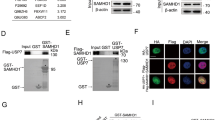
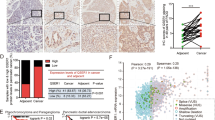
Abstract
SCML2 has been found to be highly expressed in various tumors. However, the extent to which SCML2 is involved in tumorigenesis and cancer therapy is yet to be fully understood. In this study, we aimed to investigate the relationship between SCML2 and DNA damage response (DDR). Firstly, DNA damage stabilizes SCML2 through CHK1-mediated phosphorylation at Ser570. Functionally, this increased stability of SCML2 enhances resistance to DNA damage agents in p53-positive, p53-mutant, and p53-negative cells. Notably, SCML2 promotes chemoresistance through distinct mechanisms in p53-positive and p53-negative cancer cells. SCML2 binds to the TRAF domain of USP7, and Ser441 is a critical residue for their interaction. In p53-positive cancer cells, SCML2 competes with p53 for USP7 binding and destabilizes p53, which prevents DNA damage-induced p53 overactivation and increases chemoresistance. In p53-mutant or p53-negative cancer cells, SCML2 promotes CHK1 and p21 stability by inhibiting their ubiquitination, thereby enhancing the resistance to DNA damage agents. Interestingly, we found that SCML2A primarily stabilizes CHK1, while SCML2B regulates the stability of p21. Therefore, we have identified SCML2 as a novel regulator of chemotherapy resistance and uncovered a positive feedback loop between SCML2 and CHK1 after DNA damage, which serves to promote the chemoresistance to DNA damage agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All the uncropped original western blots used in the manuscript were listed in Original Western blot images file in Supplementary materials.
References
Lecona E, Narendra V, Reinberg D. USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol Cell Biol. 2015;35:1157–68.
Lecona E, Rojas LA, Bonasio R, Johnston A, Fernandez-Capetillo O, Reinberg D. Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol. 2013;11:e1001737.
Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, et al. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. Elife. 2014;3:e02637.
Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, et al. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell. 2015;32:574–88.
Luo M, Zhou J, Leu NA, Abreu CM, Wang J, Anguera MC, et al. Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet. 2015;11:e1004954.
Adams SR, Maezawa S, Alavattam KG, Abe H, Sakashita A, Shroder M, et al. RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes. PLoS Genet. 2018;14:e1007233.
Maezawa S, Hasegawa K, Alavattam KG, Funakoshi M, Sato T, Barski A, et al. SCML2 promotes heterochromatin organization in late spermatogenesis. J Cell Sci. 2018;131:1–12.
Maezawa S, Hasegawa K, Yukawa M, Kubo N, Sakashita A, Alavattam KG, et al. Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc Nat Acad Sci USA. 2018;115:4957–62.
Grubach L, Juhl-Christensen C, Rethmeier A, Olesen LH, Aggerholm A, Hokland P, et al. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur J Haematol. 2008;81:112–22.
Qi L, Wang L, Huang J, Jiang M, Diao H, Zhou H, et al. Activated amelogenin Y-linked (AMELY) regulation and angiogenesis in human hepatocellular carcinoma by biocomputation. Oncol Lett. 2013;5:1075–9.
Shen S, Gui T, Ma C. Identification of molecular biomarkers for pancreatic cancer with mRMR shortest path method. Oncotarget. 2017;8:41432–9.
Du L, Wang L, Yang H, Duan J, Lai J, Wu W, et al. Sex comb on midleg like-2 accelerates hepatocellular carcinoma cell proliferation and metastasis by activating Wnt/beta-Catenin/EMT signaling. Yonsei Med J. 2021;62:1073–82.
Yang JJ, Huang H, Xiao MB, Jiang F, Ni WK, Ji YF, et al. Sex comb on midleg like-2 is a novel specific marker for the diagnosis of gastroenteropancreatic neuroendocrine tumors. Exp Ther Med. 2017;14:1749–55.
Fan T, Jiang G, Shi R, Yu R, Xiao X, Ke D. Construction of AP003469.4-miRNAs-mRNAs ceRNA network to reveal potential biomarkers for hepatocellular carcinoma. Am J Cancer Res. 2022;12:1484–501.
Vousden KH, Prives C. Blinded by the Light: The growing complexity of p53. Cell. 2009;137:413–31.
Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78.
Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34.
Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53.
Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1–2.
Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell cycle. 2005;4:1166–70.
Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–54.
Rodriguez J, Herrero A, Li S, Rauch N, Quintanilla A, Wynne K, et al. PHD3 regulates p53 protein stability by hydroxylating proline 359. Cell Rep. 2018;24:1316–29.
Cui D, Xiong X, Shu J, Dai X, Sun Y, Zhao Y. FBXW7 confers radiation survival by targeting p53 for degradation. Cell Rep. 2020;30:497–509.e494.
Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8.
Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Molecular cell. 2004;13:879–86.
Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–96.
Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Molecular cell. 2006;21:307–15.
Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53-Mdm2-HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262–6.
Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92.
Kategaya L, Di Lello P, Rouge L, Pastor R, Clark KR, Drummond J, et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–8.
Wang M, Zhang Y, Wang T, Zhang J, Zhou Z, Sun Y, et al. The USP7 inhibitor P5091 induces cell death in ovarian cancers with different P53 status. Cell Physiol Biochem. 2017;43:1755–66.
Song MS, Song SJ, Kim SY, Oh HJ, Lim DS. The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J. 2008;27:1863–74.
Epping MT, Meijer LA, Krijgsman O, Bos JL, Pandolfi PP, Bernards R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13:102–8.
Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36.
Tsabar M, Mock CS, Venkatachalam V, Reyes J, Karhohs KW, Oliver TG, et al. A switch in p53 dynamics marks cells that escape from DSB-induced cell cycle arrest. Cell Rep. 2020;32:107995.
Porter JR, Fisher BE, Batchelor E. p53 pulses diversify target gene expression dynamics in an mRNA half-life-dependent manner and delineate co-regulated target gene subnetworks. Cell Syst. 2016;2:272–82.
Paek AL, Liu JC, Loewer A, Forrester WC, Lahav G. Cell-to-cell variation in p53 dynamics leads to fractional killing. Cell. 2016;165:631–42.
Georgakilas AG, Martin OA, Bonner WM. p21: A two-faced genome guardian. Trends Mol Med. 2017;23:310–9.
Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ, et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol. 2016;18:777–89.
Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14.
Neizer-Ashun F, Bhattacharya R. Reality CHEK: Understanding the biology and clinical potential of CHK1. Cancer Lett. 2021;497:202–11.
Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23.
Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–43.
O’Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–27.
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6.
Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39.
Valles GJ, Bezsonova I, Woodgate R, Ashton NW. USP7 is a master regulator of genome stability. Front Cell Dev Biol. 2020;8:717.
Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–91.
Choi ES, Lee H, Sung JY, Lee CH, Jang H, Kim KT, et al. FAM188B enhances cell survival via interaction with USP7. Cell Death Dis. 2018;9:633.
Shen Y, Tu W, Liu Y, Yang X, Dong Q, Yang B, et al. TSPY1 suppresses USP7-mediated p53 function and promotes spermatogonial proliferation. Cell Death Dis. 2018;9:542.
Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J Biol Chem. 2004;279:16000–6.
Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–53.
Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, et al. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018;361:806–10.
Leung-Pineda V, Huh J, Piwnica-Worms H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–7.
Alonso-de Vega I, Martin Y, Smits VA. USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle. 2014;13:3921–6.
Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59.
Huh J, Piwnica-Worms H. CRL4(CDT2) targets CHK1 for PCNA-independent destruction. Mol Cell Biol. 2013;33:213–26.
Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70.
Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–36.
Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12.
Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88:956–65.
Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96.
Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nature Cell Biology. 2014;16:864–75.
Wang S, Zhao Y, Aguilar A, Bernard D, Yang CY. Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: progress and challenges. Cold Spring Harb Perspect Med. 2017;7:1–10.
Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–4.
Funding
QC is supported by grants from the National Key Research and Development Program of China (2018YFC1003400), the National Natural Science Foundation of China (32170698, 31770868) and the Fundamental Research Funds for the Central Universities (2042022dx0003).
Author information
Authors and Affiliations
Contributions
QC designed and supervised the study. Q-QP studied the regulation between SCML2 and CHK1. XS constructed all the cell lines and analyzed the regulation between SCML2, USP7 and p21. Q-QP and XS jointly performed the remaining experiments. D-WL performed MST assay. JG and X-QZ generated constructs and purified proteins. QC, XS, X-YZ and Q-QP wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peng, Q., Shi, X., Li, D. et al. SCML2 contributes to tumor cell resistance to DNA damage through regulating p53 and CHK1 stability. Cell Death Differ 30, 1849–1867 (2023). https://doi.org/10.1038/s41418-023-01184-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01184-3