Abstract
Glioblastoma multiforme (GBM) is an extremely aggressive brain tumor for which new therapeutic approaches are urgently required. Unfolded protein response (UPR) plays an important role in the progression of GBM and is a promising target for developing novel therapeutic interventions. We identified ubiquitin-activating enzyme 1 (UBA1) inhibitor TAK-243 that can strongly induce UPR in GBM cells. In this study, we evaluated the functional activity and mechanism of TAK-243 in preclinical models of GBM. TAK-243 significantly inhibited the survival, proliferation, and colony formation of GBM cell lines and primary GBM cells. It also revealed a significant anti-tumor effect on a GBM PDX animal model and prolonged the survival time of tumor-bearing mice. Notably, TAK-243 more effectively inhibited the survival and self-renewal ability of glioblastoma stem cells (GSCs) than GBM cells. Importantly, we found that the expression level of GRP78 is a key factor in determining the sensitivity of differentiated GBM cells or GSCs to TAK-243. Mechanistically, UBA1 inhibition disrupts global protein ubiquitination in GBM cells, thereby inducing ER stress and UPR. UPR activates the PERK/ATF4 and IRE1α/XBP signaling axes. These findings indicate that UBA1 inhibition could be an attractive strategy that may be potentially used in the treatment of patients with GBM, and GRP78 can be used as a molecular marker for personalized treatment by targeting UBA1.
Similar content being viewed by others
Introduction
Glioblastoma multiforme (GBM) is the most common primary malignant brain tumor in adults with the highest mortality rate [1]. Although comprehensive treatment regimens such as surgery combined with chemoradiotherapy are used in the clinic, tumor recurrence remains a problem as it often results in a poor prognosis. The average survival time of GBM is only 12–15 months, and the 5-year overall survival rate is <6% [2]. Glioblastoma stem cells (GSCs) are resistant to chemotherapy and radiotherapy and thus considered one of the key reasons for GBM recurrence and treatment failure [3]. Temozolomide remains the major chemotherapeutic drug for the treatment of GBM [4]. Nonetheless, GBM is one of the few solid tumors lacking targeted therapeutic drugs [5]. Therefore, it is imperative to identify new therapeutic targets for GBM, and provide more effective treatment methods and strategies.
The synthesis, correct folding, and degradation of proteins is a dynamic equilibrium known as ER proteostasis [6]. Due to the special intracranial microenvironment, GBM faces more severe environmental pressure than other tumors, which include hypoxia, oxidative stress, and nutritional deprivation, resulting in a significant increase in the rate of protein misfolding and disruption of ER proteostasis [7]. Tumor cells can activate a series of adaptive regulatory signals to promote proper protein folding, eliminate misfolded proteins, and restore proteostasis, which is collectively called unfolded protein response (UPR) [8]. Endoplasmic reticulum stress can activate UPR using three pathways: PERK/eIF2/ATF4, IRE1α/XBP1, and ATF6/ATF6f cascade signals [9]. In the inactivated state, cells block PERK, IRE1α, and ATF6 sensor receptors through the molecular chaperone protein GRP78 (also known as BiP). When endoplasmic reticulum stress is activated, the association is disrupted and the UPR signal transduction cascade commences [10, 11]. It is worth noting that UPR plays a dual function in tumor cells, i.e., if it cannot effectively control endoplasmic reticulum stress and restore proteostasis, then it will initiate apoptosis [12]. Therefore, disruption of ER homeostasis of glioblastoma may inhibit tumor cell proliferation and cause tumor cell apoptosis, which is a potential treatment strategy.
ER-associated degradation (ERAD) is the main pathway to eliminate misfolded proteins in the endoplasmic reticulum [13]. The ERAD system is comprised of many regulatory genes that are responsible for the identification of misfolded proteins, and transportation of these proteins to the cytoplasm for degradation. Once misfolded proteins are transported to the cytoplasm, these are mainly degraded through the ubiquitin-proteasome system (UPS). Ubiquitin is a polypeptide composed of 76 amino acid residues. Ubiquitin can bind to target proteins through the action of a series of enzymes, and proteins tagged with more than four ubiquitin molecules will be transported to the 26S proteasome for degradation [14]. Therefore, blocking the UPS process of glioblastoma cells may cause excessive accumulation of misfolded proteins in the endoplasmic reticulum and initiate UPR-induced apoptosis response.
In this study, we found that ubiquitin-activating enzyme 1 (UBA1) inhibitor TAK-243 could strongly induce UPR in GBM cells. UBA1 is the main ubiquitin-activating E1 enzyme and is responsible for initiating the ubiquitination cascade [15, 16]. TAK-243 is a first-in-class inhibitor of UBA1 [17] and is currently undergoing multiple phase 1 clinical trials on advanced malignancies (NCT02045095, NCT03816319). In this study, we evaluated the preclinical activity and mechanism of TAK-243 in GBM. Notably, TAK-243 also effectively inhibited the self-renewal of GSCs. We also found that GRP78 is a biomarker for the sensitivity of GBM cells to TAK-243 treatment. Therefore, targeting UBA1 may be a promising approach for GBM treatment.
Results
UBA1 inhibition suppresses GBM cell proliferation and colony formation
To evaluate the effect of UBA1 inhibitor TAK-243 on GBM cell growth, the CCK-8 was used to determine the viability of seven GBM cell lines and eight primary GBM cells. The IC50 of TAK-243 on GBM cell lines was between 15.64 and 396.3 nM (Fig. 1a), and the IC50 on primary GBM cells was between 23.42 and 936.8 nM (Fig. 1j). The results showed that TAK-243 reduced the survival of GBM cells and primary GBM cells in a concentration-dependent manner. Of note, we found that the sensitivity of different GBM cell lines and primary GBM cells to TAK-243 significantly varied.
a GBM cell lines were treated with increasing concentrations of TAK-243 (0–10 μM) for 72 h. Cell viability was then measured by CCK-8 assay. b, c Measurement of antiproliferation effects of TAK-243 by EdU incorporation assay, scale bar: 100 μm. d, e TAK-243 inhibits colony formation in U251 and LN229 cells in a dose-dependent manner. Quantitative analysis of the results of the colony formation experiment was performed. f Construction of UBA1 downregulated-LN229 cell lines. The efficiency of UBA1 downregulation was assessed by Western blotting assay. g Cell viabilities were measured after UBA1 downregulation by CCK-8 assay. h, i Assessment of antiproliferation and proapoptotic effects of UBA1 downregulation by EdU incorporation assay (scale bar: 100 μm) and TUNEL staining (scale bar: 100 μm). j Primary GBM cell lines were exposed to either 0.1% DMSO or TAK-243 for 72 h. Cell viability was then determined by CCK-8. k, i Colony formation abilities of primary cell lines were assessed by methylcellulose clonal assays (scale bar: 100 μm). All the Data are presented as means ± SD. **P < 0.01, ***P < 0.001 compared with the 0.1% DMSO treated group.
To determine the effect of TAK-243 on GBM cell proliferation, EdU incorporation assay was employed. Compared with the control group, TAK-243 treatment significantly reduced the ratio of EdU-positive cells (Fig. 1b, c). To further verify the function of the UBA1 gene in GBM cells, we constructed a UBA1 knockdown cell line. Similarly, downregulation of UBA1 expression significantly inhibited GBM cell survival and proliferation, and induced GBM cell death (Fig. 1f–i). These results suggest that UBA1 is a potential therapeutic target for GBM.
In order to observe the inhibitory effect of TAK-243 on the long-term proliferation of GBM cells, we examined the effect of TAK-243 on the colony formation ability of GBM cells. Compared with the control group, TAK-243 treatment significantly reduced the number of GBM colonies formed. Treatment using 50 and 100 nM TAK-243 resulted in a decrease in the relative number of formed U251 cell colonies to 52.33 and 13.67%, respectively (Fig. 1d). Similar results were observed in LN229 cells (Fig. 1e). Moreover, the colony formation assay showed that TAK-243 treatment significantly reduced the colony number and sizes in primary GBM cells (Fig. 1k, i). In summary, TAK-243 can significantly inhibit the survival, proliferation, and colony formation of GBM cells.
TAK-243 arrests the GBM cell cycle and induces apoptosis
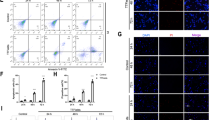
The cell cycle progression is strictly regulated by ubiquitin-mediated proteolysis of key regulators, including cyclins, which are required for cyclin-dependent kinase activity [18]. To assess the effect of UBA1 inhibition on cell cycle regulation, we used flow cytometry to detect the cell cycle distribution of GBM cells. As shown in Fig. 2a, b, the number of G2/M phase cells significantly increased, and the number of G1 phase cells decreased correspondingly. While treating with high concentrations of TAK-243 resulted in accumulation of cells both in S and G2/M phases. Consistent with these results, TAK-243 treatment caused significant changes in the expression of cell cycle regulatory proteins, Cdc2, cyclin D1, and p21, in GBM cells (Fig. 2g, h).
a, b Representative data of the cell cycle analysis of TAK-243-treated cells. U251 and LN229 cells were treated withTAK-243 at the indicated concentrations for 24 h. The cell cycle profile was evaluated using flow cytometry. c, d Cells were treated with TAK-243 for 24 h, and were stained with Annexin V/PI. Apoptosis was evaluated by flow cytometry. The total proportion of Annexin V-positive (early apoptotic) cells and Annexin V/PI-stained double-positive (late apoptotic) cells was counted as apoptotic to prepare the bar graph. e, f U251 and LN229 cells were treated with the indicated concentration of TAK-243 for 24 h. Caspase-3/7 activity was assessed. g, h Western blotting analysis of protein levels of cell cycle and cell apoptosis regulators in U251 and LN229 cells treated with TAK-243 with indicated antibodies. All the Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we tested the effect of TAK-243 on GBM cell apoptosis and found that the percentage of apoptotic cells significantly increased in TAK-243-treated groups in a concentration-dependent manner compared to the control group (Fig. 2c, d and Supplemental Fig. S1). Consistent with these data, caspase-3/7 activities were increased and the cleaved PARP level was increased in GBM cells after TAK-243 treatment (Fig. 2e−h). Taken together, these results indicate that TAK-243 treatment effectively induces GBM cell apoptosis.
Pathway annotation based on quantitative ubiquitin-modified proteome
UBA1 is the initiating enzyme of the ubiquitination cascade. As expected, TAK-243 treatment reduced polyubiquitination of global cellular proteins in a time- and dose-dependent manner (Fig. 3a). We wanted to use the quantitative ubiquitin-modified proteome to comprehensively analyze the effect of TAK-243 on the ubiquitination of GBM cells. The U251 cells were treated with vehicle or TAK-243 for 6 h. Then the proteins were extracted, enzymatically digested, ubiquitin-peptide enriched, followed by LC-MS/MS analysis. The ubiquitin-modified proteome analysis identified 478 and 244 proteins in the TAK-243 treatment group whose ubiquitination levels increased or decreased, respectively (Fig. 3b). We observed that the ubiquitination levels of PSMD1, PSMD4, NEDD8, UBE2C, USP5, c-Myc, TP53, and CDK1 were significantly changed (Fig. 3c). Gene ontology analysis showed that these differential proteins were related to functions in cellular and biological processes, biological regulation, and metabolic processes (Fig. 3d). Protein–protein interaction network analysis showed that these differentially expressed ubiquitination-modified proteins were mainly associated with ubiquitin-mediated proteolysis, proteasome, ribosome, and cell-cycle-related pathways (Fig. 3e). These results support the function and specificity of TAK-243 in reducing global protein ubiquitination.
a GBM cells were treated with increasing concentrations of TAK-243 for indicated times. Then, the whole-cell lysates were prepared and levels of global ubiquitylated proteins, UBA1 and β-actin were measured by immunoblotting. b Heatmap of the differentially ubiquitylated proteins between control and TAK-243 treatment groups. c Volcano plot showing the quantitative levels of ubiquitylated proteins between control and TAK-243 treatment groups. d Gene Ontology (GO) enrichment analysis of the differentially ubiquitylated proteins between control and TAK-243 treatment groups. e Protein−Protein interaction networks functional enrichment analysis of the differentially ubiquitylated proteins using STRING database.
TAK-243 induces ER stress and UPR in GBM cells
Ubiquitin is known to be important for proper retro-translocation of misfolded proteins from the ER to the cytosol. Failure to degrade abnormally folded and damaged proteins may cause endoplasmic reticulum stress and cell death [12, 19]. Considering the role of UBA1 as the main ubiquitin-activating enzyme, we investigated whether UBA1 inhibition induces ER stress and UPR in GBM cells. After treating U251 and LN229 cells with TAK-243, the expression levels of ER stress- and UPR pathway-related proteins were detected. We observed that TAK-243 treatment resulted in the accumulation of ER chaperone GRP78. The basal expression level of GRP78 protein in U251 cells is lower than that in LN229 cells. We observed that the TAK-243-induced GRP78 expression was more sensitive in U251 cells than in LN229 cells. TAK-243 treatment increased the phosphorylation of PERK and eIF2α, and induced the expression of ATF4 and CHOP, indicating activation of the PERK axis. TAK-243 also induced phosphorylation of IRE1αand JNK1/2, and XBP1 splicing to the active form, indicating activation of IRE1α axis (Fig. 4a). However, there was no significant change in the expression of ATF6. We further examined the effects of TAK-243 on gene expression downstream of PERK and IRE1α arms. As shown in Fig. 4c−f, the mRNA levels of GADD34 and NOXA (ATF4 target genes), and PDI-P5 and Edem1 (XBP1s target genes) were significantly increased after TAK-243 treatment.
a Representative western blot analysis showing dose-response and time course of the effects of TAK-243 on the ER stress and UPR in U251 and LN229 cells. b U251 and LN229 cells treated with 0.1% DMSO or indicated concentration of TAK-243 for 24 h were stained with ER-Tracker Red kit (scale bar: 5 μm). c−f Quantitative PCR was performed to examine the relative mRNA level of GADD34, NOXA, PDI-P5, and Edem1 after TAK-243 treatment. g Quantitative PCR was performed to assess the relative mRNA level of Bloc1s1 and Col6A1, which are downstream genes of RIDD pathway. h Western blotting showing the expression levels of p62 and LC3B proteins.
To investigate the function of UPR activation in TAK-243-induced cell death, we assessed the cytotoxicity of TAK-243 after treatment with UPR inhibitors. The results showed that PERK inhibitor GSK2606414 or IRE1α inhibitor 4μ8C could rescue TAK-243-induced cell death. In contrast, ATF6 inhibitor nelfinavir did not change TAK-243-induced cell death (Supplemental Fig. S2). Furthermore, we observed the changes of ER morphology under ER stress and found that TAK-243 treatment induced a rapid extended of the ER cisternae (Fig. 4b). ER, stress-mediated IRE1α activation induces the regulated IRE1-dependent decay (RIDD) pathway activation [20]. We found that the mRNA levels of RIDD target genes (Bloc1s1 and Col6A1) were downregulated in a dose-dependent manner after TAK-243 treatment (Fig. 4g). In summary, our data suggest that TAK-243 induced cell death through activation of the PERK and IRE1α pathways.
Autophagy is another mechanism by which accumulated or misfolded proteins undergo proteasomal degradation [21]. In this pathway, ubiquitinated proteins aggregate and are transported to autophagosomes by p62 [22]. We observed that TAK-243 induced the conversion of LC3B I to II (Fig. 4h). However, the expression of p62 was increased after TAK-243 treatment, indicating that autophagy was inefficient following TAK-243 treatment. This may be due to the decreased ubiquitination level of p62 protein caused by UBA1 inhibition, which resulted in the accumulation of p62 protein. When the autophagy response is not sufficient to compensate for the stress condition, it will activate the apoptosis signal and promote apoptosis [23, 24]. We also assessed the cytotoxicity of TAK-243 after treatment with autophagy inhibitor 3-MA. The results showed that co-treatment of LN229 or U251 cells with 3-MA increased TAK-243-induced cell death (Supplemental Fig. S3). Taken together, our results suggest that autophagy inhibition may enhance TAK-243-induced cell death.
GRP78 determines sensitivity to TAK-243 in GBM cells
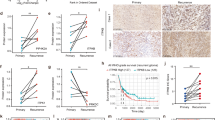
The above results showed that the sensitivity of different GBM cells to TAK-243 varied. To explore the potential mechanism of resistance to TAK-243, we examined the expression of multiple UPR-related proteins in GBM cell lines. The correlation between protein levels and the TAK-243 sensitivity was analyzed using blot quantitative values and IC50 values. There was no significant correlation between the expression level of UBA1 and the sensitivity of GBM cells to TAK-243 (Fig. 5a). Interestingly, only the expression of GRP78 was significantly related to the sensitivity of GBM cell lines (Fig. 5b). We further verified this significant correlation in primary GBM cells (Fig. 5c). Taken together, these results demonstrate that the expression levels of GRP78 are negatively correlated with the TAK-243 sensitivity in GBM cells.
a The expression levels of UBA1, GRP78, PERK, p-eIF2α, eIF2α, and IRE1a were evaluated by western blotting in GBM cell lines. b Correlation analysis of relative GRP78 levels and IC50 values of TAK-243 in GBM and primary GBM cells. c The expression levels of GRP78 in eight primary GBM cell lines. d−f LN229 and U87 cells expressed with shGRP78-1 were treated with TAK-243 for 24 h. Cell viability, apoptosis, and caspase 3/7 activity were assessed by CCK-8, Annexin V staining, and Caspase-Glo 3/7 activity assay. g The protein levels of p-PERK, PERK, XBP1s, CHOP, cleaved PARP, and cleaved caspase-3 were assessed by immunoblotting in indicated cells. h−i GRP78 overexpressed U251 cells were treated with TAK-243 for 24 h, and then cell viability, apoptosis, and caspase 3/7 activity were assessed. j The expression levels of p-PERK, PERK, XBP1s, CHOP, cleaved PARP, and cleaved caspase-3 were assessed by immunoblotting in indicated cells. All the Data are presented as means ± SD. **P < 0.01, ***P < 0.001.
To further analyze whether the high expression of GRP78 renders GBM cells resistant to TAK-243, we downregulated GRP78 expression in U87 and LN229 cells, which had high GRP78 expression, and over-expressed GRP78 in U251 cells, which had a low level of GRP78 expression (Supplemental Fig. S4). Compared with the control group, U87 and LN229 cells with GRP78 knocked down showed a significant increase in sensitivity to TAK-243 (Fig. 5d). GRP78 knockdown in U87 and LN229 also resulted in an increase in sensitivity to TAK-243-induced apoptosis and caspase-3/7 activities (Fig. 5e, f and Supplemental Fig. S5). GRP78 knockdown also enhanced TAK-243-induced expression of p-PERK, XBP1s, CHOP, cleaved-PARP, and cleaved Caspase-3 (Fig. 5g). However, overexpression of GRP78 reduced the sensitivity to TAK-243, and blocked the increase in TAK-243-induced apoptosis, and the expression of p-PERK, XBP1s, CHOP, cleaved-PARP, and cleaved Caspase-3 in U251 cells (Fig. 5h–j and Supplemental Fig. S6). These results support that GRP78 is a key factor in determining the sensitivity to TAK-243 in GBM cells.
UBA1 inhibition disrupts GSC stemness maintenance
GSCs are related to the development, resistance to radiotherapy and chemotherapy, and tumor recurrence of gliomas [25]. We further determined whether TAK-243 treatment affects GSCs viability and maintenance. Compared with the control group, TAK-243 treatment resulted in a significant decrease in the proliferation rate of GSC1 and GSC2 cells in a concentration-dependent manner (Fig. 6a). Notably, the inhibition of TAK-243 on the proliferation of GSCs was at least five times more effective than on GBM cells. Therefore, we continued to test whether the high sensitivity of GSC cells to TAK-243 is also related to the expression level of GRP78. Interestingly, the expression level of GRP78 in GSC cells was significantly lower than in GBM cell lines (Fig. 6b). Moreover, CCK-8 results also showed that the differentiated GSC cells were more resistant to TAK-243 than GSCs (Fig. 6c). When GSCs differentiated into adherent GBM cells, the expression level of GRP78 also significantly increased (Fig. 6d). These results further support that the expression level of GRP78 protein can determine the sensitivity of tumor cells to TAK-243.
a Effects of TAK-243 on cell viability in GSC1 and GSC2. b GRP78 and UBA1 expression levels were examined by immunoblotting in GSCs and GBM cells lines. c Cell viabilities were evaluated by CCK-8 assays in GSCs and differentiated GSCs. d Immunoblot analyses of GRP78 levels in GSCs and differentiated GSCs. e−g Effects of TAK-243 on GSCs neurosphere formation, scale bar: 100 μm. h In vitro limiting dilution assays of GSC1 and GSC2 treated with indicated doses of TAK-243 or DMSO. i Representative western blot analysis showing dose-response and time course of the effects of TAK-243 on the polyubiquitylation and ER stress in GSC1 and GSC2, as assessed by immunoblotting for polyubiquitin (polyUb), GRP78, p-PERK(Thr980), PERK, p-eIF2α, eIF2α, ATF4, p-IRE1a(Ser724), IRE1a, CHOP, XBP1s, and ATF6. j The effects of TAK-243 on GSC1 and GSC2 apoptosis as evaluated by Annexin V/PI staining. All the Data are presented as means ± SD. **P < 0.01, ***P < 0.001.
Next, we examined the impact of TAK-243 on GSC self-renewal in GSC1 and GSC2. The neurosphere formation assay showed that with increasing TAK-243 concentration, the inhibition on GSC1 and GSC2 neurosphere formation gradually increased (Fig. 6e–g). Compared with the control group, at a TAK-243 concentration of 20 nM, the neurosphere formation rates of GSC1 and GSC2 were 44.56 and 22.61% of the control group, respectively, indicating that TAK-243 can significantly inhibit neurosphere formation of GSCs. In vitro limiting dilution assay demonstrated that TAK-243 treatment suppressed GSCs self-renewal potential (Fig. 6h). These results indicate that TAK-243 inhibits the proliferation and the maintenance of GSCs.
We also determined the effects of TAK-243 treatment on ubiquitination of cellular proteins and the induction of UPR in GSCs. Compared with GBM cells, TAK-243 at a lower concentration rapidly reduced the level of polyubiquitin chains in GSC cells (Fig. 6i). We also observed rapid induction of ER stress and UPR following TAK-243 treatment in GSC cells. Exposure to TAK-243 resulted in the accumulation of the ER chaperone GRP78, and increased phosphorylation of PERK, eIF2α, and IRE1α. Protein levels of ATF4, CHOP, and XBP1s were also upregulated in TAK-243-treated GSC1 and GSC2 cells. Little or no changes were observed in the ATF6 axis (Fig. 6i). Moreover, TAK-243 treatment also effectively induced GSCs apoptosis, as evaluated by flow cytometry analyses using Annexin V and PI as apoptosis markers (Fig. 6j and Supplementary Figs. S7, S8). Taken together, the higher sensitivity of GSCs to TAK-243 indicates that targeting UBA1 may be a promising approach for eliminating GSCs.
TAK-243 inhibits tumor growth and prolongs the survival of tumor-bearing mice in a PDX mouse model
To determine the clinical application of this drug, we examined whether pharmacologic inhibition of UBA1 by TAK-243 suppresses tumor growth in preclinical models. We constructed a nude mouse subcutaneous GBM tumor model with LN229 cells and analyzed the size and weight of xenograft tumors. The results showed that TAK-243 treatment significantly inhibited the growth of subcutaneous tumor cells in nude mice. The tumor volume of the treatment group mice was significantly smaller than that of the control group, and the tumor weight also gradually decreased with increasing drug dose (Fig. 7a–c).
a Representative tumors isolated from the control and TAK-243-treated groups of subcutaneous tumor model. b The mean tumor volumes were assessed at the indicated number of days after tumor implantation. c Tumors isolated from control and TAK-243-treated groups were weighted. The tumor weight was analyzed statistically. d Mice bearing GSC2-derived xenograft tumor were treated with TAK-243 (10 or 20 mg/kg) or vehicle control by intraperitoneally injection twice a week for 4 weeks. Representative images of H&E staining of whole-brain sections from the control group and TAK-243 treatment group. e Kaplan−Meier survival curves of mice implanted with GSC2 cells (n = 10). The survival time of tumor-bearing mice was counted. f Representative IHC staining images of polyubiquitin (polyUb), GRP78, and Ki67 expression in GSC2-derived xenograft tumor of control and TAK-243 treatment groups, scale bar: 50 μm. All the data are expressed as means ± SD. **P < 0.01, ***P < 0.001.
Next, we examined the anti-tumor effect of TAK-243 in the PDX mouse model. Four weeks after TAK-243 administration, HE staining of tumor tissues showed that the tumor size of tumor-bearing mice in the treatment group was smaller than that in the control group (Fig. 7d). Moreover, the survival time of tumor-bearing mice was significantly prolonged after TAK-243 treatment (Fig. 7e). Pathological analysis showed that the number of Ki67-positive cells in the TAK-243 treatment group was significantly reduced, the level of global protein polyubiquitination was significantly reduced, and GRP78 expression was increased (Fig. 7f). These results indicate that TAK-243 can inhibit the proliferation of GBM cells in vivo.
Discussion
GBM is the most malignant and lethal brain tumor [1]. However, no effective treatment measures have been established to date. In this study, we evaluated the preclinical efficacy of UBA1 inhibitor TAK-243 in the treatment of GBM. We found that TAK-243 rapidly activates GBM cells to produce UPR, which in turn induces apoptosis of GBM cells, and significantly prolongs the survival time of tumor-bearing mice. Notably, TAK-243 also inhibited GSC cell proliferation and neurosphere formation. Moreover, we found that GRP78 is a key molecule that determines the sensitivity of tumor cells to TAK-243. However, our preliminary results showed that TAK-243 and temozolomide combined had no significant synergistic effect in GBM cells (data not shown).
UBA1 is the initiating enzyme in the ubiquitination cascade, responsible for initiating the transfer of ubiquitin molecules to target proteins, which are to be degraded by proteasomes [15, 16]. We observed that after TAK-243 inhibited UBA1, polyubiquitination of global cellular proteins in GBM cells was reduced. We identified changes in more than 1,000 ubiquitination modification sites using ubiquitin-modified proteome analysis. The most regulated genes upon TAK-243 treatment impact on UPS and cell death. We observed that the ubiquitination levels of PSMD1, PSMD4, NEDD8, UBE2C, USP5, c-Myc, TP53, and CDK1 were significantly changed. Blocking protein degradation will lead to the accumulation of intracellular proteins and induce ER stress, UPR, and cell death [12, 26]. When cells are unable to cope with ER stress, CHOP proteins that rely on UPR activation will regulate the expression of apoptosis-related proteins and initiate the apoptosis program [27,28,29]. It has been reported that TAK-243 has anti-proliferative activity in multiple human cancers, and regulates cell apoptosis by inducing ER stress and UPR [30,31,32]. We found that TAK-243 can activate PERK/ATF4 and IRE1α/XBP signaling axis, then induce cell apoptosis of GBM. Although UBA1 inhibition reduces the ubiquitination level of global cellular proteins, the UBA1 inhibitor induces a higher rate of cell death in malignant cells compared to normal cells. Given that multiple proteasome inhibitors have been successfully approved for clinical applications, targeting UBA1 for the treatment of tumors is also a promising potential treatment strategy.
Notably, the sensitivity of TAK-243 in different GBM cell lines significantly varies. Among the seven GBM cell lines, the highest and the lowest IC50 differ by 25-fold, and in primary cells, the difference is about 40-fold. This thus becomes an obstacle in the selection of patients who are sensitive to TAK-243 treatment. Resistance to therapy is also one of the important factors that affect the long-term survival of tumor patients and cause recurrence. However, the sensitivity to TAK-243 in GBM is not significantly correlated with the expression level of the target protein UBA1. Interestingly, we found that the expression level of GRP78 is a key factor that determines the sensitivity to TAK-243 in GBM cells. Tumor cells with high GRP78 expression are resistant to TAK-243 treatment. Knockdown or overexpression of GRP78 can affect the sensitivity to TAK-243. GRP78 is a switch that controls the initiation of UPR. When ER stress is activated, misfolded proteins bind to GRP78 and destroy its inhibitory effect, and thereby the UPR response is activated [33, 34]. UBA1 inhibition also induces the increase of GRP78 expression in GBM cells, which may be a negative feedback and self-protection mechanism of the cells. Of note, it has been reported that the sensitivity to TAK-243 in a variety of other solid tumors and hematological tumors also significantly vary [17, 30, 31], while our understanding of the mechanism that determines its sensitivity is limited. In the future, it is necessary to analyze whether GRP78 expression is related to TAK-243 sensitivity in other tumor types. GRP78 may be a biomarker for the sensitivity of TAK-243 tumor treatment, which can guide precision therapy by targeting UBA1.
GSCs have high carcinogenic potential, self-renewal ability, and multidirectional differentiation ability. GSCs are resistant to current therapeutic approaches, which is also considered one of the main causes of malignant recurrence of GBM [25, 35,36,37]. Although there has been some understanding on the signaling pathways that maintain the stem cell characteristics of GSCs, there is still no effective therapeutic target or method to eliminate GSCs. Surprisingly, we found that GSCs are sensitive to UBA1 inhibitor TAK-243 treatment that the inhibitory effect of TAK-243 on the proliferation of GSCs is at least five times more effective than on GBM cell lines. The concentration of TAK-243 to induce UPR response in GSCs cells is also much lower than in GBM cells. Interestingly, this compound could also more effectively suppress the sphere formation of GSCs than its adherent GBM cells on culture dishes. It has been reported that TAK-243 has no significant effect on the survival of normal hematopoietic stem cells [31]. We found that the sensitivity of GSCs to TAK-243 is also affected by the expression level of GRP78. Compared with GBM cell lines, the expression level of GRP78 in GSCs is much lower. Consistent with the cell sensitivity results, the expression level of GRP78 was increased significantly in differentiated GSCs, and these cells showed greater resistance to TAK-243 treatment. These results further support that GRP78 may be utilized as a biomarker for TAK-243 sensitivity. Taken together, targeting UBA1 may be a promising approach in eliminating GSCs.
In summary, our study demonstrates that the UBA1 inhibitor TAK-243 exhibits good anti-tumor activity in GBM cell lines, primary cells, and PDX animal models. Furthermore, TAK-243 more effectively blocks the survival and self-renewal ability of GSC cells than those of GBM cells. We found that GRP78 is a predictor of GBM sensitivity to TAK-243 treatment. Collectively, our findings strongly support further clinical trials of TAK-243 in GBM.
Materials and methods
Culture of cell lines and primary cell lines
The human GBM cells lines (U87, U251, A172, T98G, LN229, U118, and LN18) used in this study were purchased from Shanghai Cell Bank, Chinese Academy of Sciences. These cell lines were cultured and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Primary cell lines GBM1, GBM2, GBM3, GBM4, GBM5, GBM6, GBM7, and GBM8, isolated from tumor tissues of patients with GBM, were cultured with DMEM/F12 containing 10% FBS. These cell lines were grown in a humidified incubator containing 5% CO2 at 37 °C. Written informed consent was obtained from all of the patients, and this study was approved by the Research Ethics Committee of the Affiliated Hospital of Xuzhou Medical University.
Culture of GSCs
GSCs were isolated and purified from GBM surgical specimens and were named GSC1 and GSC2, respectively. The two GSC cell lines were cultured in NeurobasalTM medium containing basic fibroblast growth factor, EGF, B27 supplement, haprin, L-glutamine, and N2 supplement to form a GSC-rich neurosphere culture. Every three days, 1/3 volume of the medium was replaced with fresh medium. A NeuroCult Chemical Dissociation Kit was used to separate the neurosphere for cell subculture.
Antibodies and reagents
Ubiquitin (#3933), UBA1 (#4891), p21 (#2947), Cyclin D1 (#2922), CDC2 (#9116), c-Jun (#9165), cleaved PARP (#9532), LC3B (#3868), PERK (#5683), p-PERK (#3179), CHOP (#2895), ATF4 (#11815), IRE1α (#3294), eIF2α (#5324), p-eIF2α (#9721), XBP1s (#27901), GRP78 (#3177), cleaved caspase-3 (#9661), p-JNK (#9251), and β-actin (#9562) primary antibodies were purchased from Cell Signaling Technology (CST, MA, USA). Antibody against Ki-67 (RM-9106) was purchased from Thermo Fisher (Waltham, MA, USA). ATF6 (sc-166659 and JNK1 (sc-1648) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). p-IRE1α (ab243665) and p62 (ab56416) were purchased from Abcam (Burlingame, CA, USA). UBA1 inhibitor TAK-243 was obtained from CSNpharm (CSNpharm, Chicago, IL, USA). TAK-243 was dissolved in DMSO to create a 10 mmol/L solution, which was diluted to different concentrations in DMEM medium before use.
Construction and production of lentivirus
Short hairpin RNA (shRNA) against UBA1 or GRP78 gene and control non-targeting sequences were constructed in pHBLV-U6/GFP/Puro vector. The sequences of shRNA are listed in Table S1. Lentiviruses were generated by co-transfecting lentiviral vector and two packaging vectors in a 3:2:1 ratio in 293FT cells by use of PolyJet™ transfection reagent. The supernatant was collected and concentrated by ultracentrifugation after 48 h of incubation.
Construction of stable cell lines
For UBA1 or GRP78 silencing cell lines, LN229 cells were transduced with shUBA1 or shGRP78 and control lentivirus for 72 h and selected using 2.5 μg/mL puromycin. The stable cell lines were obtained by cultivating the survived cells. For overexpression of GRP78, the GRP78 gene was cloned into a pGV146 expression vector. The pGV146-GRP78 plasmid was transfected into U251 cells by electroporation, and the stably transfected cells were screened by G418.
CCK-8 assay
Cell viability was examined using a Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). GBM cell lines or primary GBM cells were seeded into a 96-well plate at 3,000 cells per well, and different concentrations of TAK-243 were added. After 72 h, 10 μL of CCK-8 solution were added to each well. The optical density at a wavelength of 450 nm (OD450) was measured using a microplate reader 2 h later. Background reading of the medium was subtracted from each well for result standardization.
EdU incorporation assay
The cell-light EdU Cell Proliferation Detection Kit (Ruibo Biotech, Guangzhou, China) was used for the detection of cell proliferation. The cells were cultured in a 96-well plate and treated with different concentrations of TAK-243. After 24 h, the cells were incubated with 50 μM EdU for 4 h and then fixed with 4% paraformaldehyde for 15 min and treated with 0.5% Triton X-100 for 20 min. The cells were incubated with 1× Apollo® reaction cocktail in the dark for 30 min and stained with DAPI for 20 min. After washing thrice with PBS, images of the cells were captured using a fluorescent inverted microscope.
Colony formation assay
U251 and LN229 cells were cultured in six-well plate at 500 cells/well. The experimental group was treated with TAK-243, and the control group was treated with DMSO. After 24 h, the cells were cultured with fresh DMEM medium with 10% FBS for 10–14 d. After washing with PBS, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cell colonies were observed, photographed, and counted.
The ability to create colonies was further verified using 3D culture with methylcellulose. GBM3 and GBM8 cells pre-treated with TAK-243 were mixed with complete methylcellulose medium at a density of 600 cells/mL. Cells were plated in the culture mixture in six-well plates and incubated at 37 °C in a humidified atmosphere with 5% CO2 for 13–16 days.
Cell cycle and apoptosis assay
U251 and LN229 cells were treated with indicated concentrations of TAK-243 and cultured for 24 h. Cells were centrifuged at 1,000 rpm for 5 min at 4 °C and fixed with 70% cold methanol overnight. After washing twice with PBS, the cells were stained with a PI solution containing RNase A for 30 min. The cells were examined by flow cytometry, and cell cycle distribution was analyzed using a flow cytometry software (Becton-Dickinson, Franklin Lakes, NJ, USA).
Cells treated with different concentrations of TAK-243 were harvested, washed twice with pre-chilled PBS, and resuspended in 200 μL of pre-chilled binding buffer. Approximately 5 μL Annexin V-FITC and 5 μL PI were added to the cell suspension, mixed gently, and incubated on ice for 10 min in the dark. The apoptotic status was assessed by flow cytometry (Becton-Dickinson, Franklin Lakes, NJ, USA).
Caspase-Glo 3/7 activity assay
GBM cells were seeded in a 96-well plate and treated with TAK-243 for 24 h. Caspase-Glo 3/7 enzymatic activities were measured according to the manufacturer’s protocol (Promega, Madison, WI, USA). Approximately 100 μL of Caspase-Glo 3/7 reagent were added to TAK-243-treated cells or blank negative control cells and mixed well. After 30 min, 200 μL of the solution were transferred to white-walled multiwell luminometer plates, and the luminescence of each group of samples was detected using a GloMax Luminometer.
Western blot analysis
Western blot analysis was performed as previously described [38]. Cells were seeded in a 6-well plate, and TAK-243 at different concentrations was added at the indicated time after the cells were attached. Total proteins were extracted, and protein concentration was determined. Approximately 50 μg total protein was separated using SDS-PAGE electrophoresis and transferred onto PVDF membrane. The membrane was blocked with 5% nonfat milk at room temperature for 1 h, incubated overnight with a specific primary antibody at 4 °C, incubated with secondary antibody the next day, and detected using ECL reagent.
Label-free quantitative ubiquitin-modified proteome
The quantitative ubiquitin-modified proteome was established and analyzed using Jingjie PTM Biolab (Hangzhou) Co., Ltd. (China, Hangzhou). Briefly, the U251 cells were treated with vehicle or TAK-243 (100 nM) for 6 h. Then the proteins were extracted, enzymatically digested, and enriched. The tryptic peptides were analyzed using liquid chromatography-mass spectrometry (LC-MS/MS). The resulting MS/MS data were processed using the Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against the human UniProt database concatenated with reverse decoy database.
Bioinformatics annotation analysis
Gene ontology (GO) annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). Cluster membership was visualized by a heat map using the “heatmap.2” function from the “gplots” R-package. All differentially expressed modified protein database accession or sequence were searched against the STRING database version 10.1 for protein−protein interactions. Interaction network form STRING was visualized in R package “networkD3”.
ER-Tracker Red staining
U251 and LN229 cells were seeded onto glass slides for overnight culture and treated with different concentrations of TAK-243 for 12 h. After fixing with 4% formaldehyde at 37 °C for 2 min, the cells were washed twice with an appropriate amount of Hanks’ balanced salt solution with Ca2+ and Mg2+ for 5 min each time. ER-Tracker Red staining solution (Beyotime Technology, Shanghai, China) was added and incubated with the cells for 30 min at 37 °C. The cells were washed twice with a cell culture medium, and ER morphology was assessed under a laser confocal microscope.
RNA extraction and real-time quantitative PCR
Total RNA was isolated from control or TAK-243-treated cells, and was reversely transcribed into cDNA. Quantitative PCR was performed using SYBR® Premix Ex TaqTM (Takara) in an Applied Bio-systems 7500 instrument. Expression values were corrected to the housekeeping gene β-actin. Primer sequences for quantitative PCR were shown in Supplementary Table S2.
In vitro cell viability and tumorsphere formation assay
GSC1 and GSC2 cells were seeded into a 96-well plate at 1,000 cells per well and treated with the indicated concentrations of TAK-243 or DMSO. Cell viability was assessed on days 0, 3, 6, 9, and 12 using Cell Titer-Glo luminescent cell viability kit (Promega, Madison, WI, USA). For the tumorsphere formation assay, GSC1 and GSC2 cells were seeded in 96-well plate at 1,000 cells per well. The cells were cultured in NeurobasalTM medium containing a certain concentration of TAK-243 or DMSO, and the formation of colony spheres was assessed under a microscope after 10–14 days. Neurospheres with more than 50 cells were scored, and the number of neurospheres in each well was counted.
In vitro limiting dilution assay
Cells were dissociated into single cells and then plated in 96-well plates at a density of 1, 5, 10, 20, or 40 cells per well, with ten replicates each. After 7 days, the presence of tumorspheres in each well was assessed. Extreme limiting dilution assays were analyzed using online software (http://bioinf.wehi.edu.au/software/elda/)
In vivo studies
Sixty-three male athymic BALB/c nude mice aged 5–6 weeks were purchased from Beijing Vital River Experimental Animal Technology Co. Ltd., China. All animal protocols were approved by the Ethics Committee of the Xuzhou Medical University (Jiangsu Province, China). Subcutaneous animal model and in situ GSC-derived xenograft model were used to evaluate the therapeutic effect of TAK-243 on GBM. For a subcutaneous tumor model, LN229 cells (1 × 106) were injected into the right flank of nude mice. When the tumor grew to a volume of about 50−100 mm3, the nude mice were randomly divided into three groups, namely, the control group (vehicle), 10 mg/kg TAK-243 treatment group, and 20 mg/kg TAK-243 treatment group. TAK-243 was injected intraperitoneally twice a week for 4 weeks. The size of the tumor was measured with a caliper every 2 days. The volume of the subcutaneous tumors was calculated using the following equation: Tumor volume = (Length × Width2)/2 (assuming a prolate shape).
For the intracranial tumor model, 5 × 104 GSC2 cells were injected into the right striatum of each nude mouse in situ with a small animal stereotaxic instrument [39]. After 5 days, tumor-bearing mice were randomly divided into three groups with 15 mice per group. Mice were treated with intraperitoneal injection of TAK-243 (10 mg/kg or 20 mg/kg) or vehicle twice a week for 4 weeks. After 30 days, five mice from each group were randomly selected and euthanized, and the brains were perfused and removed to observe the tumor size. The remaining ten mice in each group were used for survival analysis.
Hematoxylin-eosin (HE) staining and immunohistochemistry (IHC)
The whole mouse brains of the control group and the treatment groups were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and then sliced into sections with a thickness of 5 μm. The sections were fixed on glass slides and parched dry. HE staining was performed to observe the tumor size. Immunohistochemical test was used to detect the effect of TAK-243 treatment on the expression levels of global protein polyubiquitination, GRP78, and Ki67 in the mice.
Statistical analysis
Each experiment was independently performed at least three times. The figures show representative images of the results of repeated experiments. The experimental results were statistically analyzed using GraphPad Prism 6.0 (San Diego, CA, USA). The data are here presented as mean ± SD. Comparison between two samples was performed using an independent sample t-test. The Kaplan−Meier method was used for the survival analysis of the mice. The log-rank test was used to compare survival time between the two study groups. P < 0.05 was considered statistically significant.
Data availability
The datasets supporting the conclusions of this article are included within the article.
Change history
05 March 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41419-024-06574-0
References
Zhou Y, Wu W, Bi H, Yang D, Zhang C. Glioblastoma precision therapy: from the bench to the clinic. Cancer Lett. 2020;475:79–91.
Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature. 2017;547:355–9.
Sharifzad F, Ghavami S, Verdi J, Mardpour S, Mollapour Sisakht M, Azizi Z, et al. Glioblastoma cancer stem cell biology: potential theranostic targets. Drug Resist Updat. 2019;42:35–45.
Chua J, Nafziger E, Leung D. Evidence-based practice: temozolomide beyond glioblastoma. Curr Oncol Rep. 2019;21:30.
Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11:504–14.
Wang Y, Wang K, Jin Y, Sheng X. Endoplasmic reticulum proteostasis control and gastric cancer. Cancer Lett. 2019;449:263–71.
Obacz J, Avril T, Le Reste PJ, Urra H, Quillien V, Hetz C, et al. Endoplasmic reticulum proteostasis in glioblastoma-from molecular mechanisms to therapeutic perspectives. Sci Signal. 2017;10:eaal2323.
Corazzari M, Gagliardi M, Fimia GM, Piacentini M. Endoplasmic reticulum stress, unfolded protein response, and cancer cell fate. Front Oncol. 2017;7:78.
Yadav RK, Chae SW, Kim HR, Chae HJ. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19:75–88.
Markouli M, Strepkos D, Papavassiliou AG, Piperi C. Targeting of endoplasmic reticulum (ER) stress in gliomas. Pharmacol Res. 2020;157:104823.
Ibrahim IM, Abdelmalek DH, Elfiky AA. GRP78: a cell’s response to stress. Life Sci. 2019;226:156–63.
Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69:169–81.
Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, et al. BOK Is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–33.
Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11.
Barghout SH, Schimmer AD. E1 Enzymes as therapeutic targets in cancer. Pharmacol Rev. 2021;73:1–56.
Clague MJ, Heride C, Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–26.
Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat Med. 2018;24:186–93.
Bonacci T, Emanuele MJ. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020;67:145–58.
Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–35.
Go D, Lee J, Choi JA, Cho SN, Kim SH, Son SH, et al. Reactive oxygen species-mediated endoplasmic reticulum stress response induces apoptosis of Mycobacterium avium-infected macrophages by activating regulated IRE1-dependent decay pathway. Cell Microbiol. 2019;21:e13094.
Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: autophagy induction, inhibition, and selection. Autophagy. 2015;11:1956–77.
Liu WJ, Ye L, Huang WF, Guo LJ, Xu ZG, Wu HL, et al. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol Biol Lett. 2016;21:29.
Qi WC, Zhou XT, Wang JQ, Zhang K, Zhou YJ, Chen SP, et al. Cordyceps sinensis polysaccharide inhibits colon cancer cells growth by inducing apoptosis and autophagy flux blockage via mTOR signaling. Carbohydr Polym. 2020;237:116113.
Yang WY, Jiang CL, Xia WY, Ju HY, Jin SF, Liu SL, et al. Blocking autophagy flux promotes interferon-alpha-mediated apoptosis in head and neck squamous cell carcinoma. Cancer Lett. 2019;451:34–47.
Minata M, Audia A, Shi J, Lu S, Bernstock J, Pavlyukov MS, et al. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 2019;26:1893–905 e1897.
Obeng EA, Carlson LM, Gutman DM, Harrington WJ Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16.
Dufey E, Urra H, Hetz C. ER proteostasis addiction in cancer biology: novel concepts. Semin Cancer Biol. 2015;33:40–47.
Maurel M, McGrath EP, Mnich K, Healy S, Chevet E, Samali A. Controlling the unfolded protein response-mediated life and death decisions in cancer. Semin Cancer Biol. 2015;33:57–66.
Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015;5:586–97.
Best S, Hashiguchi T, Kittai A, Bruss N, Paiva C, Okada C, et al. Targeting ubiquitin-activating enzyme induces ER stress-mediated apoptosis in B-cell lymphoma cells. Blood Adv. 2019;3:51–62.
Barghout SH, Patel PS, Wang XM, Xu GW, Kavanagh S, Halgas O, et al. Preclinical evaluation of the selective small-molecule UBA1 inhibitor, TAK-243, in acute myeloid leukemia. Leukemia. 2019;33:37–51.
Liu YJ, Awadia S, Delaney A, Sitto M, Engelke CG, Patel H, et al. UAE1 inhibition mediates the unfolded protein response, DNA damage an caspase-dependent cell death in pancreatic cancer. Transl Oncol. 2020;13:100834.
Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32.
Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111.
Hattermann K, Fluh C, Engel D, Mehdorn HM, Synowitz M, Mentlein R, et al. Stem cell markers in glioma progression and recurrence. Int J Oncol. 2016;49:1899–910.
Hombach-Klonisch S, Mehrpour M, Shojaei S, Harlos C, Pitz M, Hamai A, et al. Glioblastoma and chemoresistance to alkylating agents: involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther. 2018;184:13–41.
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401.
Liu X, Chen X, Shi L, Shan Q, Cao Q, Yue C, et al. The third-generation EGFR inhibitor AZD9291 overcomes primary resistance by continuously blocking ERK signaling in glioblastoma. J Exp Clin Cancer Res. 2019;38:219.
Liu X, Chong Y, Tu Y, Liu N, Yue C, Qi Z, et al. CRM1/XPO1 is associated with clinical outcome in glioma and represents a therapeutic target by perturbing multiple core pathways. J Hematol Oncol. 2016;9:108.
Funding
The research was supported by the National Natural Science Foundation of China (No. 81972345, 81772658); The Natural Science Foundation of Jiangsu Province (BK20180104, BK20180271); Jiangsu Qing Lan Project for Xuejiao Liu; The 333 Project of Jiangsu Province (BRA2020301); and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX21_2684, KYCX20_2497, KYCX20_2508).
Author information
Authors and Affiliations
Contributions
XL, RY, and MN designed this study. GL and JY performed the main experimental procedures in vitro. RW and LS conducted the animal assays in vivo. XZ, WZ, XZ, YW, and HL carried out partial experiments. YS and CW performed the statistical analysis. XL and MN wrote this paper. All the authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The experimental protocol for animal studies was reviewed and approved by ethics committee of Xuzhou Medical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Stephanou
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-024-06574-0
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, G., Yu, J., Wu, R. et al. RETRACTED ARTICLE: GRP78 determines glioblastoma sensitivity to UBA1 inhibition-induced UPR signaling and cell death. Cell Death Dis 12, 733 (2021). https://doi.org/10.1038/s41419-021-04023-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-04023-w
This article is cited by
-
Emerging mechanisms of the unfolded protein response in therapeutic resistance: from chemotherapy to Immunotherapy
Cell Communication and Signaling (2024)
-
Chimeric antigen receptor T cells targeting cell surface GRP78 efficiently kill glioblastoma and cancer stem cells
Journal of Translational Medicine (2023)
-
Knockdown of ENTPD5 inhibits tumor metastasis and growth via regulating the GRP78/p-eIF-2α/CHOP pathway in serous ovarian cancer
Journal of Ovarian Research (2022)
-
The anterior gradient homologue 2 (AGR2) co-localises with the glucose-regulated protein 78 (GRP78) in cancer stem cells, and is critical for the survival and drug resistance of recurrent glioblastoma: in situ and in vitro analyses
Cancer Cell International (2022)