiple DNA metabolic processes including DNA replication, DNA repair, cell cycle regulation, and maintenance of telomeres [86]. Similar to the RGG/RG motifs, the OB-fold can also act as a PAR reader. Evidence supported that the OB-fold recognizes iso-ADP-ribose and bind to PAR chain [87, 88].
PIN domains
PIN domains are named after the amino terminus of the PilT protein and contain approximately 130 amino acids in length. Proteins possessing PIN domains are present in all kingdoms of life and act in a metal-iron-dependent way, usually through magnesium or manganese ion [89]. The PIN domains are evolutionarily conserved and recognize single-stranded DNA or RNA. A certain number of studies also confirm that the PIN domains bind to PAR with relatively high affinity, and this interaction is required for the rapid recruitment of the related protein to DNA break sites in response to DNA damage [90].
KR-rich and SR repeats motifs
Some of the most highly enriched PAR readers such as lysine- and arginine rich (KR-rich) motifs and Serine/Arginine repeats (SR repeats) do not contain a classic ADP-ribose binding domain. However, they have repeats of positively charged residues [91]. The positively charged arginine residues contribute to a strong electrostatic interaction with negatively charged PAR chains. Therefore, their affinity for both RNA and PAR is based on the positive charges associated with the arginine rich repeats.
CCCH zinc finger domains
Zinc finger proteins are commonly regard as DNA-binding transcription factors. However, Cys–Cys–Cys–His (CCCH) zinc finger proteins are capable of binding to target RNA and regulate RNA metabolism such as mRNA splicing, polyadenylation, export, translation and so on. The CCCH zinc finger domains consist of three conserved cysteines followed by a histidine that coordinated with a zinc ion, maintaining the structural stability [92]. So far, over fifty CCCH zinc finger proteins have been found in in humans and mice. Increasing functions of CCCH zinc finger proteins are being revealed such as regulation of cell differentiation and tumor growth, cytokine production, immune cell activation and immune homeostasis [93, 94].
Collectively, several modules have been identified to recognize ADP-ribose and play pivotal roles in the transduction of ADP-ribose signals, however, it appears that only macrodomains can interact with MAR. Hence more domains functions as readers of MAR are still to be further discovered.
MARylation of protein
PTM is a common tool to media a rapid change of the cellular status via adding chemical moieties. Protein was firstly identified as the substrate of ADP-ribosylation, subsequently various protein targets were discovered. As a highly conserved PTM, MARylation of proteins either directly alters protein activity or impacts on their interactions with other molecules. Unlike serine residues as the major acceptor sites for PARylation [95], the major MARTs are capable of modifying a variety of amino acid residues of modified proteins involving arginine, cysteine, tyrosine and histidine [96] in addition to serine [97]. Similar with other types of PTM, MARylation is also in a reversible pattern and the macrodomains are key modules that regulates MARylation.
Functions of protein MARylation
Conjugation of mono-ADP-ribose to proteins plays crucial signaling functions in various biological processes including RNA biology, cancer, cell-cycle, DNA damage repair, inflammatory response, virus infection and so on (Table 3).
DNA damage
The functions of PARylation in response to genotoxic stress have been so far well-established. However, recent studies provide novel insights into roles of MARylation in DNA damage repair. PARP3 is a DNA-dependent PARP and specific functions in cellular response to DNA strand breaks. PARP3 is able to be activated by DNA nicks, breaks and gaps possessing 5′ phosphorylated ends. The non-homologous end joining (NHEJ) is a classical double-strand breaks repair pathway in two DNA termini are directly ligated and active throughout whole cell cycle. PARP3 is shown to be efficiently recruited to DNA damage sites and to interact with different substrates belonging to the NHEJ pathway [98]. Moreover, PARP3 regulate mitotic progression and DNA damage repair by modifying the mitotic spindle components NuMa and the histone H2B, respectively [99, 100]. PARP3 also binds to PARP1 and activates PARP1, even in the absence of DNA, leading to synthesis of PAR chain [101]. Interestingly, PARP3 even modifies itself following their DNA-dependent activation [102]. PARP9 combines with E3 ligase Dtx3L to form heterodimer that is localized DNA damage sites and contributes to DNA repair response [103, 104]. Interesting, PARP9/DTX3L complex makes a hydbrid MAR-ubiquitin modification which also supports the connection between ubiquitination and ADP-ribosylation [105]. PARP10 has been the first identified MARylating enzyme and possesses multiple functions based on modification of its protein substrates [106]. A study showed that PARP10 regulates DNA damage hypersensitivity via binding to proliferating cell nuclear antigen (PCNA), a machinery component of DNA replication [107]. Consistently, PARP10 knockdown lead to sensitivity enhancement to DNA damage. PARP14 can also bind with PCNA to promote DNA replication and genomic stability in human cell lines [108]. While the opposite outcomes were shown in BRCA-deficient cells upon replication stress, in this study, PARP14 interacted with MRE11, a nuclease for replication forks degradation, to form a complex based on its catalytic activity and subsequent promoted genomic instability and DNA injury hypersensitivity [109]. These contradictory outcomes appear to be due to the diverse functions of PARP14-specific protein targets, supporting multiple roles of MARylation in DNA damage regulation. Notably, although lack of direct experimental evidence, PARP4 actually contains a BRCT domain which responsible for interacting with DNA damage repair proteins [110], indicating the potential role of PARP4 in DNA repair. Additionally, Sirt6 is recruited to DNA break sites and improves DNA repair by activation of PARP1 in MARylation-dependent way, and this action also strongly indicates the potential crosstalk between MAR and PAR [15]. KRAB-associated protein 1 (KAP1), a nuclear corepressor protein, is the other modified target of Sirt6. SIRT6 MARylates KAP1 to promote gene stability in response to DNA damage [111]. Current research has expanded the understanding of MARTs in DNA repair, however, less is known about the rest ones upon this context and further study needs to address this.
Cancers
The cancer-linked roles of MARylation and MARTs is relatively well-established. PARP6, a mono-ADP-ribose generating enzyme, directly MARylates Chk1 kinas which involved in regulating centrosome functions to promote tumorigenesis in breast cancer [112]. Correspondingly, PARP6 specific inhibitor AZ0108 induced the multipolar spindle phenotype to result in apoptosis of breast cancer cells [112]. In breast and colon cancer cells, PARP7 down-regulates oncogenic transcription factors such as HIF-1α, c-MYC, and estrogen receptor to modulate the War-burg effect and tumorigenesis. Mechanistically, PARP7 promotes these protein substrates ubiquitination and proteasomal degradation based on its MARylation activity [113]. Consistently, deletion of PARP7 in breast and colon cancer xenografts promotes tumorigenesis. This evidence also highlighted the crosstalk between MARylation and ubiquitination. In addition, PARP7 was reported to MARylate androgen receptor (AR) on its multiple cysteine residues to restrain AR-signaling in prostate cancer cells [114, 115]. Given the crucial role of AR prostate cancer, the PARP7-AR axis may be a potential therapeutic strategy for prostate cancer. However, in ovarian cancer, PARP7 plays the opposite role. As was reported, PARP7 MARylates a-tubulin, a cytoskeletal protein, to destabilize microtubules and facilitates tumor growth and motility, and PARP7 knockdown results in inhibition of tumor growth, migration and invasion [116]. These inconsistent data demonstrated the cancer-specific effects of PARP7. In general, MARylation mediated by PARP7 leads to a negative regulation of protein substrates and the multi-faceted effects probably due to the biofunctions of different targets and their downstream signaling. Notably, PARP9-Dtx3L complex not only regulates DNA repair but also exists in prostate cancer cells and possesses oncogenic capacity [117]. This complex inhibits interferon regulatory factor 1 (IRF1), a tumor suppressor, to promote the proliferation of different prostate cancer cell lines [117]. Similar in diffuse large B-cell lymphoma, PARP9 as a oncogenic co-factor represses the STAT1-IRF1 pathway in MARylation-dependent manner and upregulates two protooncogenes IRF2 and B-cell CLL/lymphoma (BCL)-6 levels [118]. Furthermore, PARP10 was reported to MARylate polo-like kinase 1 (PLK1), a kinase that relevant to tumor progression, to restrain its activity and oncogenic function in hepatocellular carcinoma [119]. The epithelial-mesenchymal transition (EMT) contributes to migration and invasion of tumor. PARP12 act as a tumor suppressor that controls hepatocellular carcinoma (HCC) metastasis via regulating the EMT process [120]. These novel functions occur through stabilization of the transcription factor FHL2, resulting in reduced levels of transforming growth factor beta 1 (TGF-β1), a EMT facilitator increasing mesenchymal marker levels and reducing epithelial marker levels. Integrin α7 is a cell surface adhesion molecule which regulates cell growth, migration and adhesion. MARylation of integrin α7 induced by ARCT1 modulate EMT and, accordingly, alteration of ARCT1 expression significantly changed the EMT markers levels [121, 122]. Given that poly-ADP-ribosyl transferases inhibitors as anticancer agents have successfully entered clinical practice [123], supporting the potential clinic uses of mono-ADP-ribosyl transferases inhibitors. Together, although strong evidence has supported the novel therapeutic strategies by targeting Mono-ADP-ribosyl transferases, further study still needs to identify the cancer-associated roles of MARylation and MAR-related enzymes.
Virus infection
MARylation plays a complex role in antiviral response through modifying host and viral proteins. Interferons (IFNs), designated types FIN-I, FIN-II and FIN-III, mediate the rapid host response to viral infection and play key roles in the innate immune system. PARP11 MARylates the E3 ligase β-transduction repeat-containing protein to promote IFN-I receptor subunit 1 ubiquitination and degradation, leading to suppression of antiviral capacity [124]. Inhibiting PARP11 pharmacologically could stabilize IFN-I receptor subunit 1 and enhance IFN-I pathway transduction to improve the resistant of mice to viral infection [124]. PARP14 functions as the main antiviral PARP against coronavirus infection and plays a crucial role in the induction of interferon [125]. TBK1 is an important kinase that drives IFN-I generation. It was reported that PARP7 MARylated TBK1 to restrain its phosphatase activity and inhibited IFN-I-induced innate response against viral infection [126]. STAT1 functions as activator of IFN signaling. PARP14 induces MARylation of STAT1 to suppresses IFN signaling which reversed by PARP9 via interacting with PARP14 [127]. PARP12, an interferon-stimulated MARTs, localizes Golgi and is characterized by its anti-viral function. During stress stimulation, PARP12 reversibly translocate from the trans-Golgi network to stress granules triggered by PARP1 [128, 129]. NS1 and NS2 are both nonstructural proteins of Zika virus (ZIKV), and responsible for the host antibody response and viral immune evasion strategies. PARP12 possesses anti-ZIKV activity via initially MARylating NS1 and NS2 to induce their proteasome-mediated degradation in a PARP-dependent way [130]. Chikungunya virus (CHIKV) is a mosquito-borne virus and encodes a nonstructural polyprotein (nsP) followed by cleaved into four components (nsP1-nsP4). PARP10 has been demonstrated to MARylate nsP2 to prevent multiple protein processing and replication of CHIKV [131, 132]. Stress granules (SGs), localized in the cytoplasm, is closely related to viral infections and innate immunity response. PARP10 MARylates and recruits GAPDH into SGs without affecting its dehydrogenase activity [133]. Besides, PARP12, PARP13 and PARP15 were also localized in SGs [134], and PARP12, PARP14 and PARP15 were defined as components of heat shock-induced SGs [135], suggesting the potential roles of MARylation in SGs-linked stress responses. Collectively, targeting PARPs is a potential strategy for antiviral response and will become a research hotspot. So far, there are only a few substrates of MARylation that have been identified. Therefore, more relevant substrates from host and virus need to be defined.
Inflammatory responses
Inflammation response is an essential immune reaction while uncontrolled injury will lead to a range of inflammation-related diseases. Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) is a nuclear transcription factor that plays a key role in inflammation and triggers transcription of pro-inflammatory cytokines such as interleukins (ILs), interferons (IFNs) and tumor necrosis factor (TNFs). PARP10 negatively modifies NF-kB signaling by MARylating NEMO, an important component of this pathway, upon the stimulation of IL-1β or TNF-a [136]. Macrophage activation is divided into pro-inflammatory and anti-inflammatory phenotypes. Pro-inflammatory macrophage activation participates in pathogenesis of inflammation-associated disorders. It was reported that PARP9 and PARP14 cross-regulated macrophage and possessed opposing functions in macrophages activation and arterial lesions [127]. Mechanistically, PARP14 MARylates Glu657 and Glu705 of signal transducer and activator of transcription 1 (STAT1) that implicated as a regulator of pro-inflammatory mediators to inhibit Try701 phosphorylation, and which suppressed by PARP9. These findings also suggest interplay between MARylation and phosphorylation in inflammatory diseases mediated by pro-inflammatory macrophage activation. In cancers, PARP14 plays an immune suppressive role via regulation of IL-4 and IFNs-γ signaling pathways [137]. RBN012759, a selective PARP14 inhibitor, reverses protumor gene expression in macrophages and mediates inflammatory response in human tumor explants [137]. Human neutrophil peptide 1 (HNP-1) is a crucial component of innate immunity response. ARTC1 MARylates HNP-1 on its arginine residue mediated multi-functions including inhibition of cytotoxic and anti-microbial roles while enhancement of IL-8 release and neutrophil recruitment [138].
Lipid metabolism
Liver X receptors (LXRs) are nuclear receptors that responsible for the regulation of cholesterol, lipid, and glucose metabolism. It was reported that PARP7 could MARylate LXR-α and LXR-β subunits to enhance LXRs-dependent transactivation which inhibited by ADP-ribosyl hydrolase, MACROD1 [139]. As the downstream targets of LXRs, Sterol regulatory-element binding proteins (SREBPs) are responsible for lipid synthesis and PARP7 acts as a positive regulator of SREBP1 level via LXR pathway [139]. Downregulation of PARP10 level is associated with an increasing of fatty acid oxidation [140]. Furthermore. PARP10 knockdown results in a reduction of secreted apolipoprotein B, a main carrier of triglyceride-rich lipoproteins and low-density lipoprotein cholesterol [141].
Cell cycle
The mitotic kinase Aurora-A is a critical player in centrosome function and mitosis. It was reported that PARP10 regulated the transition from the G2-phase to mitosis by MARylation of Aurora-A [142]. Depletion of PARP10 affects Aurora-A recruitment in centrosomes and exhibited a delayed progression in the G2/M phase. RAN is a GTPase and responsible for nucleocytoplasmic trafficking and mediation of mitosis [143]. RAN was identified as a MARylated substrates of PARP10 that partially revealing the mechanisms by which PARP10 regulates cell-cycle [54]. Moreover, ARTC1 can regulate mitosis though MARylating platelet-derived growth factor-BB (PDGF-BB) in human cell line [144].
RNA biology
MARylation of RNA-regulatory proteins can affect their function and regulate RNA biology such as translation, transcription. Ribosomes are the key components participating in mRNA translation. A recent research showed that PAPP16-mediated MARylation of ribosomal proteins including RPL24 and RPS6 significantly restrained polysome assembly, mRNA loading and translation [145]. Elongation factor 2 (EF2) is a GTPase that participates in ribosome translocation. MARylation of EF2 suppressed its activity and inhibited protein synthesis even though the mechanism of this inhibitory effect is unknown [146]. Argonaute (Ago) proteins are other important ingredients of the RNA-induced silencing complex (RISC) which regulates mRNAs translation. PARP13 functioned in these processes via modifying Ago2 to induce translational repression [128]. In transcriptional aspect, it was found that PARP7 played an important role in transcription regulation via MARylating aryl hydrocarbon receptor (AHR) a ligand-activated transcription factor acting as a negative feedback regulator of transcription [147], and the PARP7-induced effects can be reversed by mono-ADP-ribosylase MACROD1 [148].
Endoplasmic reticulum (ER) stress
Protein kinase RNA-like ER kinase (PERK) and inositol-requiring enzyme 1 (IRE1α) are two unfolded protein response-associated kinases that play key roles in ER stress. And a research demonstrated PERK and IRE1α were the substrates of PARP16, an ER transmembrane protein [149]. As reported, PARP16-induced MARylation of PERK and IRE1α increased their kinase activity and promoted downstream events [149]. Knockdown of PARP16 made cells more sensitive to ER stress and reduced cell viability. GRP78/BiP is a key ER chaperone and also an identified target of ARTC1. ARTC1 inhibited GRP78/BiP activity in MARylation-dependent way and led to reduction of protein synthesis and protein flux into ER in response to ER stress [150].
Other new functions
MARylation also play roles in other physiological and pathological processes including autophagy, insulin secretion and so on. Some studies demonstrated that PARP10 and PARP12 could both interact with p62, a ubiquitin receptor associated with autophagy [151, 152], suggesting the their potential role of in autophagy. However, it is still lacking sufficient evidence to prove these functions are based on MARylation of their substrate p62. In addition, PARP10-mediated MARylation can directly regulate the kinase activity of glycogen synthase kinase 3β (GSK3β), a well-known enzyme playing important role in metabolism, immunity, cell proliferation and tumorigenesis [153]. Glutamate dehydrogenase (GDH) is documented to enhance glutamate and glutamine metabolism to promote insulin secretion. Sirt4 represses GDH activity by MARylation to reduce insulin release in response to glucose and amino acids [154].
MARylation of DNA
ADP-ribosylation of DNA was first reported in 2001 [155]. This study showed that pierisin-1, an ARTC enzyme from the cabbage butterfly, is able to MARylate double-stranded DNA (dsDNA) at the N2 position of guanine [155]. Notably, the pierisin-1-mediated modification is irreversible. In 2021, tightly regulated reversible DNA ADP-ribosylation systems were reported in a variety of bacteria that control for instance DNA replication and growth [156]. In Mammalian, there are three DNA-associated PARPs including PAPR1-3 which commonly harbor a tryptophan-glycine-arginine (WGR) domain responsible for DNA binding. PARP1 and PARP2 are able to PARylate DNA to generate PAR-DNA adducts. However, among these three enzymes, PARP3 is the only DNA MARTs which catalyzes attachment of a single ADP-ribose onto phosphorylated DNA ends. PARP3 shows MARylation activity on both 3′ - and 5′ -terminal phosphorylated groups at double- and single-strand DNA (ssDNA) break ends, whereby the 5’-termini presents the preferred target [25, 157]. An in vitro experiment also demonstrated that PARP3 catalyzed MARylation at the 5′-phosphate terminal of gapped DNA, which can serve as a substrate of DNA ligases for ligation with dsDNA and also serve as primer for PARP1- or PARP2-induced PARylation [158]. Just recently, two studies showed that PARP14 could also MARylates both ssDNA with a phosphate group at the termini and nascent dsDNA [159]. Of note, MARylation of DNA ends is a reversible process. Several cellular known hydrolases such as PARG, MACROD2, TARG1 and ARH3 can remove the mono-ADP-ribose moiety covalently attached to DNA, and PARG is most efficient in removing ADP-ribosylation on DNA, supporting that DNA termini MARylation serve as a transient mark [25, 157]. Currently, (patho)physiological roles of DNA ADP-ribosylation in eukaryotes are not clear yet but there are many suggestions that it exists.
Functions of DNA MARylation
PARP3 is stimulated by DNA double-strand breaks (DSBs) and associated with the cellular response to DNA damage. Co-immunoprecipitation studies of PARP3 identified various proteins that are involved in NHEJ DNA damage repair to induce the direct re-ligation of DNA lesions [98, 160]. In accord, punished data support the function of PARP3-dependent MARylation in this type of DNA damage repair [161, 162]. Given that PAR-DNA complex breakage sites are reported to be prevented from degradation by nucleases [163], together with the initial single ADP-ribose added by PARP3 can be extended by PARP1 and PARP2, it is reasonable to propose PARP3-mediated DNA MARylation may play a protective role by shielding breakage sites until repair proteins fully recruited during DNA damage. The involvement of PARP3 in chromatin compaction regulation and chromosomal rearrangements in cells also supports its participation in the DNA repair mechanisms [164, 165]. In opposite, PARP14 bound with nascent DNA to promote MRE11-mediated fork degradation and induced gene instability, as above mentioned. For now, MARylation of DNA ends likely functions as a modification for protection of targeted DNA from nucleases-mediated uncontrolled degradation and a signal for recruitment of DNA repair-associated proteins. However, the biochemical characterization of ADP-ribosylation at DNA breakage sites is still largely unclear and the function of this novel DNA modification in cells remains to be unraveled.
MARylation of RNA
RNA has so far been demonstrated to serve as a substrate for MARylation catalyzed by MARTs involving PARP10, PARP11, PARP12, PARP14, PARP15 and a PARP like enzyme -TRPT1 [159, 166]. Some other MARTs such as PARP7 and PARP12 have been predicted to bind phosphorylated RNA ends [167]. PARP7, PARP12 and PARP13 contain zinc finger domains of the CCCH-type while PARP10 and PARP14 contain multiple RRMs. However, it was reported the catalytic domains of PARP4, PARP6, PARP13 and PARP14 as well as full-length PARP3 and PARP16 had no activity on the modification of 5’-phosphorylated RNA [24]. In contrary, a recent research proved evidence for first time single-stranded RNA (ssRNA)-binding ability and RNA MARylation capacity of both PARP14 and its fragment containing a KH domain which functions as a sequence-specific RNA-binding module [159]. PARP14 is not able to modify RNA substrates with unmodified ends but induce MARylation on a phosphorylated termini [159]. PARP10 can also MARylate phosphorylated ssRNA ends with a preference for 5′-terminal phosphate rather than 3’-terminal phosphate [24]. However, PARP10 is not able to modify ssDNA. Interestingly, the full-length PARP10 mortifies RNA ends with less efficient activity than its catalytic domain polypeptide alone that probably due to an auto-inhibitory pattern involving an interaction of N-terminal sequences with the catalytic domain. In addition, PARP11 and PARP15 are also capable of catalyzing phosphorylated ssRNA. TRPT1, known for its essential function in the fungal tRNA splicing pathway, transfers ADP-ribose moiety to 5’-phosphorylated ends of RNA to form a 5’-cap structure, independent of oligomer length [24]. Importantly, a recent study presented that RNA MARylation occurs in human cells and is further confirmed to be conducted by TRPT1, PARP10, PARP11, PARP12, and PARP15 [166]. Similar to MARylated DNA, this non-canonical modification of RNA in cells can be reversed by cellular hydrolases including TARG1, PARG, ARH3, and MacroD1/2 indicating high dynamicity of this modification [24, 166]. Interestingly, a very recent study provides evidence that ADP-ribosylated RNA can be further ubiquitylated to produce a potential dual ubiquitin-ADP-ribose- modification [168].
Functions of RNA MARylation
RNA functions are heavily dependent on post-transcriptional modifications. Before translated and degraded, pre-mRNAs have to undergo multiple processing processes involving 5′ capping, splicing, 3′ polyadenylation, and then generate mature mRNAs. To data, little is known about the role of RNA MARylation as a newly identified post-transcriptional regulator. Recently, a study in mammalian cells demonstrated that various cellular stressors such as IFN, MG132, Arsenite and H2O2 could regulate the level of MARylated RNA [166]. Furthermore, MAR-capped target mRNA is prevented from XRN1-induced degradation but not allowed by translation [166]. This exciting evidence indicates a potential mechanism that MAR acts as a novel mRNA cap and protects modified mRNA from degradation in cellular stress conditions, and then this modification is removed by ADP-ribosyl hydrolases until get rid of cellular stress. Two recent studies showed that PARP14 binds to cyclin D1 mRNA 3′UTR to regulate cell-cycle progression and interacts with tissue factor (TF) mRNA 3’UTR to regulate its mRNA degradation [169, 170]. Given that PARP14 functions as a RNA binding protein and possesses MARTs activity, a possibility is that PARP14 selectively MARylates mRNA 3′UTR in order to affect mRNA stability. Similarly, PARP13, a catalytically inactive enzyme, binds to a region in cellular TRAILR4 3’UTR to increase cell sensitivity to TRAIL-mediated apoptosis [171]. Moreover, PARP16, function as a RNA binding protein, modulates the mRNA stability of amyloid precursor protein (APP), the precursor of beta-amyloid (Aβ) and prevents modified APP from degradation in Alzheimer’s disease (AD) mice [172]. Notably, different from other MARylated mRNAs with significant inhibition of their translation in cellular stresses, PARP16-mediated modification promotes mRNA stability and increased APP protein level [172]. It is probably due to the chronic and degenerative pathological processes of AD, compared to acute stress conditions, and the neuronal damage role of PARP16 in AD which exacerbates APP production. Additionally, a recent study showed that PARP12 could interact with SARS-CoV-2 RNA and depletion of PARP12 led to viral SARS-CoV-2 RNA levels increased, indicating the antiviral function of RNA MARylation [173]. Generally, MARylation provides a novel potential mechanism for the post-transcriptional regulation of target gene expression, especially in response to cellular stresses. In this process, different MARTs catalyze different sites such as 3′UTR or 5′UTR. The main biological effects triggered by this modification are prevention of MARylated mRNA from degradation which is similar with that of DNA, and inhibition of target gene translation during acute cellular stresses. Given the identification of RNA MARylation is rather recent, it is still lack of sufficient in vivo data about RNA MARylation. Future studies in cells are necessary to sufficiently reveal RNA MARylation’s roles and impactions such as mRNA stability, localization and translation. It would also be interesting to identify specific MAR-capped RNAs, such as mRNA, transfer RNA and ribosomal RNA and so on.
Detection of ADP-ribosylated substrates
Currently, techniques for MARylated substrates detection, especially endogenous MARylation are scarce, as a result, its regulation and specific sites remain largely unclear. Based on the ADP-ribosylation reaction, cofactor radiolabelled NAD+ as substrate is used to identify ADP-ribosylated substrates such as protein and nucleic acids. This method has high sensitivity and no effects on enzyme catalysis [24]. In addition, biotin-labelled NAD+ and NAD+ analogues have also used to label MARylated targets [153]. However, these modified NAD+ methods are limited to in vitro assays. In mammalian cells experiments, the first MAR-specific rabbit polyclonal antibody was produced in 1992 and targeted MARylated Sulfolobus acidocaldarius eEF2, an earliest known protein substrate of this modification [174]. Functional domains that bind to ADP-ribose have been used as alternatives to antibodies for ADP-ribosylation detection [175]. For example, Af1521 macrodomain interacts with MARylated and PARylated substrates while macro2/macro3 macrodomain prefer to recognize MARylated proteins [54, 176]. Encouragingly, the first protein site-specific antibodies that have very recently gotten available [177]. Furthermore, the toolkit for detection of ADP-ribosylation on DNA has been established in human cells [178]. Some groups have also developed agents with high affinity and specificity for MARylated RNA [24, 179]. Additionally, recent labeling methods including a clickable aminooxy alkyne (AO-alkyne) probe, enzymatic labeling of terminal ADP-ribose (ELTA) and N6 -propargyl adenosine (N6pA) tag provide a suite of techniques for ADP-ribosylation detection, although not always specifically targeting MARylation [180,181,182,183].
Progress and challenges of MARTs inhibitors
So far, therapeutic benefits of PARPs inhibitors mainly have come from inhibitors of PARP1 and other nucleus PAPRs for cancer treatment [184]. However, selective inhibitors targeting MARTs are very limited. Recent advances in the understanding of MARTs structures and bioactivities lead to some progress in development of MARTs inhibitors. PARP3 modulates mitotic progression and DNA damage repair as mentioned above. A specific inhibitor of PARP3 known as ME0328 is in early-phase development, and has been proved to promote the chemotherapy agent vinorelbine cytotoxicity [185, 186], supporting the potential combination strategies of MARTs inhibitor with other therapeutic modalities. PARP7 also play key roles in several types of cancers. RBN-2397, the inhibitor specific for PARP7, can enhance type I IFN signaling to induce antitumor immunity dependent on CD8 T cells [187]. Different from nucleus PARPs inhibitors commonly tested as combination partners for chemotherapies [188], RBN-2397 administered alone represents complete and durable inhibition of tumor growth [187]. Now RBN-2397 is in Phase I clinical trials for solid tumors treatment (https://clinicaltrials.gov/ct2/show/NCT04053673). In 2016, Venkannagari et al identified OUL35 as a selective and potent inhibitor for PARP10. OUL35 reverses PARP10-induced human cell death and sensitizes cells to DNA damage [189]. In the follow-up study, Holechek et al designs PARP10/PARP14 selective inhibitors based on OUL35 that possess good metabolic stability against mouse liver microsomes [190]. Kirby group identifies a potent and selective PARP11 inhibitor ITK7 [191]. This study demonstrates that ITK7 inhibits PARP11 auto-MARylation and causes PARP11 to dissociate from the nuclear envelope [191]. Mariko et al. perform high-throughput screening and an immunoradiometric assay to identify PAPR14 inhibitors via competing for binding with NAD+ [192]. In 2021, the first highly potent and selective inhibitor of PARP14 RBN012759 has been reported. RBN012759 enables to reverse IL-4-driven protumor gene expression in macrophages and induces an inflammatory gene signature in kidney cancer tumor explants [137]. Subsequently, evidence shows that RBN012759 reduce stress granule assembly in ovarian cancer cells [193]. Wang et al characterizes epigallocatechin-3-gallate (EGCG), a major flavonoid of green tea, as a potential inhibitor of PARP16 [194]. Given that PARP16 play an important role in ER stress, EGCG has been shown to promote the ER stress-mediated cancer cells apoptosis [194].
In general, the development of clinical inhibitors of the known MARTs is still lagged behind. The efficacies of some inhibitors are not well-documented, and data about the safety and resistance of MARTs inhibitors are almost blank. Further research needs to evaluate their characteristics in vivo and even in patients. Moreover, current inhibitors of PARPs are mainly used for cancer therapy due to their roles in DNA repair pathway. Given the multiple functions of PARPs and MARTs in various diseases, MARTs inhibitors may also have potential effects on non-oncological indications such as inflammation, viral infection, lipid metabolic diseases. Notably, better screening techniques, reliable biomarkers of response and refined structural information will also transfer benefits to design and discovery of novel MARTs inhibitors. Addition to MARTs, the ADP-ribosyl hydrolases that remove MAR could also serve as targets for drugs development.
Future perspectives
MARylation is a regulatory modification on protein or nucleic acids, and participates in modulation of various cellular pathways and plays crucial roles in health condition and multiple diseases. However, it is still in the initial stage and remains largely unknown. Therefore, there are several important issues that are required to be further addressed.
To well understand biological activities and mechanisms
Compare to the functions of PARylation is well documented, we do not yet well understand the roles of MARylation in pathogenic and physiological conditions. Further studies are needed to address the roles of MARylation and MARylation-associated enzymes such as MARTs and MAR hydrolases in health and disease. At the same time, as a regulatory modification, the relevant molecular and biochemical mechanisms of MARylation on protein and nucleic acids, the mechanisms underlying their functions are also required to revealed. Furthermore, identification of novel MARylated target will contribute to better understanding of this modification. With the list of substrates growing, MARylation would become a most exciting cellular function.
To increase in vivo tests
So far, most data about MARylation and MARTs derive from in vitro tests by commonly using recombinant components, remaining a vital issue that the relevance in vivo are unknown. Moreover, different MARTs show diverse patterns of tissue distribution, and might possess relevant biofunctions in specific organizations. Thence, more experiments in cells and animals are urgently in need to confirm previous hypothesis and provide better understanding biological influences of MARylation.
To develop specific MARylation detection methods
Although significant progress is made in developing techniques for ADP-ribosylated targets detection and analysis, it is still not enough, especially for MARylated molecules. Given the biological importance of MARylation is beginning to emerge, further development and refinement of MAR-specific methods will provide a tremendous progress in the field. Reliable tools will contribute to identify and functional assessment of specific sites of MARylated substrates, as well as advance the understanding of MARylation’s functions and mechanisms.
To pay attention on nucleic acids MARylation
Unlike protein, nucleic acid MARylation is newly identified and in a more emerging field that leaving some objectives of further research about MAR-RNA and MAR-DNA to achieve. One is to further confirm the existence of MARylation on nucleic acids, in particular RNA, in vivo. The other one is to reveal the regulatory mechanisms of MARylation-associated enzymes to understand their substrates and substrate recognition pattern. In addition, it is required to comprehensively elaborate the outcomes of MARylation on nucleic acids and their downstream events. Better addressing all the remaining problems will enormously advance the deeply understanding of MARylation in response to physiological and pathological signals.
To identify undiscovered enzymes regulating MARylation reactions
Following rapid advances in the MARylation field, increasing ARTs and hydrolyses members have been identified. However, we are still far from having a clear image of MARyaltion system and there are probably remaining undiscovered enzymes modulating MARylation reactions. Identification of novel enzymes possessing a substrate MARylation activity will contribute to understanding of this extraordinary modification and also provide potential targets of specific inhibitors for human therapy. Remarkably, to find out the human homologs of MARylation-relating enzymes in bacteria, fungi or viruses may be an effective strategy. For instance, TRPT1 family was initially found in Saccharomyces cerevisiae [195] and its human version referred to as TRPT1. Furthermore, it is also a feasible method to explore the MARylation activity from those known enzymes.
To reveal the potential connection between MARylation and PARylation
Growing evidence support that MARylation has close association with PARylation. As above, an established PAR transferase PAPR1 is a target of Sirt6 [15]. Under oxidative stress, Sirt6 activates PAPR1 in MARylation-dependent manner to regulate DNA repair pathway [15]. In addition, MARylation functions as a second wave of PARP1 pathway via recruitment of a MARyationn reader RNF114 in response to DNA damage [177]. Besides genomic stability, PARP1 play roles in various cellular processes thus this co-modulatory effect mediated by MARylation and PARylation may exists in diverse pathophysiology conditions. Moreover, with improvement of detection techniques, we may uncover more biological significances and mechanisms behind the crosslink between MAR and PAR.
Data availability
All data are available in the manuscript; expression constructs are available on request.
References
Schuller M, Ahel I. Beyond protein modification: the rise of non-canonical ADP-ribosylation. Biochem J. 2022;479:463–77.
Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43.
Pascal JM. The comings and goings of PARP-1 in response to DNA damage. DNA Repair. 2018;71:177–82.
Lüscher B, Bütepage M, Eckei L, Krieg S, Verheugd P, Shilton BH. ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem Rev. 2018;118:1092–136.
Weixler L, Schäringer K, Momoh J, Lüscher B, Feijs KLH, Zaja R. ADP-ribosylation of RNA and DNA: from in vitro characterization to in vivo function. Nucleic Acids Res. 2021;49:3634–50.
Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Adv Exp Med Biol. 1997;419:35–43.
Bazan JF, Koch-Nolte F. Sequence and structural links between distant ADP-ribosyltransferase families. Adv Exp Med Biol. 1997;419:99–107.
Suskiewicz MJ, Prokhorova E, Rack JGM, Ahel I. ADP-ribosylation from molecular mechanisms to therapeutic implications. Cell. 2023;186:4475–95.
Groslambert J, Prokhorova E, Ahel I. ADP-ribosylation of DNA and RNA. DNA Repair. 2021;105:103144.
Hu QD, Lu H, Huo K, Ying K, Li J, Xie Y, et al. A human homolog of the yeast gene encoding tRNA 2′-phosphotransferase: Cloning, characterization and complementation analysis. Cell Mol Life Sci. 2003;60:1725–32.
Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426.
Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013;4:2240.
Bheda P, Jing H, Wolberger C, Lin H. The substrate specificity of sirtuins. Annu Rev Biochem. 2016;85:405–29.
Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–90.
Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–6.
Schuller M, Raggiaschi R, Mikolcevic P, Rack JGM, Ariza A, Zhang YG, et al. Molecular basis for the reversible ADP-ribosylation of guanosine bases. Mol Cell. 2023;83:2303–2315.e6.
Mashimo M, Kato J, Moss J. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair. 2014;23:88–94.
Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–13.
Wang H, Norén A. Metabolic regulation of nitrogen fixation in Rhodospirillum rubrum. Biochem Soc Trans. 2006;34:160–1.
Rack JGM, Ariza A, Drown BS, Henfrey C, Bartlett E, Shirai T, et al. (ADP-ribosyl)hydrolases: structural basis for differential substrate recognition and inhibition. Cell Chem Biol. 2018;25:1533–1546.e12.
Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27:5534–43.
Danhauser K, Alhaddad B, Makowski C, Piekutowska-Abramczuk D, Syrbe S, Gomez-Ospina N, et al. Bi-allelic ADPRHL2 mutations cause neurodegeneration with developmental delay, ataxia, and axonal neuropathy. Am J Hum Genet. 2018;103:817–25.
Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, Ahel I. Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife. 2017;6:1–20.
Munnur D, Bartlett E, Mikolčević P, Kirby IT, Rack JGM, Mikoč A, et al. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 2019;47:5658–69.
Munnur D, Ahel I. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017;284:4002–16.
Beijer D, Agnew T, Rack JGM, Prokhorova E, Deconinck T, Ceulemans B, et al. Biallelic ADPRHL2 mutations in complex neuropathy affect ADP ribosylation and DNA damage response. Life Sci Alliance. 2021;4:1–15.
Yingfei W, No Soo K, Jean-Francois H, HoChul K, Karen KD, Shaida AA, et al. Poly (ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (Parthanatos). Sci Signal. 2011;4:ra20.
Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. J Biol Chem. 2012;287:42419–27.
Rosenthal F, Feijs KLH, Frugier E, Bonalli M, Forst AH, Imhof R, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–7.
Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–20.
Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–22.
Illuzzi G, Fouquerel E, Amé JC, Noll A, Rehmet K, Nasheuer HP, et al. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res. 2014;42:7776–92.
James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, et al. First-in-class chemical probes against poly(ADP-ribose) glycohydrolase (PARG) inhibit DNA repair with differential pharmacology to olaparib. ACS Chem Biol. 2016;11:3179–90.
Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:82–86.
Sharifi R, Morra R, Denise Appel C, Tallis M, Chioza B, Jankevicius G, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–37.
Shao L, Jing W, Wang L, Pan F, Wu L, Zhang L, et al. LRP16 prevents hepatocellular carcinoma progression through regulation of Wnt/β-catenin signaling. J Mol Med. 2018;96:547–58.
Meng YG, Han WD, Zhao YL, Huang K, Si YL, Wu ZQ, et al. Induction of the LRP16 gene by estrogen promotes the invasive growth of Ishikawa human endometrial cancer cells through the downregulation of E-cadherin. Cell Res. 2007;17:869–80.
Lombardo B, Esposito D, Iossa S, Vitale A, Verdesca F, Perrotta C, et al. Intragenic deletion in macrod2: A family with complex phenotypes including microcephaly, intellectual disability, polydactyly, renal and pancreatic malformations. Cytogenet Genome Res. 2019;158:25–31.
Jones R, Cadby G, Blangero J, Abraham L, Whitehouse A, Moses E. MACROD2 gene associated with autistic-like traits in a general population sample. Psychiatr Genet. 2014;24:241–8.
Crawford K, Oliver PL, Agnew T, Hunn BHM, Ahel I. Behavioural characterisation of macrod1 and macrod2 knockout mice. Cells. 2021;10:1–24.
Palazzo L, Thomas B, Jemth AS, Colby T, Leidecker O, Feijs KLH, et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem J. 2015;468:293–301.
Kulikova VA, Nikiforov AA. Role of NUDIX hydrolases in NAD and ADP-ribose metabolism in mammals. Biochem. 2020;85:883–94.
Perraud AL, Shen B, Dunn CA, Rippe K, Smith MK, Bessman MJ, et al. NUDT9, a member of the Nudix hydrolase family, is an evolutionarily conserved mitochondrial ADP-ribose pyrophosphatase. J Biol Chem. 2003;278:1794–801.
O’Sullivan J, Tedim Ferreira M, Gagné JP, Sharma AK, Hendzel MJ, Masson JY, et al. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun. 2019;10:1–14.
Palazzo L, Daniels CM, Nettleship JE, Rahman N, McPherson RL, Ong SE, et al. ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 2016;283:3371–88.
Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–80.
Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–76.
Fischbach A, Krüger A, Hampp S, Assmann G, Rank L, Hufnagel M, et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018;46:804–22.
Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci Signal. 2011;4:20.
Andrabi SA, Umanah GKE, Chang C, Stevens DA, Karuppagounder SS, Gagné JP, et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci USA. 2014;111:10209–14.
Feijs KLH, Forst AH, Verheugd P, Lüscher B. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol. 2013;14:443–451.
Rack JGM, Perina D, Ahel I. Macrodomains: structure, function, evolution, and catalytic activities. Annu Rev Biochem. 2016;85:431–54.
Lehmann LC, Hewitt G, Aibara S, Leitner A, Marklund E, Maslen SL, et al. Mechanistic insights into autoinhibition of the oncogenic chromatin remodeler ALC1. Mol Cell. 2017;68:847–859.e7.
Forst AH, Karlberg T, Herzog N, Thorsell AG, Gross A, Feijs KLH, et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure. 2013;21:462–75.
Barkauskaite E, Jankevicius G, Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol Cell. 2015;58:935–46.
Ahel D, Hořejší Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3.
Dukić N, Strømland Ø, Elsborg JD, Munnur D, Zhu K, Schuller M, et al. PARP14 is a PARP with both ADP-ribosyl transferase and hydrolase activities. Sci Adv. 2023;9:1–18.
He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, et al. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–64.
Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagné JP, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci USA. 2011;108:14103–8.
Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17:692–9.
Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly (ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–40.
Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–5.
Pellegrino S, Altmeyer M. Interplay between ubiquitin, SUMO, and poly(ADP-Ribose) in the cellular response to genotoxic stress. Front Genet. 2016;7:63.
Matsumoto Y, Rottapel R. PARsylation-mediated ubiquitylation: lessons from rare hereditary disease Cherubism. Trends Mol. Med. 2023;29:390–405.
Hu K, Wu W, Li Y, Lin L, Chen D, Yan H, et al. Poly(ADP ‐ribosyl)ation of BRD 7 by PARP 1 confers resistance to DNA ‐damaging chemotherapeutic agents. EMBO Rep. 2019;20:e46166.
Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 2015;29:157–70.
Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–9.
Matsumoto Y, La Rose J, Lim M, Adissu HA, Law N, Mao X, et al. Ubiquitin ligase RNF146 coordinates bone dynamics and energy metabolism. J Clin Investig. 2017;127:2612–25.
Wu H, Li Y, Zhang Q, Wang H, Xiu W, Xu P, et al. Crocetin antagonizes parthanatos in ischemic stroke via inhibiting NOX2 and preserving mitochondrial hexokinase-I. Cell Death Dis. 2023;14:50.
Zhou ZD, Chan CHS, Xiao ZC, Tan EK. Ring finger protein 146/Iduna is a Poly (ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adhes Migr. 2011;5:463–71.
Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85.
Li GY, McCulloch RD, Fenton AL, Cheung M, Meng L, Ikura M, et al. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc Natl Acad Sci USA. 2010;107:9129–34.
Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, et al. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat Struct Mol Biol. 2010;17:241–3.
Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27:3793–803.
Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG Motif. Mol Cell. 2013;50:613–23.
Ozdilek BA, Thompson VF, Ahmed NS, White CI, Batey RT, Schwartz JC. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017;45:7984–96.
Chowdhury MN, Jin H, The RGG. motif proteins: Interactions, functions, and regulations. Wiley Interdiscip Rev RNA. 2023;14:e1748.
Isabelle M, Gagné JP, Gallouzi IE, Poirier GG. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci. 2012;125:4555–66.
Altmeyer M, Toledo L, Gudjonsson T, Grøfte M, Rask MB, Lukas C, et al. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol Cell. 2013;52:206–20.
Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DAQ, Gomez-Herreros F, et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–14.
Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–62.
Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA damage response. Nat Cell Biol. 2012;14:318–28.
Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-vaites L, Liang AC, et al. A systematic analysis of factors localized to damaged chromatin reveals PARP-dependent recruitment of transcription factors. Cell Rep. 2016;11:1486–1500.
Krietsch J, Caron MC, Gagné JP, Ethier C, Vignard J, Vincent M, et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–301.
Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–7.
Wu Y, Lu J, Kang T. Human single-stranded DNA binding proteins: guardians of genome stability. Acta Biochim Biophys Sin. 2016;48:671–7.
Zhang F, Shi J, Bian C, Yu X. Poly(ADP-Ribose) mediates the BRCA2-dependent Early DNA damage response. Cell Rep. 2015;13:678–89.
Zhang F, Chen Y, Li M, Yu X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc Natl Acad Sci USA. 2014;111:7278–83.
Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45:6995–7020.
Zhang F, Shi J, Chen SH, Bian C, Yu X. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015;43:10782–94.
Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27.
Hall TMT. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–73.
Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol. 2017;17:130–43.
Maeda K, Akira S. Regulation of mRNA stability by CCCH-type zinc-finger proteins in immune cells. Int Immunol. 2017;29:149–55.
Suskiewicz MJ, Palazzo L, Hughes R, Ahel I. Progress and outlook in studying the substrate specificities of PARPs and related enzymes. FEBS J. 2021;288:2131–42.
Haft JW, Atluri P, Ailawadi G, Engelman DT, Grant MC, Hassan A, et al. Mapping physiological ADP-ribosylation using activated ion electron transfer dissociation. Cell Rep. 2020;110:697–700.
Bonfiglio JJ, Leidecker O, Dauben H, Longarini EJ, Colby T, San Segundo-Acosta P, et al. An HPF1/PARP1-based chemical biology strategy for exploring ADP-ribosylation. Cell. 2020;183:1086–1102.e23.
Rulten SL, Fisher AEO, Robert I, Zuma MC, Rouleau M, Ju L, et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45.
Grundy GJ, Polo LM, Zeng Z, Rulten SL, Hoch NC, Paomephan P, et al. PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B Glu2. Nat Commun. 2016;7:12404.
Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci USA. 2011;108:2783–8.
Loseva O, Jemth AS, Bryant HE, Schüler H, Lehtiö L, Karlberg T, et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–60.
Gagné JP, Ethier C, Defoy D, Bourassa S, Langelier MF, Riccio AA, et al. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair. 2015;30:68–79.
Yang CS, Jividen K, Spencer A, Dworak N, Ni L, Oostdyk LT, et al. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol Cell. 2017;66:503–516.e5.
Yan Q, Xu R, Zhu L, Cheng X, Wang Z, Manis J, et al. BAL1 and its partner E3 ligase, BBAP, link poly(ADP-Ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol Cell Biol. 2013;33:845–57.
Zhu K, Suskiewicz MJ, Hloušek-Kasun A, Hervé M, Andreja M, Vincent A, et al. DELTEX E3 ligases ubiquitylate ADP-ribosyl modification on protein substrates. Sci Adv. 2022;40:eadd4253.
Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69.
Nicolae CM, Aho ER, Vlahos AHS, Choe KN, De S, Karras GI, et al. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J Biol Chem. 2014;289:13627–37.
Nicolae CM, Aho ER, Choe KN, Constantin D, Hu HJ, Lee D, et al. A novel role for the mono-ADP-ribosyltransferase PARP14/ARTD8 in promoting homologous recombination and protecting against replication stress. Nucleic Acids Res. 2015;43:3143–53.
Dhoonmoon A, Nicolae CM, Moldovan GL. The KU-PARP14 axis differentially regulates DNA resection at stalled replication forks by MRE11 and EXO1. Nat Commun. 2022;13:5063.
Yu X, Christiano CSC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–43.
Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011.
Wang Z, Grosskurth SE, Cheung T, Petteruti P, Zhang J, Wang X, et al. Pharmacological inhibition of PARP6 triggers multipolar spindle formation and elicits therapeutic effects in breast cancer. Cancer Res. 2018;78:6691–702.
Zhang L, Cao J, Dong L, Lin H. TiPARP forms nuclear condensates to degrade HIF-1α and suppress tumorigenesis. Proc Natl Acad Sci USA. 2020;117:13447–56.
Yang CS, Jividen K, Kamata T, Dworak N, Oostdyk L, Remlein B, et al. Androgen signaling uses a writer and a reader of ADP-ribosylation to regulate protein complex assembly. Nat Commun. 2021;12:2705.
Kamata T, Yang CS, Paschal BM. PARP7 mono-ADP-ribosylates the agonist conformation of the androgen receptor in the nucleus. Biochem J. 2021;478:2999–3014.
Palavalli Parsons LH, Challa S, Gibson BA, Nandu T, Stokes MS, Huang D, et al. Identification of PARP-7 substrates reveals a role for marylation in microtubule control in Ovarian cancer cells. Elife. 2021;10:e60481.
Bachmann SB, Frommel SC, Camicia R, Winkler HC, Santoro R, Hassa PO. DTX3L and ARTD9 inhibit IRF1 expression and mediate in cooperation with ARTD8 survival and proliferation of metastatic prostate cancer cells. Mol Cancer. 2014;13:125.
Camicia R, Bachmann SB, Winkler HC, Beer M, Tinguely M, Haralambieva E, et al. Bal1/artd9 represses the anti-proliferative and pro-apoptotic IFNγ-STAT1-IRF1-p53 axis indiffuse large B-cell lymphoma. J Cell Sci. 2013;126:1969–80.
Tian L, Yao K, Liu K, Han B, Dong H, Zhao W, et al. PLK1/NF-κB feedforward circuit antagonizes the mono-ADP-ribosyltransferase activity of PARP10 and facilitates HCC progression. Oncogene. 2020;39:3145–62.
Shao C, Qiu Y, Liu J, Feng H, Shen S, Saiyin H, et al. PARP12 (ARTD12) suppresses hepatocellular carcinoma metastasis through interacting with FHL2 and regulating its stability. Cell Death Dis. 2018;9:856.
Leutert M, Menzel S, Braren R, Rissiek B, Hopp AK, Nowak K, et al. Proteomic characterization of the heart and skeletal muscle reveals widespread arginine ADP-ribosylation by the ARTC1 ectoenzyme. Cell Rep. 2018;24:1916–1929.e5.
Song GL, Jin CC, Zhao W, Tang Y, Wang YL, Li M, et al. Regulation of the RhoA/ROCK/AKT/catenin pathway by arginine-specific ADP-ribosytransferases 1 promotes migration and epithelial-mesenchymal transition in colon carcinoma. Int J Oncol. 2016;49:646–56.
Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) polymerase inhibitors: recent advances and future development. J. Clin Oncol. 2015;33:1397–406.
Guo T, Zuo Y, Qian L, Liu J, Yuan Y, Xu K, et al. ADP-ribosyltransferase PARP11 modulates the interferon antiviral response by mono-ADP-ribosylating the ubiquitin E3 ligase β-TrCP. Nat Microbiol. 2019;4:1872–84.
Grunewald ME, Chen Y, Kuny C, Maejima T, Lease R, Ferraris D, et al. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019;15:e1007756.
Yamada T, Horimoto H, Kameyama T, Hayakawa S, Yamato H, Dazai M, et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol. 2016;17:687–94.
Iwata H, Goettsch C, Sharma A, Ricchiuto P, Goh WWB, Halu A, et al. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat Commun. 2016;7:12849.
Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-Ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–99.
Catara G, Grimaldi G, Schembri L, Spano D, Turacchio G, Lo Monte M, et al. PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci Rep. 2017;7:14035.
Li L, Zhao H, Liu P, Li C, Quanquin N, Ji X, et al. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci Signal. 2018;11:eaas9332.
Krieg S, Pott F, Eckei L, Verheirstraeten M, Bütepage M, Lippok B, et al. Mono-ADP-ribosylation by ARTD10 restricts Chikungunya virus replication by interfering with the proteolytic activity of nsP2. bioRxiv. 2020;01:896977.
Krieg S, Pott F, Potthoff L, Verheirstraeten M, Bütepage M, Golzmann A, et al. Mono-ADP-ribosylation by PARP10 inhibits Chikungunya virus nsP2 proteolytic activity and viral replication. Cell Mol Life Sci. 2023;80:72.
Mayo E, Fabrizio G, Scarpa E, Stilla A, Dani N, Chiacchiera F, et al. ARTD10/PARP10 induces ADP-ribosylation of GAPDH and recruits GAPDH into cytosolic membrane-free cell bodies when overexpressed in mammalian cells. Challenges. 2018;9:22.
Zhu H, Tang YD, Zhan G, Su C, Zheng C. The critical role of PARPs in regulating innate immune responses. Front Immunol. 2021;12:12556.
Karlberg T, Klepsch M, Thorsell AG, Andersson CD, Linusson A, Schüler H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J Biol Chem. 2015;290:7336–44.
Verheugd P, Forst AH, Milke L, Herzog N, Feijs KLH, Kremmer E, et al. Regulation of NF-κB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat Commun. 2013;4:1683.
Schenkel LB, Molina JR, Swinger KK, Abo R, Blackwell DJ, Lu AZ, et al. A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants. Cell Chem Biol. 2021;28:1158–1168.e13.
Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, et al. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci USA. 2002;99:8231–5.
Bindesbøll C, Tan S, Bott D, Cho T, Tamblyn L, MacPherson L, et al. TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and co-activates liver X receptors. Biochem J. 2016;473:899–910.
Márton J, Fodor T, Nagy L, Vida A, Kis G, Brunyánszki A, et al. PARP10 (ARTD10) modulates mitochondrial function. PLoS ONE. 2018;13:e0187789.
Shen X, Wang W, Wang L, Houde C, Wu W, Tudor M, et al. Identification of genes affecting apolipoprotein B secretion following siRNA-mediated gene knockdown in primary human hepatocytes. Atherosclerosis. 2012;222:154–7.
Di Paola S, Matarese M, Barretta ML, Dathan N, Colanzi A, Corda D, et al. PARP10 mediates mono-ADP-ribosylation of aurora-A regulating G2/M transition of the cell cycle. Cancers. 2022;14:24.
Dasso M. The Ran GTPase: theme and variations. Curr Biol. 2002;12:502–8.
Saxty BA, Yadollahi-Farsani M, Upton PD, Johnstone SR, MacDermot J. Inactivation of platelet-derived growth factor-BB following modification by ADP-ribosyltransferase. Br J Pharmacol. 2001;133:1219–26.
Challa S, Khulpateea BR, Nandu T, Camacho CV, Ryu KW, Chen H, et al. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell. 2021;184:4531–4546.e26.
Chen KC, Xie H, Cai Y. Modes of action of ADP-ribosylated elongation factor 2 in inhibiting the polypeptide elongation cycle: a modeling study. PLoS ONE. 2013;8:e66446.
MacPherson L, Tamblyn L, Rajendra S, Bralha F, McPherson JP, Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41:1604–21.
Ahmed S, Bott D, Gomez A, Tamblyn L, Rasheed A, Cho T, et al. Loss of the mono-ADP-ribosyltransferase, tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J Biol Chem. 2015;290:16824–40.
Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK-and IRE1α-mediated unfolded protein response. Nat Cell Biol. 2012;14:1223–30.
Fabrizio G, Di Paola S, Stilla A, Giannotta M, Ruggiero C, Menzel S, et al. ARTC1-mediated ADP-ribosylation of GRP78/BiP: A new player in endoplasmic-reticulum stress responses. Cell Mol Life Sci. 2015;72:1209–25.
Kleine H, Herrmann A, Lamark T, Forst AH, Verheugd P, Lüscher-Firzlaff J, et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun Signal. 2012;10:28.
Welsby I, Hutin D, Gueydan C, Kruys V, Rongvaux A, Leo O. PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation. J Biol Chem. 2014;289:26642–57.
Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, et al. ARTD10 substrate identification on protein microarrays: Regulation of GSK3β by mono-ADP-ribosylation. Cell Commun Signal. 2013;11:5.
Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–54.
Takamura-Enya T, Watanabe M, Totsuka Y, Kanazawa T, Matsushima-Hibiya Y, Koyama K, et al. Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein pierisin-1 from cabbage butterfly. Proc Natl Acad Sci USA. 2001;98:12414–9.
Schuller M, Butler RE, Ariza A, Tromans-Coia C, Jankevicius G, Claridge TDW, et al. Molecular basis for DarT ADP-ribosylation of a DNA base. Nature. 2021;596:597–602.
Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, et al. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation. Nucleic Acids Res. 2018;46:2417–31.
Belousova EA, Ishchenko AA, Lavrik OI. DNA is a new target of Parp3. Sci Rep. 2018;8:4176.
Suskiewicz MJ, Munnur D, Strømland Ø, Yang J-C, Easton LE, Chatrin C, et al. Updated protein domain annotation of the PARP protein family sheds new light on biological function. Nucleic Acids Res. 2023;51:8217–36.
Rouleau M, McDonald D, Gagné P, Ouellet ME, Droit A, Hunter JM, et al. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401.
Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P, et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 2014;42:5616–32.
Langelier MF, Riccio AA, Pascal JM. PARP-2 and PARP-3 are selectively activated by 5’ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42:7762–75.
Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, et al. Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 2016;44:9279–95.
Ukraintsev A, Kutuzov M, Belousova E, Joyeau M, Golyshev V, Lomzov A, et al. PARP3 affects nucleosome compaction regulation. Int J Mol Sci. 2023;24:9042.
Day TA, Layer JV, Patrick Cleary J, Guha S, Stevenson KE, Tivey T, et al. PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA. Nat Commun. 2017;8:15110.
Weixler L, Feijs KLH, Zaja R. ADP-ribosylation of RNA in mammalian cells is mediated by TRPT1 and multiple PARPs. Nucleic Acids Res. 2022;50:9426–41.
Bock FJ, Todorova TT, Chang P. RNA regulation by Poly (ADP-Ribose) polymerases. Mol Cell. 2015;58:959–69.
Zhu K, Suskiewicz MJ, Chatrin C, Strømland Ø, Dorsey BW, Aucagne V, et al. DELTEX E3 ligases ubiquitylate ADP-ribosyl modification on nucleic acids. Nucleic Acids Res. 2024;52:801–15.
O’Connor MJ, Thakar T, Nicolae CM, Moldovan GL. PARP14 regulates cyclin D1 expression to promote cell-cycle progression. Oncogene. 2021;40:4872–83.
Bilal Iqbal M, Johns M, Cao J, Liu Y, Yu SC, Hyde GD, et al. PARP-14 combines with tristetraprolin in the selective posttranscriptional control of macrophage tissue factor expression. Blood. 2014;124:3646–55.
Todorova T, Bock FJ, Chang P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun. 2014;5:10.
Wang J, Cheng Q, Zhang Y, Hong C, Liu J, Liu X, et al. PARP16-mediated stabilization of amyloid precursor protein mRNA exacerbates Alzheimer’s disease pathogenesis. Aging Dis. 2023;14:1458–71.
Lee S, Lee YS, Choi Y, Son A, Park Y, Lee KM, et al. The SARS-CoV-2 RNA interactome. Mol Cell. 2021;81:2838–2850.e6.
Siegmund KD, Klink F. Production of an antiserum specific to the ADP-ribosylated form of elongation factor 2 from archaebacteria and eukaryotes. FEBS Lett. 1992;312:139–42.
Gibson BA, Conrad LB, Huang D, Kraus WL. Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry. 2017;56:6305–16.
Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci USA. 2009;106:4243–8.
Longarini EJ, Dauben H, Locatelli C, Wondisford AR, Smith R, Muench C, et al. Modular antibodies reveal DNA damage-induced mono-ADP-ribosylation as a second wave of PARP1 signaling. Mol Cell. 2023;83:1743–1760.e11.
Tromans-Coia C, Sanchi A, Moeller GK, Timinszky G, Lopes M, Ahel I. TARG1 protects against toxic DNA ADP-ribosylation. Nucleic Acids Res. 2021;49:10477–92.
Silverman RB, Holladay MW. Assessment of intracellular auto-modification levels of ARTD10 using mono-ADP-ribose-specific macrodomains 2 and 3 of murine Artd8. Methods Mol Biol. 2007;1813:41–63.
Morgan RK, Cohen MS. Detecting protein ADP-ribosylation using a clickable aminooxy probe. Methods Mol Biol. 2017;1608:71–77.
Morgan RK, Cohen MS. A clickable aminooxy probe for monitoring cellular ADP-ribosylation. ACS Chem Biol. 2015;10:1778–84.
Ando Y, Elkayam E, McPherson RL, Dasovich M, Cheng SJ, Voorneveld J, et al. ELTA: enzymatic labeling of terminal ADP-ribose. Mol Cell. 2019;73:845–856.e5.
Westcott NP, Fernandez JP, Molina H, Hang HC. Chemical proteomics reveals ADP-ribosylation of small GTPases during oxidative stress. Nat Chem Biol. 2017;13:302–8.
Rose M, Burgess JT, O’Byrne K, Richard DJ, Bolderson E. PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol. 2020;8:564601.
Sharif-Askari B, Amrein L, Aloyz R, Panasci L. PARP3 inhibitors ME0328 and olaparib potentiate vinorelbine sensitization in breast cancer cell lines. Breast Cancer Res Treat. 2018;172:23–32.
Lindgren AEG, Karlberg T, Thorsell AG, Hesse M, Spjut S, Ekblad T, et al. PARP inhibitor with selectivity toward ADP-ribosyltransferase ARTD3/PARP3. ACS Chem. Biol. 2013;8:1698–703.
Gozgit JM, Vasbinder MM, Abo RP, Kunii K, Kuplast-Barr KG, Gui B, et al. PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity. Cancer Cell. 2021;39:1214–1226.e10.
Teyssonneau D, Margot H, Cabart M, Anonnay M, Sargos P, Vuong NS, et al. Prostate cancer and PARP inhibitors: progress and challenges. J Hematol Oncol. 2021;14:51.
Venkannagari H, Verheugd P, Koivunen J, Haikarainen T, Obaji E, Ashok Y, et al. Small-molecule chemical probe rescues cells from mono-ADP-ribosyltransferase ARTD10/PARP10-induced apoptosis and sensitizes cancer cells to DNA damage. Cell Chem Biol. 2016;23:1251–60.
Holechek J, Lease R, Thorsell AG, Karlberg T, McCadden C, Grant R, et al. Design, synthesis and evaluation of potent and selective inhibitors of mono-(ADP-ribosyl)transferases PARP10 and PARP14. Bioorg Med Chem Lett. 2018;28:2050–4.
Kirby IT, Kojic A, Arnold MR, Thorsell AG, Karlberg T, Vermehren-Schmaedick A, et al. A potent and selective PARP11 inhibitor suggests coupling between cellular localization and catalytic activity. Cell Chem Biol. 2018;25:1547–1553.e12.
Yoneyama-Hirozane M, Matsumoto Sichi, Toyoda Y, Saikatendu KS, Zama Y, Yonemori K, et al. Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation. Biochem Biophys Res Commun. 2017;486:626–31.
Challa S, Nandu T, Kim HB, Gong A, Renshaw CW, Li W-C, et al. A PARP14/TARG1-regulated RACK1 MARylation cycle drives stress granule dynamics in ovarian cancer cells. bioRxiv. 2023;10:562273.
Wang J, Zhu C, Song D, Xia R, Yu W, Dang Y, et al. Epigallocatechin-3-gallate enhances ER stress-induced cancer cell apoptosis by directly targeting PARP16 activity. Cell Death Discov. 2017;3:17034.
Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. A 2’-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem. 1997;272:13203–10.
Sanderson DJ, Cohen MS. Mechanisms governing PARP expression, localization, and activity in cells. Crit Rev Biochem Mol Biol. 2020;55:541–54.
Di Girolamo M, Fabrizio G. Overview of the mammalian ADP-ribosyl-transferases clostridia toxin-like (ARTCs) family. Biochem Pharmacol. 2019;167:86–96.
Rack JGM, Palazzo L, Ahel I. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. 2020;34:263–84.
Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329:18–25.
Funding
This work was supported by the National Natural Science Foundation of China (No. U20A20407), Fund for Outstanding Young Scholars of Shenyang Pharmaceutical University (YQ202101) and Basic Research Projects of Liaoning Provincial Department of Education (LJKQZ20222363).
Author information
Authors and Affiliations
Contributions
Hao Wu and Jincai Lu both developed concept for this review. All co-authors participated in preparing the first version of the manuscript, tables and the figure. All co-authors provided constructive feedback to the preparation of the article, and explicitly approved its final content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, H., Lu, A., Yuan, J. et al. Mono-ADP-ribosylation, a MARylationmultifaced modification of protein, DNA and RNA: characterizations, functions and mechanisms. Cell Death Discov. 10, 226 (2024). https://doi.org/10.1038/s41420-024-01994-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-024-01994-5