Abstract
Glioma cell sensitivity to temozolomide (TMZ) is critical for effective treatment and correlates with patient survival, although mechanisms underlying this activity are unclear. Here, we reveal a new mechanism used by glioma cells to modulate TMZ sensitivity via regulation of SORBS2 and DDR1 genes by super-enhancer RNA LINC02454. We report that LINC02454 activity increases glioma cell TMZ sensitivity by maintaining long-range chromatin interactions between SORBS2 and the LINC02454 enhancer. By contrast, LINC02454 activity also decreased glioma cell TMZ sensitivity by promoting DDR1 expression. Our study suggests a bivalent function for super-enhancer RNA LINC02454 in regulating glioma cell sensitivity to TMZ.
Similar content being viewed by others
Introduction
Glioma is the most common malignant brain tumor [1]. Glioblastoma (GBM), which accounts for 56.6% of all gliomas, has the highest rate of recurrence and a 5.6% 5-year survival rate [2]. Temozolomide (TMZ) is a chemotherapy drug widely used clinically as GBM treatment. However, development of TMZ resistance is a major problem that severely reduces drug efficacy. Regulation of GBM TMZ sensitivity is reportedly associated with genome repair systems in tumor cells, including O6-methylguanine-DNA methyltransferase (MGMT)-mediated demethylation, the base excision repair (BER) system, mismatch repair (MMR) and other activities [3, 4]. Previous studies also indicate that activities of glioma stem cells and cell autophagy alter TMZ sensitivity [5, 6]. Nonetheless, these discoveries have not yet been translated into novel, clinically-relevant approaches to GBM TMZ resistance.
Following development of chromosome conformation capture techniques, several groups reported that gene regulation activities based on 3D chromatin structure play a role in glioma cell sensitivity to TMZ [7,8,9]. In this process, gene regulatory elements, such as enhancers or silencers, exert long-range control of target genes via chromatin loops. Enhancer RNAs (eRNAs) are a subset of non-coding RNAs transcribed from an enhancer locus, and some are defined as long non-coding RNAs (lncRNAs) based on their length. eRNAs reportedly regulate enhancer function and thus play important roles in control of gene expression [10,11,12]. Other studies indicate that eRNAs function by increasing chromatin accessibility and binding to transcription factors [13,14,15]. However, few studies have assessed whether eRNAs function in a 3D gene regulatory network in a way that would alter glioma cell sensitivity to TMZ.
Super-enhancers (SEs) are large clusters of transcriptional enhancers that drive gene expression and often play key roles in cancer progression [16]. In this study, we identified a SE-derived long non-coding RNA, LINC02454, that regulates glioma TMZ sensitivity through maintenance of 3D chromatin structure. Our findings suggest that SE activity at the LINC02454 locus increases glioma cell sensitivity to TMZ by regulating expression of SORBS2 (Sorbin and SH3 domain containing 2) via long-range chromatin interactions, and that LINC02454 maintains this chromatin loop. By contrast, we also show that LINC02454 can reduce glioma cell TMZ sensitivity by promoting expression of DDR1 (Discoidin domain receptor 1). Although other lncRNAs have been reported to function in glioma TMZ sensitivity [17, 18], our study reveals a new mechanism by which LINC02454 and a corresponding SE co-regulate glioma sensitivity to TMZ by altering 3D chromatin structure and through bivalent LINC02454 activities.
Materials and methods
Cell culture
U251 and U87 human glioblastoma cell lines were obtained from the National Infrastructure of Cell Line Resource (China). U251 and U87 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Bioind) and 1% penicillin/streptomycin (Gibco) at 37 °C, in 90% humidity and 5% CO2.
Chromatin Immunoprecipitation and sequencing
ChIP-seq was carried out as reported with minor modifications [19]. 107 cells were crosslinked 10 min with 1% formaldehyde (Thermo Fisher Scientific) at room temperature and the reaction was terminated by a 5 min incubation with a solution containing glycine (0.125 M) (Solarbio). After centrifugation at 4 °C, cells were washed twice with ice-cold PBS. Nuclear DNA precipitates were harvested in Farnham lysis buffer [50 mM (pH 7.5) tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail]. Nuclear DNA was then sonicated as a nuclear lysate (50 mM Tris-HCL,10 mM EDTA,1% SDS,1× protease inhibitor cocktail) to shear chromatin and obtain 200–500-bp DNA fragments. Sonicated lysates were immunoprecipitated at 4 °C with H3K27ac antibody (Abcam) and protein A/G agarose beads (Thermo Fisher Scientific) to enrich H3K27ac-bound DNA fragments. Purified ChIP DNA was used to prepare Illumina sequencing libraries and sequenced using an Illumina HiSeq 4000 system. Data analysis was performed using the GRCh37/hg19 reference genome. Super-enhancers were determined according to the Rank Ordering of Super Enhancers (ROSE) algorithm by using H3K27ac peaks [20]. In short, we used bowtie2 [21] to align ChIP-seq data to the human hg19 reference genome with default parameters. Then we used macs2 [22] to call peak with parameters “-f BAMPE -g hs”. We used ROSE [20] to identify SE with parameters “-g HG19 -s 20000 -t 2000”.
RNA-seq and data analysis
Total RNA was extracted from glioma cells. The Illumina HiSeq 4000 platform was used to sequence barcoded RNA-seq libraries. HISAT2 [23] was used to map clean reads to the human reference genome (GRCh37/hg19). HTSeq [24] was used to calculate transcript abundance. DESeq2 [25] was used for read count normalization and to regularize log transformations. DAVID [26] (Database for Annotation, Visualization and Integrated Discovery) was used for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Quantitative real-time PCR
Total RNA was extracted from glioma cells using TRIzol reagent (Life Technologies). cDNAs were obtained using the PrimerScript™ RT reagent Kit with gDNA Eraser (TaKaRa) for reverse transcription, according to the manufacturer’s instructions. The qPCR reaction was performed using HieffTM qPCR SYBR Green Master Mix (YEASEN) in the BioRad CFX Connect Real-Time system. Target gene expression levels were normalized to GAPDH and relative expression was quantified using the 2−ΔΔCt method [27]. Primer sequences used in this study are shown in Table S1.
Luciferase reporter assays
We PCR-amplified regions of the LINC02454 SE showing the most significant H3K27ac enrichment (hg19_chr12:65,995,412–65,998,341) and cloned them into the pGL3 promoter vector (Promega). We then transfected 293 T cells with that plasmid plus the pRL-TK Renilla luciferase control vector (Promega) using Lipofectamine 3000 (Invitrogen). Cells were then harvested for a luciferase assay. Luciferase activity was measured on the SpectraMax i3x Multifunctional enzyme label instrument (Molecular Devices) using the dual luciferase Reporter Assay System (Promega).
CRISPR/Cas9-mediated deletions
To delete the LINC02454 SE, U251 glioma cells were transfected with plasmids containing Cas9 and guide RNAs targeting the SE region. Cell clones were genotyped, and two clones (KO#1 and KO#2) with homozygous deletion of the LINC02454 SE were used for analyses. sgRNA sequences were designed using web-based tools at https://zlab.bio/guide-design-resources and are listed in Table S2.
Lactate Dehydrogenase (LDH) assay
LDH in culture medium was evaluated using a LDH cytotoxicity kit (Promega). Briefly after cells were treated with TMZ or transfected with LNAs, siRNAs or overexpression plasmids, culture media were collected and LDH activity was assayed based on the manufacturer’s instructions. OD values at 490 nm were measured using a microplate reader. Relative TMZ cytotoxicity of each group was calculated as a percentage of the total amount of LDH released relative to a positive control included in the kit.
Caspase 3/7 activity measurements
A Caspase-Glo 3/7 kit (Promega) was used to evaluate caspase 3/7 activities in U251 glioma cells based on the manufacturer’s instructions. Briefly, glioma cells in each group were harvested and incubated with 100 μL Caspase-Glo 3/7 reagent for 1 h in the dark. Luminescence was then measured at 485/530 nm using a microplate reader. Relative caspase 3/7 activity was calculated to evaluate fold-changes in samples treated with TMZ or transfected with LNAs, siRNAs, or overexpression plasmids relative to controls.
Chromosome conformation capture (Capture-C) assay
Capture-C was performed as reported with minor modifications [28]. In brief, U251 glioma cells (1 × 107) were harvested and used to generate a standard 3 C library with DpnII (New England BioLabs). Then 3 C library DNA was sheared to 200–300-bp fragments by sonication. Pre-capture DNA libraries were prepared using the NEBNext DNA Library Kit (New England BioLabs) according to the manufacturer’s instructions. Biotinylated “bait” probes and streptavidin beads (Invitrogen) were used to enrich the “bait” and linked chromatin loci. Barcoded Capture-C libraries were sequenced as 150-bp paired-end reads using the Illumina HiSeq 4000 platform. Capture-C data were analyzed based on the pipeline proposed by Davies et al [28]. Two biotinylated DNA oligonucleotides were designed to match both ends of the H3K27ac enrichment region at the LINC02454 locus in U251 and U87 glioma cells with the following sequence (5′-3′):
5′-Biotin-
GATCCCGAGAGCTTCCCGTGTGGGGGCGGAGGGTGGGGCAGAGCAGGATGTGTGCTTGGGTTTCACTTGGAAAAAACATACATCATATTGCAATATAATTTACTTTGAGATTTCAATTTGG-3′
5′-Biotin-
ACAATTGGCTGGAATCACAAGTGGCTTCTTTGCTCACTCTAGCGGTACTTCAGCTTTCATAAACAGCCGGTTCCTGGAGCTGTAAGGTTCCAGCACATCGGGGATTTCCCTTTGTTAGATC-3′
Gene knock-down by LNAs (Locked Nucleic Acids)
Briefly, to knock-down LINC02454, cells were collected and placed in 6-well plates. Cells were then transfected with LNAs (QIAGEN) using Lipofectamine 3000 (Invitrogen) based on the manufacturer’s instructions. Seventy-two hrs later, cells were collected for RNA extraction, and KD efficiency was determined by qRT-PCR. LNA sequences are listed in Table S3.
Gene overexpression
Candidate cDNAs were cloned into the pCDH-CMV-MCS-EF1-Puro expression vector (System Biosciences), and plasmids were used to transfect U251 cells with Lipofectamine 3000 (Invitrogen). Stably transfected cells were selected in media containing puromycin (2.5 μg/ml), and overexpression efficiency was determined by qRT-PCR.
Gene knock-down by siRNA
Glioma cells were transfected with target and control siRNAs using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. KD efficiency was determined by qRT-PCR 48–72 h after transfection. siRNAs were designed and synthesized by RiboBio Biotechnology Co., LTD., and the sequences are shown in Table S3.
Chromatin isolation by RNA purification (ChIRP)-qPCR
ChIRP was performed as reported [29]. Antisense DNA probes complementary to LINC02454 were biotin-labeled and ordered from Sangon Biotech. 107 cells were cross-linked and fixed with 3% formaldehyde (Thermo Fisher Scientific), and the reaction was terminated by incubation with a solution containing glycine (0.125 M) (Solarbio). Cells were washed twice with PBS and lysed in lysis buffer. Cell lysates were then sonicated and incubated with biotinylated antisense DNA probes at 37 °C for 4 h. RNA and DNA from ChIRP samples were quantitated by qRT-PCR. qRT-PCR primer sequences are listed in Table S4, and the antisense DNA probe sequence is listed in Table S5.
CRISPRa-mediated gene overexpression
A stable CRISPRa cell line was generated using lentiMPH v2 plasmid (Addgene). 293 T cells were transfected with pMDG.2, psPAX2 and lentiMPH v2 plasmid (Addgene). Lentiviral particles were then collected and used to transduce U251 cells, which were selected in 200 μg/ml hygromycin (Solarbio). sgRNAs were designed using an online tool (https://portals.broadinstitute.org/gppx/crispick/public) and ordered from Sangon Biotech, and their sequences are shown in Table S6. sgRNAs were cloned into lentiSAM v2 plasmid (Addgene) using BsmBI (New England Biolabs). Lentiviral particles of pMDG.2, psPAX2 and lentiSAM v2 plasmid were generated and used to transduce U251 cells, which were selected in 6 μg/ml blasticidin (Solarbio) for about 7 days.
Gene knock-down by shRNA
shRNAs were cloned into pSUPER-puro vector and used to transfect glioma cells with Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. Cells stably expressing shRNA were selected in puromycin (2.5 μg/ml) and KD efficiency was determined by qRT-PCR. qPCR primer sequences are shown in Supplementary Table S1. shRNAs were designed using an online tool (www.invivogen.com/sirnawizard/design.php) and synthesized by Sangon Biotech. shRNAs sequences are shown in Table S3.
Survival data analysis
Glioma patients’ survival data were obtained from TCGA, and patient samples were divided into two groups based on gene expression levels. Samples with expression higher or lower than the median were marked as “high expression” or “low expression”, respectively. A p < 0.001 indicates a significant survival difference between the two groups.
Statistical analysis
Data represent means ± S.E.M. Statistical analysis was performed using Student’s t-test. * p < 0.05, ** p < 0.01, *** p < 0.001.
Published data used in this study
The following published datasets were used in this study. SRR444436 for U87 H3K27ac ChIP-seq [30]; SRR13238369 and SRR13238368 for NHA H3K27ac ChIP-seq [31]; SRR3627718 for U87 ATAC-seq [32]; SRR5583266 for U87 BRD4 ChIP-seq [33]; SRR444453 for U87 MED1 ChIP-seq [30]; and PRJNA479416 for RNA-seq of U251 cells treated for 0, 4, 9,12, and 16 days with TMZ [34].
Results
Identification of glioma-specific SE lncRNAs associated with glioma TMZ sensitivity
To identify enhancer lncRNAs that may regulate glioma cell TMZ sensitivity, we focused on lncRNAs derived from glioma-specific SEs. To identify such SEs, we used H3K27ac ChIP-seq data from glioma lines U251 and U87 and a normal human astrocyte line (NHA) (Fig. 1A, B). We identified 1094 and 1174 SEs in U251 and U87 cells, respectively, and 355 of those were common to both lines (Fig. 1C). Moreover, 34 SEs were identified in all three cell lines (Fig. 1D). Among SEs found in glioma lines, 321 were specific to U251 and U87 lines (Fig. 1D), while 207 SEs were specific to the NHA line (Fig. 1D). We then used data from the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases to assess transcript levels of genes adjacent to glioma-specific SEs. That analysis revealed significant changes in transcription of 52% of glioma-specific SE-associated genes, and 42.51% of those were significantly upregulated in glioma relative to normal brain tissue (Fig. S1A). Analysis of glioma-specific, SE-associated genes using the Kyoto Encyclopedia of Genes and Genomes (KEGG) indicated enrichment in focal adhesion and MAPK pathway factors (Fig. S1B), many previously linked to cancer cell proliferation, migration, apoptosis and chemoresistance [35,36,37].
A Ranking of enhancers based on the H3K27ac signal at enhancers in glioma (U251 and U87) and astrocyte (NHA) lines. B H3K27ac signal of GBM-specific SE in U251, U87 and NHA cells. C Number of SEs in U251 and U87 cells. D Number of SEs in U251 and U87 cells compared to the number in NHA. Volcano plots of genes differentially expressed (FD ≥ 2 or ≤−2, p < 0.05) in U251 (E) and U87 (F) lines treated with TMZ (1 mM, 72 h), as compared with Ctrls. Number of up-regulated (G) and down-regulated (H) genes in U251 and U87 cells treated with TMZ (1 mM) for 72 h, as compared with Ctrls. I Heat map shows clustering of differentially expressed lncRNAs in U251 and U87 cells treated with TMZ (1 mM, 72 h), as compared to untreated Ctrl cells.
We then performed RNA-seq in TMZ-treated (1 mM, 72 h) U251 and U87 cells to monitor lncRNAs potentially associated with glioma TMZ sensitivity. That analysis identified 712 and 1220 genes (fold-change ≥ 2, p < 0.05) significantly up-regulated (Fig. 1E, F) and 1274 and 700 genes significantly down-regulated (fold-change ≤ −2, p < 0.05) in U251 and U87 cells, respectively (Fig. 1E, F). Eighty genes were up-regulated in both U251 and U87 lines, while 74 were down-regulated (Fig. 1G, H). Among genes that changed consistently in both cell lines, 14 were lncRNAs (Fig. 1I), and 2 of those, LINC02454 and MSRB3-AS1, were located at glioma-specific SEs (Fig. 2A). Based on data from TCGA and GTEx databases, LINC02454 expression in GBM samples was significantly higher than that in normal brain tissue (Fig. 2B), while MSRB3-AS1 expression was comparable in GBM samples and normal brain samples (Fig. 2C), prompting our focus on LINC02454.
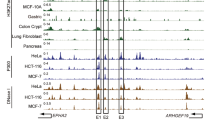
A Two lncRNAs that changed consistently in both TMZ-treated U251 and U87 cells (TMZ_diff_lncRNA) were located at glioma-specific SEs. Expression of LINC02454 (B) and MSRBS-AS1 (C) in GBM (166 cases) and normal brain (1157 cases) tissues. Gene Expression data are from TCGA and GTEx databases, ***p < 0.001. D qRT-PCR analysis of LINC02454 RNA levels in TMZ-treated (1 mM, 72 h) and Ctrl cells. Data represent means ± S.E.M. of three independent experiments. ***p < 0.001, compared with Ctrl cells. E, F RNA-seq of read counts of LINC02454 in Ctrl U251 and U87 cells treated with TMZ (1 mM, 72 h). G Analysis of survival data from glioma patients based on high and low LINC02454 expression. H ATAC-seq and H3K27ac and BRD4 and MED1 ChIP of the LINC02454 SE in glioma cells and astrocytes (NHA).
We next performed qRT-PCR analysis of U251 and U87 lines treated 72 h with TMZ (1 mM) and untreated controls. Relative to corresponding controls, TMZ-treated cells showed significantly upregulated LINC02454 (Fig. 2D), consistent with RNA-seq data (Fig. 2E, F). Moreover, analysis of patient survival indicated that high LINC02454 expression in tumor samples was significantly correlated with low survival probability (Fig. 2G). ChIP analysis also showed significant enrichment of H3K27ac signals at the LINC02454 locus in U251 and U87 glioma cells relative to NHA astrocytes (Fig. 2H), suggesting that locus is a glioma-specific SE. In addition, ChIP-seq showed enrichment of SE-enriched transcriptional coactivators BRD4 and MED1 at the LINC02454 locus in U87 glioma cells (Fig. 2H). Finally, ATAC-seq analysis indicated an open chromatin status of LINC02454-associated SE regions in glioma cells (Fig. 2H).
LINC02454 knock-down increases glioma cell sensitivity to TMZ
To assess whether LINC02454 activity regulates glioma cell sensitivity to TMZ, we transfected U251 cells with locked nucleic acids (LNAs) to knock-down (KD) LINC02454. qRT-PCR analysis of control and KD cells confirmed LINC02454 downregulation to approximately a third of levels seen in control cells (Fig. 3A). We then evaluated lactate dehydrogenase (LDH) release and caspase 3/7 activity as indicators of cytotoxicity in both control and LINC02454 KD U251 cells treated for various times with TMZ. Relative to TMZ-treated controls, LINC02454 KD cells treated with TMZ (1 mM) showed significantly increased LDH release (Fig. 3B, C). Specifically, TMZ cytotoxicity in LINC02454 KD cells increased to ~1.6-fold that of controls at 24 h (Fig. 3B) and to ~1.9-fold of controls at 48 h (Fig. 3C). Caspase 3/7 activities also markedly increased in TMZ-treated LINC02454 KD relative to control glioma cells at 48 and 72 h time points (Fig. 3D, E). By contrast, LINC02454 overexpression (OE) in U251 cells significantly decreased LDH release by 48 h of TMZ treatment relative to TMZ-treated controls (Fig. S2A, B). Overall, these findings suggest that LINC02454 plays a role in TMZ sensitivity in glioma cells.
A qRT-PCR was used to assess LNA KD efficiency in control (Ctrl) and LINC02454 KD (KD) U251 glioma cells. Data represent means ± S.E.M. of three independent experiments. ***p < 0.001 compared with Ctrl. B, C LDH release levels from Ctrl and LINC02454 KD cells treated with 1 mM TMZ for 24 h and 48 h. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, ***p < 0.001 compared with Ctrl. D, E Caspase 3/7 activity in Ctrl and LINC02454 KD cells treated with 1 mM TMZ for 48 h and 72 h. Data represent means ± S.E.M. of three independent experiments. **p < 0.01 compared with Ctrl. F Volcano plot depicting changes in gene expression in LINC02454 KD and Ctrl cells. G Volcano plot depicting changes in gene expression in LINC02454 overexpression (OE) and Ctrl cells. Functional analysis of genes differentially expressed in LINC02454 KD (H) and LINC02454 OE (I) cells relative to Ctrl cells.
RNA-seq analysis of LINC02454 KD and OE glioma cells revealed global changes in mRNA levels (Fig. 3F, G). Relative to controls, LINC02454 KD cells showed significant down-regulation of 550 genes (fold-change ≤ - 2, p < 0.05) and up-regulation of 443 (fold-change ≥ 2, p < 0.05) (Fig. 3F). Moreover, relative to controls, LINC02454 OE cells showed significant down-regulation of 409 genes (fold-change ≤ −2, p < 0.05) and up-regulation of 302 (fold-change ≥ 2, p < 0.05) (Fig. 3G). KEGG analysis of LINC02454 KD glioma cells indicated that genes differentially expressed in LINC02454 KD relative to control cells were significantly enriched in cancer progression-associated signaling pathways like MAPK and Rap1 (Fig. 3H). MAPK signaling is reportedly associated with regulation of glioma TMZ resistance [37]. Genes differentially expressed in LINC02454 OE cells were significantly enriched in cell adhesion and signaling factors associated with ECM receptor interactions, which was found to be associated with TMZ resistance (Fig. 3I) [38]. Overall, these results suggest that LINC02454 functions in glioma cell sensitivity to TMZ.
Knock-out of the LINC02454 super-enhancer decreases glioma cell sensitivity to TMZ
Previous studies have reported coordinated functions of enhancers and corresponding enhancer lncRNAs in regulating physiological cellular processes [39, 40]. ChIP-seq data indicated H3K27ac enrichment at the LINC02454 locus in glioma cells, which was identified as the LINC02454 super-enhancer (LINC02454 SE) (Fig. 4A). We then assessed enhancer activity using a luciferase reporter assay and found that the H3K27ac-enriched region has robust enhancer activity (Fig. 4B).
A A schematic showing the deleted LINC02454 SE region and knock-out screening primers. B Luciferase reporter assay of Ctrl (reporter without the enhancer) and SE (reported plus LINC02454 SE region) constructs performed in 293 T cells. Data represent means ± S.E.M. of three independent experiments, ***p < 0.001 compared with Ctrl. C, D LDH release levels in Ctrl and LINC02454 SE KO (lines KO#1 and KO#2) cells treated with 1 mM TMZ for 24 h and 48 h. Data represent means ± S.E.M. of three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 compared with Ctrl. Caspase 3/7 activity in Ctrl and LINC02454 SE KO (lines KO#1 and KO#2) cells treated with 1 mM TMZ for 48 h (E) and 72 h (F). Data represent means ± S.E.M. of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared with Ctrl. G Volcano plot depicting changes in gene expression in LINC02454 SE KO and control Ctrl cells.
To investigate LINC02454 SE function, we used the CRISPR/Cas9 system to knock out (KO) the H3K27ac enrichment region at that locus in U251 cells (Fig. 4A), resulting in KO#1 and KO#2 lines (Fig. S3A, B). We also evaluated LDH release and caspase 3/7 activity in TMZ-treated WT controls, KO#1 and KO#2 U251 lines. Compared with TMZ (1 mM)-treated WT controls, we observed a significant decrease in LDH release in TMZ (1 mM)-treated KO#1 and KO#2 cells (Fig. 4C, D). Specifically, TMZ cytotoxicity in KO#1 and KO#2 cells decreased to 40–50% of controls at 24 h TMZ treatment (Fig. 4C) and to 60–70% of controls at 48 h (Fig. 4D). Moreover, relative to control TMZ-treated U251 cells, caspase 3/7 activities markedly decreased in TMZ-treated KO#1 and KO#2 cells at 48 and 72 h time points (Fig. 4E, F), suggesting an overall decrease in glioma TMZ sensitivity in LINC02454 SE KO cells.
RNA-seq analysis of WT controls and KO#1 and KO#2 U251 cells indicated global changes in mRNA levels in KO relative to control cells (Fig. 4G). Specifically, transcript levels of 332 genes significantly changed (fold-change ≥ 2 or ≤ −2, p < 0.05), including 188 down-regulated (fold-change ≤ −2, p < 0.05) and 144 up-regulated (fold-change ≥ 2, p < 0.05) in KO relative to WT U251 cells (Fig. 4G). Gene Ontology (GO) analysis showed down-regulated genes in cells were significantly enriched in categories related to cell adhesion, positive regulation of cell migration and regulation of phosphatidylinositol 3-kinase signaling associated with cancer progression (Fig. S3C). GO analysis of up-regulated genes in KO cells indicated significant enrichment in cancer progression-associated processes such as regulation of cell proliferation and cell death and Ras signaling (Fig. S3D). Down-regulated genes were significantly enriched in the PI3K-Akt signaling pathway, which is reportedly associated with glioma TMZ resistance (Fig. S3E) [41]. These results suggest that LINC02454 SE KO induces transcriptional changes in several genes associated with glioma cell sensitivity to TMZ.
LINC02454 SE activity increases glioma cell sensitivity to TMZ by maintaining SORBS2 expression via 3D chromatin structure
We then performed Capture-C, using a previously reported protocol [28], in U251 cells to define the 3D chromatin structure-mediated gene regulation network of the LINC02454 SE, using the 972-bp H3K27ac enrichment region as Capture-C bait (Fig. 5A). Chromatin interactions between the LINC02454 SE and target genes are shown in Circos plots [42] (Fig. 5B), in which lines represent interactions detected in two replicates (Fig. 5B). This analysis identified 34 genes in chromatin interaction regions potentially regulated by the LINC02454 SE (Fig. 5B).
A The 972-bp Capture-C bait region and H3K27ac signals near the bait region. B Circos plot showing genome-wide intra- and inter-chromatin interactions, indicated by curves extending from the bait region. Intra- and inter-chromatin interactions are in blue and red, respectively. C Counts of genes that interact with the LINC02454 SE (Capture-C), genes downregulated in LINC02454 SE KO relative to Ctrl cells (KO_down), genes upregulated in LINC02454 OE relative to Ctrl cells (OE_up), and genes downregulated in LINC02454 KD compared with Ctrl cells (KD_down). D qRT-PCR analysis of SORBS2 transcripts in Ctrl and LINC02454 SE KO (lines KO#1, KO#2) cells. Data represent means ± S.E.M. of three independent experiments, ***p < 0.001 compared with Ctrl. E qRT-PCR verification of SORBS2 overexpression in LINC02454 SE KO cells. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, ***p < 0.001. F Caspase 3/7 activity in Ctrl, LINC02454 SE KO (lines KO#1, KO#2), and SORBS2 OE/LINC02454 SE KO (SORBS2) cells treated with 1 mM TMZ for 48 h. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, ***p < 0.001. G qRT-PCR analysis of SORBS2 transcript levels in Ctrl and LINC02454 KD cells. Data represent means ± S.E.M. of three independent experiments. ***p < 0.001 compared with Ctrl. H ChIRP-qPCR analysis of LINC02454 probe enrichment efficiency in Ctrl and LINC02454 KD cells. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. ChIRP-qPCR analysis of LINC02454 binding to the LINC02454 SE (I) and the SORBS2 locus (J) in Ctrl and LINC02454 KD cells. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Combined analysis of both Capture-C and RNA-seq data in both SE KO cells and in LINC02454 KD or OE cells indicated SORBS2 downregulation in LINC02454 KD and LINC02454 SE KO cells and upregulation in LINC02454 OE cells (Fig. 3F, G, Fig. 4G and Fig. 5C). Capture-C data also indicated a long-range interaction of SORBS2, which is located on chromosome 4, with the LINC02454 SE (Fig. 5C). Consistent with RNA-seq results, qRT-PCR analysis confirmed down-regulation of SORBS2 expression in LINC02454 SE KO cells (Fig. 5D). We then transfected LINC02454 SE KO lines with SORBS2 OE vectors to establish stable lines and verified overexpression efficiency qRT-PCR (Fig. 5E). SORBS2 OE in LINC02454 SE KO cells significantly increased TMZ sensitivity in that line relative to LINC02454 SE KO cells (Fig. 5E, F). We then assessed caspase 3/7 activities in TMZ-treated (1 mM, 48 h) control and LINC02454 SE KO lines and in SE KO cells overexpressing SORBS2. Relative to the LINC02454 SE KO line, caspase 3/7 activities markedly increased in SORBS2-overexpressing SE KO cells (Fig. 5F). Enhancer lncRNAs reportedly maintain interactions of corresponding enhancers with target genes [39, 40]. Accordingly, our RNA-seq data indicated parallel changes in expression of SORBS2 and LINC02454 (Fig. 5C), and qRT-PCR data showed significant SORBS2 downregulation in LINC02454 KD cells (Fig. 5G).
We next performed ChIRP-qPCR in control and LINC02454 KD U251 cells to assess binding of LINC02454 to the LINC02454 SE and to the SORBS2 locus. LINC02454 RNA was effectively captured in control cells, while LINC02454 RNA enrichment significantly decreased in LINC02454 KD cells (Fig. 5H). LINC02454 RNA was significantly enriched at the LINC02454 SE in control cells, while those enrichment levels decreased in LINC02454 KD cells (Fig. 5I). Moreover, LINC02454 RNA was significantly enriched at the SORBS2 locus in control cells but significantly decreased in LINC02454 KD cells (Fig. 5J). These results indicate that LINC02454 binds to the LINC02454 SE and the SORBS2 locus and suggest that LINC02454 mediates interactions between the LINC02454 SE and that locus. Previous studies have shown that enhancer lncRNAs activate target genes by interacting with Mediator complexes to create long-range chromatin loops [43, 44]. Mediator subunit MED1 was significantly enriched at SORBS2 genomic loci interacting with the LINC02454 SE (Fig. S4A), and MED1 KD decreased SORBS2 transcript levels (Fig. S4B). These results suggest that MED1 and LINC02454 together may mediate interaction of the SORBS2 locus with the LINC02454 SE.
Analysis of gene expression data from TCGA and GTEx databases showed significant downregulation of SORBS2 transcripts in GBM and low grade glioma (LGG) relative to normal control samples (Fig. S5A, B), suggesting that SORBS2 functions as a tumor suppressor in this context, an activity reported by others [45]. To verify SORBS2 function in regulating glioma cell sensitivity to TMZ, we performed SORBS2 OE followed by qRT-PCR analysis and confirmed a 170-fold upregulation in SORBS2 transcript levels in glioma cells (Fig. S5C). We then assessed LDH release and caspase 3/7 activity in control and SORBS2 OE glioma cells treated with TMZ. Compared with TMZ (1 mM)-treated control glioma cells, we observed significantly increased LDH release in similarly treated SORBS2 OE cells (Fig. S5D). TMZ cytotoxicity in SORBS2 OE cells significantly increased by 48 h of treatment (Fig. S5D). Consistently, caspase 3/7 activities in TMZ-treated SORBS2 OE cells markedly increased by the 48 h time point, relative to comparably treated control cells (Fig. S5E). These results suggest overall that SORBS2 upregulation increases glioma cell sensitivity to TMZ.
LINC02454 functions bivalently to regulate glioma cell sensitivity to TMZ
Knock-out of the LINC02454 SE in glioma cells downregulates LINC02454 levels (Fig. 6A). However, LINC02454 KD in glioma cells increased TMZ sensitivity (Fig. 3B–E), an outcome opposite to the decrease in TMZ sensitivity seen in LINC02454 SE KO glioma cells. These findings suggest that LINC02454 may regulate TMZ sensitivity in glioma cells by mechanisms other than maintaining chromatin interaction between the LINC02454 SE and SORBS2.
A qRT-PCR analysis of LINC02454 expression of Ctrl and LINC02454 SE KO (KO#1, KO#2) cells. Data represent means ± S.E.M. of three independent experiments, ***p < 0.001 compared with Ctrl. B Venn diagram showing intersection of three gene sets: KD_up (genes up-regulated in LINC02454 KD cells), OE_down (genes down-regulated in LINC02454 OE cells), TMZ diff_genes (genes differentially expressed in U251 cells after TMZ treatment for 4, 9, 12, and 16 days, compared with untreated U251 cells). C Venn diagram showing intersection of three gene sets: KD_down (genes downregulated genes in LINC02454 KD relative to Ctrl cells), OE_up (genes upregulated genes in LINC02454 OE relative to Ctrl cells), TMZ diff_genes (genes differentially expressed in U251 cells treated with TMZ for 4, 9, 12, and 16 days, compared with untreated U251 cells). Fold-change in transcript levels of 32 genes in LINC02454 KD (D) and LINC02454 OE (E) compared with Ctrl cells. F qRT-PCR analysis of DDR1 mRNA levels in Ctrl and DDR1-CRISPRa cells (CRISPRa). Data represent means ± S.E.M. of three independent experiments, ***p < 0.001 compared with Ctrl. G qRT-PCR of DDR1 mRNA levels in Ctrl and DDR1-CRISPRa cells transfected with LNA. Data represent means ± S.E.M. of three independent experiments, **p < 0.01, ***p < 0.001. H LDH release levels in Ctrl and DDR1-CRISPRa cells treated with LNA and 1 mM TMZ for 48 h. Data represent means ± S.E.M. of three independent experiments. *p < 0.05, ***p < 0.001. I Caspase 3/7 activity in Ctrl and DDR1-CRISPRa cells treated with LNA and 1 mM TMZ for 48 h. Data represent means ± S.E.M. of three independent experiments, *p < 0.05, **p < 0.01. J ChIRP-qPCR analysis of LINC02454 binding to the DDR1 locus in Ctrl and LINC02454 KD cells. Data represent means ± S.E.M. of three independent experiments, **p < 0.01, ***p < 0.001.
To identify other genes regulated by LINC02454 that govern glioma cell sensitivity to TMZ, we analyzed RNA-seq data of LINC02454 KD and OE U251 glioma cells and U251 glioma cells treated with TMZ (50 µM) for 0, 4, 9,12, and 16 days [34] (Fig. 6B, C). That analysis indicated that transcript levels of 5569 genes changed with TMZ treatment. Among them, 11 were upregulated in LINC02454 KD glioma cells and downregulated in LINC02454 OE cells (Fig. 6B), while 21 were downregulated in LINC02454 KD glioma cells and upregulated in LINC02454 OE cells (Fig. 6C). Ranking these 32 genes based on fold-change in LINC02454 KD or OE glioma cells, DDR1 ranked in the top 3 both in LINC02454 KD and OE glioma cells (Fig. 6D, E).
Gene expression data from TCGA and GTEx databases showed significant DDR1 up-regulation in GBM and LGG relative to normal samples (Fig. S6A, B), suggesting DDR1 has an oncogenic function. To confirm this function, we used the CRISPRa system to activate DDR1 in U251 glioma cells. qRT-PCR data showed that CRISPRa-induction of DDR1 significantly upregulated DDR1 transcripts in control glioma cells and glioma cells that were transduced with LINC02454 LNAs (Fig. 6F, G). DDR1 upregulation decreased LDH release from glioma cells treated 48 h with TMZ (1 mM) (Fig. S6C). DDR1 upregulation also significantly decreased caspase 3/7 activities in glioma cells treated 48 h with TMZ (1 mM) (Fig. S6D). We then evaluated LDH release and caspase 3/7 activity in control glioma cells and in CRISPRa glioma cells transduced with LINC02454 LNAs and treated with TMZ. Compared with TMZ (1 mM, 48 h)-treated control glioma cells transduced with LINC02454 LNA, we observed a significant decrease in LDH release in TMZ (1 mM, 48 h)-treated CRISPRa cells (Fig. 6H). Consistently, caspase 3/7 activities markedly decreased in 48 h TMZ (1 mM)-treated CRISPRa cells relative to control glioma cells transduced with LINC02454 LNA (Fig. 6I). These data indicate that DDR1 upregulation blocks the decrease in TMZ sensitivity seen in LINC02454 KD glioma cells. ChIRP-qPCR results also showed significant enrichment of LINC02454 at the DDR1 locus in control but not LINC02454 KD glioma cells (Fig. 6J), suggesting that LINC02454 binds to that locus and regulates DDR1 transcription.
Discussion
There are several reports that lncRNAs play important roles in progression of several tumor types, including glioma [46,47,48,49,50], and function to regulate cancer cell proliferation, invasion, chemoresistance and metastasis [46, 51,52,53]. Some eRNAs are lncRNAs that function with a corresponding enhancer to regulate gene expression. Transcription of eRNAs is highly correlated with enhancer activity and corresponding promoter-driven gene transcription [10,11,12, 54]. For example, the eRNA KLK3e reportedly promotes chromatin interaction between its enhancer locus and KLK2 to activate KLK2 transcription in prostate cancer cells [55]. Lai et al. found that enhancer lncRNA functioned to establish or maintain chromatin interactions between enhancers and promoters, and lncRNA KD eliminated chromatin loops and those enhancer-promoter interactions [43].
Here, we show that the SE lncRNA LINC02454 has bivalent and opposing functions in regulating glioma cell TMZ sensitivity. On one hand, LINC02454 activity enhanced SORBS2 expression by maintaining 3D chromatin interactions via the LINC02454 SE, increasing glioma cell sensitivity to TMZ. On the other, LINC02454 promoted DDR1 expression to decrease glioma cell sensitivity to TMZ. Dynamic equilibrium of both of these functions may govern glioma progression by modulating responses to TMZ treatment, although further studies are needed to define LINC02454 activities in specific contexts. Here, we also identified another lncRNA, MSRB3-AS1, as a potential SE lncRNA associated with TMZ sensitivity. However, MSRB3-AS1 KD in glioma cells did not significantly alter their TMZ sensitivity (Fig. S7A–E). Compared with control, no significant change of LDH release was detected in MSRB3-AS1 knock-down glioma cells that were treated with TMZ (1 mM) for 24 h and 48 h (Fig. S7B, C). Similarly, no significant change of caspase 3/7 acitivities were detected in MSRB3-AS1 knock-down glioma cells that were treated with TMZ (1 mM) for 48 h and 72 h, relative to control (Fig. S7D, E).
In addition to synergizing with enhancers, lncRNAs reportedly also inhibit enhancer/target gene chromatin interactions. The lncRNA Haunt and its corresponding enhancer show opposing activities in regulating HOXA genes: Haunt inhibits chromatin interaction between its enhancer locus and the HOXA locus, downregulating HOXA gene expression [56]. In our study, 443 transcripts were significantly upregulated in LINC02454 KD glioma cells. However, none of their gene loci showed a robust Capture-C signal with the LINC02454 enhancer. By contrast, the SORBS2 locus, which encodes a gene downregulated in LINC02454 KD glioma cells, showed significant interaction with the LINC02454 enhancer based on Capture-C data. Although further studies are needed to rule out the possibility that LINC02454 inhibits chromatin interactions, our study suggests a synergistic function of LINC02454 and the LINC02454 SE in regulating SORBS2 expression.
An important finding reported here is that SORBS2 and DDR1 are two major genes regulated by LINC02454, and that both regulate glioma cell sensitivity to TMZ. SORBS2 encodes an RNA binding protein that regulates the actin cytoskeleton and cell movement [57]. SORBS2 has been reported to function as a tumor suppressor in hepatocellular carcinoma, gastric cancer, pancreatic cancer, clear cell renal cell carcinoma and ovarian cancer [45, 58,59,60,61]. Mechanistically, SORBS2 protein stabilizes mRNAs with tumor suppressor function by binding to their 3′UTR [45, 60, 61]. Findings reported here support the idea that SORBS2 expression is regulated by LINC02454 and the LINC02454 SE through long-range chromatin loops mediated by 3D chromatin structure. High levels of SORBS2 protein in glioma cells may stabilize mRNAs associated with tumor suppression and increase glioma cell TMZ sensitivity. DDR1 is a receptor tyrosine kinase that reportedly functions in development and progression of several cancer types [62,63,64]. In addition, DDR1 reportedly regulates tumor cell chemoresistance [65, 66]. We found that LINC02454 KD downregulated DDR1 transcription and increased glioma cell TMZ sensitivity, consistent with a previous study of GBM showing that inhibition of DDR1 combined with radiochemotherapy including TMZ increased TMZ sensitivity and prolonged patient survival [67]. DDR1 functions physiologically by activating MAPK, PI3K/Akt and other signaling pathways [68], many of which reportedly regulate TMZ resistance of glioma cells [37, 41]. In our study, KEGG analysis shown that LINC02454 KD in glioma cells induced significant changes in MAPK signaling (Fig. 3H). Thus, LINC02454 may regulate DDR1 and alter glioma cell TMZ sensitivity through the MAPK pathway, although further studies are required for confirmation. Our findings also indicate that SORBS2 and DDR1 could be therapeutic targets to enhance glioma cell TMZ sensitivity. Also, modulation of corresponding regulatory mechanisms, such as long-range chromatin interactions at the SORBS2 locus and via an enhancer mediated by LINC02454, could also enhance TMZ efficacy.
In conclusion, our study shows that LINC02454 increases glioma cell TMZ sensitivity by maintaining chromatin interactions between the SORBS2 gene and a LINC02454 enhancer and at the same time can decrease TMZ sensitivity of glioma cells by promoting DDR1 expression. These represent novel mechanisms used by LINC02454 to regulate glioma cell TMZ sensitivity in two different contexts.
Data availability
All sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE229600.
References
Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–33.
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;20:iv1–86.
Cen L, Carlson BL, Pokorny JL, Mladek AC, Grogan PT, Schroeder MA, et al. Efficacy of protracted temozolomide dosing is limited in MGMT unmethylated GBM xenograft models. Neuro Oncol. 2013;15:735–46.
Atkins RJ, Ng W, Stylli SS, Hovens CM, Kaye AH. Repair mechanisms help glioblastoma resist treatment. J Clin Neurosci. 2015;22:14–20.
Yamada R, Nakano I. Glioma stem cells: their role in chemoresistance. World Neurosurg. 2012;77:237–40.
Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–57.
Chen X, Zhang M, Gan H, Wang H, Lee JH, Fang D, et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nature Commun. 2018;9:2949.
Chen B, He A, Bi J, Sun S, Ma Y, Wang W, et al. Long-range gene regulation network of the MGMT enhancer modulates glioma cell sensitivity to temozolomide. J Genet Genom. 2021;48:946–9.
Zhang L, He A, Chen B, Bi J, Chen J, Guo D, et al. A HOTAIR regulatory element modulates glioma cell sensitivity to temozolomide through long-range regulation of multiple target genes. Genome Res. 2020;30:155–63.
De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384.
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7.
Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–23.
Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–17.
Zhao Y, Wang L, Ren S, Wang L, Blackburn PR, McNulty MS, et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 2016;15:599–610.
Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47.
Jiang C, Shen F, Du J, Fang X, Li X, Su J, et al. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed Pharmacother. 2018;97:844–50.
Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother Pharmacol. 2018;81:671–8.
Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–9.
Grytten I, Rand KD, Nederbragt AJ, Storvik GO, Glad IK, Sandve GK. Graph peak caller: calling ChIP-seq peaks on graph-based reference genomes. PLoS Comput Biol. 2019;15:e1006731.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15.
Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Davies JO, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods. 2016;13:74–80.
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78.
Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67.
Wang J, Huang TY, Hou Y, Bartom E, Lu X, Shilatifard A, et al. Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci Adv. 2021;7:eabg4126.
Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–6.
Xu L, Chen Y, Mayakonda A, Koh L, Chong YK, Buckley DL, et al. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci USA. 2018;115:E5086–95.
Rabe M, Dumont S, Alvarez-Arenas A, Janati H, Belmonte-Beitia J, Calvo GF, et al. Identification of a transient state during the acquisition of temozolomide resistance in glioblastoma. Cell Death Dis. 2020;11:19.
Zhou J, Yi Q, Tang L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: a focused review. J Exp Clin Cancer Res. 2019;38:250.
Ullah R, Yin Q, Snell AH, Wan L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 2022;85:123–54.
Xu P, Zhang G, Hou S, Sha LG. MAPK8 mediates resistance to temozolomide and apoptosis of glioblastoma cells through MAPK signaling pathway. Biomed Pharmacother. 2018;106:1419–27.
Zeng H, Xu N, Liu Y, Liu B, Yang Z, Fu Z, et al. Genomic profiling of long non-coding RNA and mRNA expression associated with acquired temozolomide resistance in glioblastoma cells. Int J Oncol. 2017;51:445–55.
Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MC, et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood. 2019;134:239–51.
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–31.
Shi F, Guo H, Zhang R, Liu H, Wu L, Wu Q, et al. The PI3K inhibitor GDC-0941 enhances radiosensitization and reduces chemoresistance to temozolomide in GBM cell lines. Neuroscience. 2017;346:298–308.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501.
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5.
Lv Q, Dong F, Zhou Y, Cai Z, Wang G. RNA-binding protein SORBS2 suppresses clear cell renal cell carcinoma metastasis by enhancing MTUS1 mRNA stability. Cell death & disease. 2020;11:1056.
Zong Y, Zhang Y, Hou D, Xu J, Cui F, Qin Y, et al. The lncRNA XIST promotes the progression of breast cancer by sponging miR-125b-5p to modulate NLRC5. Am J Transl Res. 2020;12:3501–11.
Wang J, Gu J, You A, Li J, Zhang Y, Rao G, et al. The transcription factor USF1 promotes glioma cell invasion and migration by activating lncRNA HAS2-AS1. Biosci Rep. 2020;40:BSR20200487.
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. 2019;36:645–59.e648.
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25.
Shi T, Guo D, Xu H, Su G, Chen J, Zhao Z, et al. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol Biol Rep. 2020;47:2723–33.
David A, Zocchi S, Talbot A, Choisy C, Ohnona A, Lion J, et al. The long non-coding RNA CRNDE regulates growth of multiple myeloma cells via an effect on IL6 signalling. Leukemia. 2021;35:1710–21.
Gong X, Zhu Z. Long Noncoding RNA HOTAIR Contributes to progression in hepatocellular carcinoma by sponging miR-217-5p. Cancer Biother Radiopharm. 2020;35:387–96.
Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun. 2020;11:5513.
Kim TK, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol. 2015;7:a018622.
Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111:7319–24.
Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA Gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16:504–16.
Roignot J, Soubeyran P. ArgBP2 and the SoHo family of adapter proteins in oncogenic diseases. Cell Adh Migr. 2009;3:167–70.
Taieb D, Roignot J, Andre F, Garcia S, Masson B, Pierres A, et al. ArgBP2-dependent signaling regulates pancreatic cell migration, adhesion, and tumorigenicity. Cancer Res. 2008;68:4588–96.
Tong Y, Li Y, Gu H, Wang C, Liu F, Shao Y, et al. HSF1, in association with MORC2, downregulates ArgBP2 via the PRC2 family in gastric cancer cells. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1104–14.
Han L, Huang C, Zhang S. The RNA-binding protein SORBS2 suppresses hepatocellular carcinoma tumourigenesis and metastasis by stabilizing RORA mRNA. Liver Int. 2019;39:2190–203.
Zhao L, Wang W, Huang S, Yang Z, Xu L, Yang Q, et al. The RNA-binding protein SORBS2 suppresses metastatic colonization of ovarian cancer by stabilizing tumor-suppressive immunomodulatory transcripts. Genome Biol. 2018;19:35.
Li W, Ping Z, Xuemei G, Hongjuan M, Yi H, Xiaoli L, et al. Chlorogenic acid regulates the proliferation and migration of high-grade serous ovarian cancer cells through modulating the miR199a5p/DDR1 axis. Acta Biochim Pol. 2022;69:855–64.
Han Q, Xiao F, Ma L, Zhou J, Wang L, Cheng H, et al. DDR1 promotes migration and invasion of breast cancer by modulating the Src-FAK signaling. Neoplasma. 2022;69:1154–64.
Zhang X, Hu Y, Pan Y, Xiong Y, Zhang Y, Han M, et al. DDR1 promotes hepatocellular carcinoma metastasis through recruiting PSD4 to ARF6. Oncogene. 2022;41:1821–34.
Ambrogio C, Darbo E, Lee SW, Santamaria D. A putative role for Discoidin Domain Receptor 1 in cancer chemoresistance. Cell Adh Migr. 2018;12:394–7.
Lai SL, Tan ML, Hollows RJ, Robinson M, Ibrahim M, Margielewska S, et al. Collagen induces a more proliferative, migratory and chemoresistant phenotype in head and neck cancer via DDR1. Cancers. 2019;11:1766.
Vehlow A, Klapproth E, Jin S, Hannen R, Hauswald M, Bartsch JW, et al. Interaction of Discoidin Domain Receptor 1 with a 14-3-3-Beclin-1-Akt1 complex modulates glioblastoma therapy sensitivity. Cell Rep. 2019;26:3672–83.e3677.
Mehta V, Chander H, Munshi A. Complex roles of discoidin domain receptor tyrosine kinases in cancer. Clin Transl Oncol. 2021;23:1497–510.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 32130018, 32170598, 82073119); the National Key R&D Program of China (Grant No. 2020YFA803700); the Natural Science Foundation of Tianjin City of China (Grant No. 22JCYBJC00600, 20JCQNJC01880).
Author information
Authors and Affiliations
Contributions
TS, LZ, and WL designed the research; TS, DG, YZ, WW, JB, AH, SF, GS, XZ, ZZ, YS, SS, PL, ZZ, and JS performed the experiments; TS, DG and WW analyzed the data; TS, LZ, and WL wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
No human subjects or animals were used in these studies.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Bertrand Joseph
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, T., Guo, D., Zheng, Y. et al. Bivalent activity of super-enhancer RNA LINC02454 controls 3D chromatin structure and regulates glioma sensitivity to temozolomide. Cell Death Dis 15, 6 (2024). https://doi.org/10.1038/s41419-023-06392-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-06392-w