Abstract
Elevated levels of PDLIM3 expression are frequently detected in sonic hedgehog (SHH) group of medulloblastoma (MB). However, the possible role of PDLIM3 in MB tumorigenesis is still unknown. Here, we found that PDLIM3 expression is necessary for hedgehog (Hh) pathway activation in MB cells. PDLIM3 is present in primary cilia of MB cells and fibroblasts, and such cilia localization is mediated by the PDZ domain of PDLIM3 protein. Deletion of PDLIM3 significantly compromised cilia formation and interfered the Hh signaling transduction in MB cells, suggesting that PDLIM3 promotes the Hh signaling through supporting the ciliogenesis. PDLIM3 protein physically interacts with cholesterol, a critical molecule for cilia formation and hedgehog signaling. The disruption of cilia formation and Hh signaling in PDLIM3 null MB cells or fibroblasts, was significantly rescued by treatment with exogenous cholesterol, demonstrating that PDLIM3 facilitates the ciliogenesis through cholesterol provision. Finally, deletion of PDLIM3 in MB cells significantly inhibited their proliferation and repressed tumor growth, suggesting that PDLIM3 is necessary for MB tumorigenesis. Our studies elucidate the critical functions of PDLIM3 in the ciliogenesis and Hh signaling transduction in SHH-MB cells, supporting to utilize PDLIM3 as a molecular marker for defining SHH group of MB in clinics.
Similar content being viewed by others
Introduction
Medulloblastoma (MB) is the most common malignant brain tumor in children. Human MB consists of at least four main groups: WNT, SHH, group 3 and group 4 [1]. Each group is characterized by distinct transcriptional and mutational signatures, epigenetic profiles and clinical features. The SHH group accounts for approximately 30% of human MB, which is further classified into four clinically and cytogenetically distinct subgroups including SHH α, SHH β, SHH γ and SHH δ. Infant SHH tumors are mainly distributed across SHH β and SHH γ, whereas SHH δ are primarily composed of adults [2]. Previous studies showed that SHH-MB originates from cerebellar granule neuron precursors (GNPs). Normal proliferation and differentiation of GNPs is supported by the SHH pathway in the developing cerebellum [3, 4]. However, overactivation of SHH pathway in GNPs leads to MB formation [5, 6].
SHH pathway signaling plays a crucial role in normal development of many tissues and organs [7, 8]. Aberrant activation of the SHH pathway is associated with many human malignancies such as MB and basal cell carcinoma. The key players of the Hh pathway, including SHH ligand, Patched1 (Ptch1, the antagonizing receptor), Smoothened (Smo, a G protein-coupled-like receptor), Suppressor of fused (Sufu) and the glioma-associated transcriptional factor (Gli1, Gli2 and Gli3). Activation of the SHH pathway is triggered by the binding of SHH ligand with Ptch1, resulting in the release and activation of Smo. Activated Smo transduces SHH signaling across the cytomembrane to activate Gli1/2 that involve dissociating Gli from a suppressive complex containing Sufu. The above molecular events mainly occur in primary cilia, a microtubule-based projection of the cell membrane that generally functions in vertebrates as a sensor of various extracellular signaling pathways, including the SHH pathway [9].
PDZ and LIM Domain 3 (PDLIM3) belongs to a large family of proteins containing an N-terminal PDZ domain and a C-terminal LIM domain. PDZ domains (~90 amino acids) are small protein-protein interaction domains widely conserved from yeast to human, which typically interact with the C-terminal peptide motif of partner proteins [10, 11]. Recent studies revealed that PDZ domains also bind to phospholipids and cholesterol [12, 13]. The LIM domain is defined by a cysteine-rich consensus, which is also considered as a protein-interaction module [14]. However, recognition motifs for the LIM domain have not been established. PDLIM3 is expressed at high levels in skeletal muscle and is believed to be involved in the organization of actin-filament arrays within muscle cells [15, 16]. It was previously reported that PDLIM3 binds the spectrin-like repeats of α-actinin-2 and co-localizes with α-actinin-2 at the Z lines of skeletal muscle [17]. Interestingly, PDLIM3 was found associated with SHH group MB based on transcriptomic profiling of human MB [18, 19]. However, it is still unknown whether PDLIM3 plays a role in the tumorigenesis of SHH-MB.
Here, our studies reveal that PDLIM3 is upregulated in SHH group MB in human and mice. PDLIM3 is required for SHH pathway activation in MB cells. We further show that PDLIM3 can bind with cholesterol via the PDZ domain. Moreover, PDLIM3 localizes in primary cilia and promotes ciliogenesis by tethering cholesterol in the cilium. Finally, we found that deletion of PDLIM3 significantly represses MB cell proliferation and inhibits MB growth. Our findings demonstrate the critical functions of PDLIM3 in SHH signaling and tumorigenesis of SHH group MB.
Material and methods
Animals
Ptch1 heterozygous mice (Ptch1+/− mice) and wild type mice (C57BL/6 mice) were purchased from the Institute of Model Animals at Nanjing University. Math1-Cre Mice, Ptch1fl/fl mice, Cas9 mice and CB17/SCID mice were purchased from the Jackson’s laboratory, and maintained in the Laboratory Animal Facility (LAF) at Fox Chase Cancer Center. All experiments were performed in accordance with procedures approved by the laboratory animal committee at Soochow University and Fox Chase Cancer Center.
Cell culture
NIH3T3 and 293T cells were purchased from American Type Culture Collection. Both cells were cultured in DMEM containing 10% fetal bovine serum (FBS, Gibco) and 1% Pen/Strep. All cell lines were regularly tested for mycoplasma infection (MycoAlert Mycoplasma Detection Kit, Lonza).
Primary mouse embryonic fibroblasts (MEFs) were prepared from embryos (E10) of wild type mice, Ptch1fl/fl mice or SmoM2 mice as described in our previous studies [20]. Dissociated MEFs were cultured in DMEM supplemented with 20% FBS, 1 mmol/L sodium pyruvate, 2 mmol/L L-glutamine, 1 g/L glucose, 100 U/mL penicillin/streptomycin, and 1% nonessential amino acids (all from Life Technologies, unless noted otherwise). GNPs were isolated from wild-type C57BL/6 mice, and MB cells were isolated from Ptch1+/− mice or Math1-Cre/ Ptch1fl/fl mice. The cerebellar or tumor tissue was digested with papain solution to obtain a single-cell suspension, followed by gradient centrifugation at 35% and 65% Percoll. 35–65% of the cells at the interface were suspended in Dulbecco’s PBS (DPBS) with 0.5% BSA. Cells were then suspended in NB-B27 (Neurobasal with 1 mM sodium pyruvate, 2 mM L-glutamine, B27 supplement and 1% Pen/Strep) (Invitrogen) and plated on a poly D-Lysine (PDL) coated coverslips.
Immunofluorescence and western blotting
For immunofluorescence, cells were fixed for 15 mins with 4% paraformaldehyde (PFA), permeabilized in 0.1% Triton X-100 for 10 min. Cells were then blocked for 1 hr with PBS containing 0.1% Triton X-100 and 1% BSA, and incubated with primary antibodies overnight at 4 °C, and incubated with secondary antibodies for 2 hrs at room temperature. Cells were counterstained with DAPI and mounted with Fluoromount-G before being visualized using a Nikon microscopy (Eclipse Ti).
For western blot analysis, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Solarbio) supplemented with protease inhibitors (5 mg/mL PMSF and cocktails) and then centrifuged at 12,000 × g for 15 mins at 4 °C. Total lysate containing equal amount of protein were separated by SDS-PAGE gel and subsequently transferred onto PVDF membrane. Membranes were then subjected to probe with antibodies. Uncropped western blots are provided in Supplemental Material.
For beads binding assay, cell lysates or synthesized protein (20 μg) were mixed with beads covalently coated with cholesterol (50 μl, Echelon Biosciences) and incubated overnight at 4 °C. The beads were then washed with lysis buffer, pelleted and re-suspended in 30 μl protein sample buffer and subjected to western blot analysis.
Primary antibodies used in this study include: anti-PDLIM3 (1:1000 for western blotting, NOVUS), anti-GFP (1:500 for immunofluorescent staining, Invitrogen; 1:1000 for western blotting, Clontech), anti-Arl13b (1:500, Proteintech), anti-β-tubulin (1:5000, Proteintech), anti-Ki67 (1:500, Abcam), anti-Gli1 (1:1000, CST), anti-His (1:1000, Proteintech), anti-HA (1:1000, Proteintech) and anti-NeuN (1:200, abcam).
SHH treatment and cholesterol treatment
SHH-CM were prepared from HEK293-shh cells (stably transfected with secreting form of SHH) as previously described [21]. HEK293 cells were seeded in 10 cm dishes and were grown to ~90% confluency. Culture medium was replaced with fresh DMEM medium and incubated for 24 hrs. SHH-enriched conditioned medium was then collected, filtered (pore size, 0.22 μm) and stored at −80 °C in small aliquots. For SHH treatment experiments, NIH3T3 cells were seeded in 6-well plates (106 cells/ml/well). After cell density reached ~80% confluency, NIH3T3 cells were starved for 24 hrs in the presence of 0.5% fetal bovine serum (FBS). 50% SHH-CM was added into cell culture for 24 hrs before collection for western blot or Q-PCR.
For cholesterol treatment experiments, NIH3T3 cells were plated on the coverslips in 3.5 cm plates at 4 ×105 cells per well and were grown to 60~70% confluency. Cells were starved in the presence of 0.5% FBS for 24 hrs, and then incubated with 50 μM water-soluble cholesterol (WSC, C27H46O, Sigma) in 0.5% FBS for 24 hrs. Cells were fixed with 4% paraformaldehyde for 30 mins at room temperature, and stained with freshly prepared 2 mg/mL filipin (Sigma-Aldrich) in PBS for 2 hrs at room temperature.
Cilia induction
For primary cilia studies, NIH3T3 cells or MB cells were plated on coverslips (4 ×105 cells per coverslip) and were grown to 60~70% confluency. Cells were then starved in the presence of 0.5% FBS for 24 hrs. Cilia were detected by immunofluorescence with an antibody specific for Arl13b, and ciliated cells were visualized and counted under a Nikon microscopy system (Nikon Eclipse Ti, Japan), equipped with a NIS-elements software (NIS-Elements BR 4.50). The cilia length was measured by the NIS-Elements software.
Tryptophan (Trp) fluorescence quenching assays
PDLIM3 was used at a final concentration of 2 µM in the buffer: 120 mM NaCl, 20 mM NaH2PO4/Na2HPO4, pH 7.4 and 1% DMSO, with addition of WS cholesterol at various concentrations. Data were collected with sample size of 800 µL, in triplicate. All experiments were carried out using Photon Technology International (PTI) spectrophotometer with 4 × 10 mm quartz cuvettes. The excitation wavelength was set to 290 nm with 1 nm band-width and the emission wavelength was set to 337 nm (where Trp has the highest fluorescence intensity), with 6 nm light pass-width. The sample was pre-incubated for 10 mins prior to measurement. Fluorescence was measured continuously for 60 s. The average fluorescence intensity over the cholesterol concentrations was plotted to determine the binding affinity between PDLIM3 and cholesterol.
Deletion of PDLIM3 using the CRISPR technique
To delete PDLIM3 gene in NIH3T3 cells by CRISPR, the exon 3 of PDLIM3 was selected for gRNA design. Single-guide RNA (gRNAs) for PDLIM3 genome editing, were designed using the online CRISPR Design Tool (http://crispr.mit.edu/). The candidate gRNAs were inserted into a pLKO-puro construct. NIH3T3 cells were co-infected with a lentivirus carrying gRNA and a lentivirus with Cas-9. Monoclones were sorted by flow cytometry and cultured on 96-well plates. Two weeks later, surviving cells were harvested to detect the PDLIM3 deletion by sequencing. Sequences of gRNAs for PDLIM3 deletion are as following:
PDLIM3-Cas9-1: AATATCCAAGACGCGCTCCA
PDLIM3-Cas9-2: AGCTATTGATGGCTTTGGTA
To delete PDLIM3 gene in MB cells, we crossed Math1-Cre/Ptch1fl/fl mice with Cas-9 mice, to generate Math1-Cre/Ptch1fl/fl/Cas-9 mice in which tumor cells universally express Cas-9. Tumor cells isolated from Math1-Cre/Ptch1fl/fl/Cas-9 mice, were infected with a lentivirus carrying mcherry-tagged gRNAs specific for PDLIM3 (as above), or an empty vector as a control. At 48 hrs following the infection, tumor cells were harvested to verify the deletion of PDLIM3 by western blotting. To investigate the tumorigenesis of MB cells after PDLIM3 knockout, CB17/SCID mice were intracranially transplanted with PDLIM3 knockout cells or control cells (infected with an empty vector), 5 × 105 cells/recipient mouse.
Protein synthesis and purification
PDLIM3 (1-316aa) or PDZ (1-218aa) was cloned into a pET28-MHL vector, resulting in the N-terminal His-fusion protein, which were used to transfect BL21 (DE3) cells. Transfected cells were cultured at 15 °C for 24 hrs upon induction with 0.25 mM Isopropyl-beta-D-thiogalactopyranoside (IPTG). Cell lysates were harvested and purified by affinity chromatography on Ni-nitrilotriacetate resin (Qiagen). The proteins were further purified by Superdex75 or Superdex200 gel-filtration column (GE Healthcare, NJ) at 4 °C, which was concentrated to 40~80 μM in a buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl and 1 mM DTT, and then stored at −80 °C. Protein concentrations were determined using theoretical molar extinction coefficients at 280 nm.
Lentiviral production and PDLIM3 knockdown
HEK293 cells were seeded in 10 cm dishes and transfected with pMD2.G, psPAX2 (Addgene) to produce lentiviruses carrying PDLIM3 shRNAs and scrambled shRNA (Genechem, GIEL0133053), pLV-AcGFP-PDLIM3, pLUT Gli1-HA or pLV-gRNA-mcherry plasmids, in accordance with the standard protocol. After 48 hrs in culture, lentiviruses-enriched media was collected, filtered (pore size, 0.22 μm) and stored at −80 °C.
Sequences for shRNAs are as following:
Scrambled shRNA: TTCTCCGAACGTGTCACGT
PDLIM3 shRNA1: ACTGTAACCTCAACCTCAA
PDLIM3 shRNA3: GGCTCTATTCAACTAGCAA
q-PCR
Total RNA was isolated from cells using the TRizol reagent (Thermo Fisher Scientific), and cDNA was synthesized using oligo (dT) and Superscript II reverse transcriptase (Takara). Q-PCR reactions were performed in triplicate using SYBR qPCR Master Mix and ABI 7500 Real-Time PCR Detection System. Primers specific to GAPDH, PDLIM3, GLI1, Sfrp1and Ptch2 are available upon request.
Statistical analysis
All graphs in the paper were prepared using GraphPad Prism 8. Data (from at least 3 biological replicates) were analyzed for statistical significance using Student’s t test unless otherwise stated. Difference in the survival of CB17/SCID mice after cell transplantation was assessed using the Kaplan-Meier survival analysis and the mantel-Cox log-rank test was used to assess the difference between the survival of two cohorts of mice. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
Results
PDLIM3 expression correlates with SHH pathway activation in MB cells and GNPs
To assess PDLIM3 mRNA expression in human MB, we interrogated the microarray expression data of a cohort of 223 human MB samples (Pfister-223-MAS5.0-u133p2) (Fig. 1A), and a cohort of 763 human patient MB samples (GSE85217) (Fig. 1B, C). As shown in Fig. 1A, B, levels of PDLIM3 are significantly upregulated in SHH-MB, compared with the other three groups including WNT, group 3 and group 4 (p < 0.001). We further examined PDLIM3 expression levels in subgroups of Shh-MB (223 samples). High expression levels of PDLIM3 were observed across all four subgroups of SHH-MB (Fig. 1C). However, PDLIM3 expression in SHH-MB appears to be subgroup-dependent, with the highest expression in SHH-δ, and lowest expression in SHH-γ (p < 0.05). Consistent with previous reports [18, 19], these data suggest that PDLIM3 expression is particularly associated with SHH group MB. However, no significant correlation was found between the expression levels of PDLIM3 and the survival of SHH-MB patients (data not shown).
Box/dot plots showing the expression levels of PDLIM3 in WNT, SHH, Group 3 and Group 4 subgroup of human MB (A Pfister-223-MAS5.0-u133p2; B, C GSE85217), which were analyzed using the on-line tool from R2: Genomics Analysis and Visualization Platform (http://r2platform.com). The black dots represent the PDLIM3 expression levels of each tumor sample. A, B PDLIM3 expression levels across 4 different groups of human MB. C PDLIM3 expression levels across 4 subtypes of SHH-MB. GLI1 and PDLIM3 proteins (D) and mRNAs (E) in MB cells from Math1-Cre/Ptch1fl/fl mice at 8 weeks of age, and cerebellar GNPs from wild type mice at postnatal day 6 (P6), examined by western blotting and q-PCR, respectively. β-tubulin protein expression was used as a loading control. Gli1 and PDLIM3 protein and mRNA expressions in cerebellar GNPs isolated from wild type mice at designated ages, examined by western blotting (F), and q-PCR (G). MB cells and GNPs were freshly isolated from rom Math1-Cre/Ptch1fl/fl mice at 8 weeks of age (H and J), and from wild type mice at P6 (I and K), respectively. H, I PDLIM3 mRNA expression in differentiated cells (TAG1+) and their undifferentiated counterparts (TAG1−), examined by q-PCR. Time course of PDLIM3 mRNA expression in MB cells (J) and GNPs (K) after being cultured in vitro.
We next examined mRNA and protein expression of PDLIM3 in mouse MB cells and their normal counterparts, cerebellar GNPs. As expected, levels of Gli1 protein, a target protein of SHH pathway, were significantly increased in MB cells compared with GNPs (Fig. 1D), confirming the overactivation of SHH pathway in MB cells. Elevated expression levels of PDLIM3 protein and mRNA were detected in MB cells compared with GNPs (Fig. 1D, E). We then harvested GNPs in developing cerebella at various stages from postnatal day 1 (P1) to day 13 (P13), and examined Gli1 and PDLIM3 proteins by Western blotting. As shown in Fig. 1F, Gli1 protein levels in GNPs progressively increased from P1 to P7. After this, the level of Gli1 protein gradually declined until P13. These expression patterns are consistent with the temporal activation of the SHH pathway observed in developing cerebella [3, 4]. In parallel with Gli1 expression, PDLIM3 protein levels increased in the GNPs from P1 to P7, and then declined until P13. Similar patterns of PDLIM3 mRNA expression were detected in GNPs from developing cerebella (Fig. 1G). These data indicate that PDLIM3 expression correlates with SHH pathway activation in MB cells and GNPs from developing cerebella.
We previously reported that GNPs and MB cells can undergo terminal differentiation in vivo and in vitro [22, 23]. Differentiated GNPs and MB cells express TAG1 protein, in which SHH pathway activation is suppressed, as in our previous studies [22, 23] and in supplementary fig. S1. Reduced levels of PDLIM3 were detected in differentiated MB cells and GNPs, purified from tumor tissues and cerebella (Fig. 1H, I). PDLIM3 mRNA expression also decreased significantly as they differentiated in vitro (Fig. 1J, K). These data suggest that the expression of PDLIM3 mRNA and protein is downregulated in differentiated MB cells and GNPs as SHH pathway is repressed in these cells.
Collectively, these data confirm that PDLIM3 expression is associated with SHH pathway activation in MB cells or GNPs, suggesting that PDLIM3 may be involved in the regulation of SHH signaling transduction.
PDLIM3 is required for SHH pathway activation
To investigate the possible role of PDLIM3 in regulating SHH pathway activation, we utilized NIH3T3 cells that are commonly used to study SHH signaling transduction. NIH3T3 cells were cultured for 24 hrs in the absence or presence of SHH-containing culture medium (SHH-CM). Comparable levels of PDLIM3 mRNA expression were detected under both conditions (Fig. 2A), suggesting that PDLIM3 is not a direct target gene of SHH pathway. To examine the function of PDLIM3 in SHH signaling transduction, we knocked out PDLIM3 in NIH3T3 cells using the CRISPR technique (Fig. 2B). No cell death or apoptosis was observed in NIH3T3 cells after PDLIM3 deletion (data not shown). Wild type NIH3T3 cells or PDLIM3-null NIH3T3 cells were cultured for 24 h in the absence or presence of SHH-CM. As shown in Fig. 2C, basal levels of Gli1 protein expression were repressed in NIH3T3 cells after PDLIM3 deletion. As expected, Gli1 protein expression was elevated in wild type NIH3T3 cells following treatment with SHH-CM. However, SHH-CM failed to increase Gli1 protein levels in PDLIM3-null NIH3T3 cells. These data suggest that PDLIM3 deletion in NIH3T3 cells compromises SHH pathway activation. We further examined mRNA expression of Hh pathway target genes, including GLI1, PTCH2 and SFRP1, in NIH3T3 cells. As expected, upregulation of GLI1, PTCH2 or SFRP1 was detected in NIH3T3 cells cultured in SHH-CM, compared with control CM. However, no significant alterations in the expression of SHH pathway target genes were observed in PDLIM3-null cells following SHH-CM treatment (Fig. 2D–F). These results demonstrate that PDLIM3 is necessary for SHH pathway activation in NIH3T3 cells.
A mRNA expression of Gli1 and PDLIM3 in NIH3T3 cells after treatment with SHH-CM for 24 hrs, examined by q-PCR. B, C PDLIM3 protein in NIH3T3 cells (control) or in two lines of NIH3T3 cells after PDLIM3 knockout by CRISPR (PDLIM3 null cells, KO-1 and KO-2), examined by western blotting (B). Gli1 and PDLIM3 proteins in NIH3T3 cells (control, KO-1 and KO-2) in the presence/absence of SHH-CM, examined by western blotting (C). mRNA expression of SHH pathway genes including Gli1 (D), Ptch2 (E) and Sfrp1 (F) in control NIH3T3 cells and PDLIM3 null cells (KO-1 and KO-2), examined by q-PCR. PDLIM3 and Gli1 proteins in NIH3T3 cells infected with a construct encoding GFP-PDLIM3 or GFP alone as a control, examined by western blotting (G), which were quantified in H.
We then overexpressed PDLIM3 in NIH3T3 cells by transfection with a vector encoding GFP-PDLIM3 or an empty GFP vector as a control. No significant difference in the level of GLI1 protein was observed between PDLIM3-overexpressing NIH3T3 cells and control cells without SHH treatment (Fig. 2G). Upon SHH treatment, levels of GLI1 proteins were significantly elevated in NIH3T3 cells transfected with PDLIM3, compared with control cells (Fig. 2G, H). These data suggest that forced expression of PDLIM3 augments SHH pathway activation in NIH3T3 cells.
In summary, all the above data demonstrate that PDLIM3 is required for SHH pathway activation and PDLIM3 enhances SHH signaling transduction in NIH3T3 cells.
PDLIM3 regulates SHH signaling transduction above the level of Gli1
We next sought to define which component of SHH pathway is regulated by PDLIM3. For this purpose, we examined whether PDLIM3 deficiency affects the SHH signaling in mouse embryonic fibroblasts (MEFs) after activating SHH pathway at different levels by means of SHH treatment, PTCH1 deletion, SMO activation, SUFU deletion or GLI1 overexpression (Fig. 3A). We prepared MEFs from wild type mice, which were then transfected with a construct carrying shRNAs specific for PDLIM3, or a scrambled shRNA (Fig. 3B). Transfected MEFs were cultured in SHH-CM for 24 h. Consistent with our findings in Fig. 2C–F, the expression of GLI1 and PTCH2 mRNAs was significantly downregulated in PDLIM3-deficient MEFs, compared with MEFs transduced with scrambled shRNA (Fig. 3C), suggesting that PDLIM3 knockdown inhibits SHH pathway activation in MEFs induced by SHH treatment. We then prepared MEFs from mice carrying conditional alleles of Ptch1 (Ptch1fl/fl mice) [24], or from mice carrying a conditional allele of constitutively activated Smo (SmoM2 mice) [25], and generated Ptch1−/− MEFs and SmoM2 MEFs by infection with a viral vector encoding Cre recombinase, as in our previous studies [20]. Ptch1−/− MEFs and SmoM2 MEFs were then transfected with PDLIM3 shRNA or scrambled shRNA. As shown in Fig. 3D, E, the expression levels of GLI1 and PTCH2 were significantly depressed in Ptch1−/− MEFs or SmoM2 MEFs after PDLIM3 knockdown. Repressed SHH signaling by PDLIM3 knockdown was also observed in SUFU−/− MEFs (Fig. 3F). These data suggest that PDLIM3 deficiency compromises the SHH pathway activation induced by Ptch1 deletion, Smo activation or Sufu deletion.
A A schematic diagram of SHH pathway. In the cilia, SHH ligand binds to its negative receptor Ptch1, releasing and activating Smo, which consequently activates Gli1. The activation of Gli1 is normally repressed by Sufu. Activated Gli1 then translocates to the cell nucleus to induce the transcription of hedgehog pathway target genes such as Gli1, Gli2 and Ptch2. The functions of PDLIM3 were examined in MEFs after activating hedgehog pathway at different levels (in red boxes). B PDLIM3 protein expression in MEFs cells from wild type mice, after infection with shRNAs specific for PDLIM3 (shPD#1 or shPD#3) or scrambled shRNA (shCtrl), examined by western blotting. MEFs were prepared from wild type mice (C and G), Ptch1fl/fl mice (D), SmoM2 mice (E) and Sufu mutant mice (F). mRNA expression of Gli1 and Ptch2 in MEFs in the presence/absence of SHH-CM (C), following infection with Cre recombinase or control infection (D, E) or in Sufu mutant MEFs, examined by q-PCR. Wild type MEFs were infected with a lentivius carrying Gli1 expressing vector, or an empty vector as a control. mRNA expression of Gli2 and Ptch2 in MEFs after forced expression of Gli1 or an empty vector, examined by q-PCR (G).
Finally, we transfected wild type MEFs with a construct encoding Gli1, which were then transfected with PDLIM3 shRNAs or scrambled shRNA as above. Comparable levels of GLI1 and PTCH2 mRNA expression were found in Gli1-overexpressed MEFs transduced with PDLIM3 shRNAs or scrambled shRNA (Fig. 3G), suggesting that forced expression of Gli1 overrides the regulation of PDLIM3 on SHH pathway in MEFs.
Together these observations suggest that PDLIM3 affects SHH signaling at a level upstream from Gli1. Ptch1, Smo and Sufu regulate hedgehog signaling mainly in primary cilia. PDLIM3 deficiency counteracts the activating effects of Ptch1 deletion, Smo activation or Sufu deletion, on Hh pathway, implying that PDLIM3 may influence ciliatransduced formation and/or stability.
PDLIM3 is localized to the cilia of NIH3T3 cells and MB cells
We next examined the intracellular distribution of PDLIM3 by introducing a construct encoding a GFP-fused PDLIM3 or an empty GFP vector into NIH3T3 cells. As shown in Fig. 4A, GFP fluorescence was distributed throughout the cytoplasm and nuclei of NIH3T3 cells after transfection with GFP alone. However, the GFP signal was enriched in stress fibers (phalloidin+) of NIH3T3 cells transfected with the PDLIM3-GFP construct (Supplementary Fig. S2), in agreement with previous studies revealing the cytoskeletal localization of PDLIM3 [26, 27]. Importantly, PDLIM3-GFP protein was also detected in the cilia (Arl13b+) of NIH3T3 cells (Fig. 4A, B), whereas no ciliary GFP was found in NIH3T3 cells transfected with GFP alone (Fig. 4A, B). Similarly, GFP signal was found in the cilia of MB cells after transfection with the PDLIM3-GFP construct, but no ciliary GFP was present in MB cells transfected with the GFP alone (Fig. 4C, D). These data reveal the cilia localization of PDLIM3 in NIH3T3 cells and MB cells.
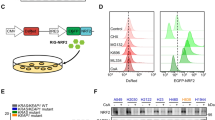
A, B Cilia of NIH3T3 cells transduced with GFP (control) or PDLIM3-GFP, were examined by immunofluorescence (ARL13B in red). Boxed regions were magnified at the right. The percentage of cells with GFP + cilia in ciliated cells was quantified in B. Scale bars: 5 μm. C, D Cilia of MB cells from Math1-Cre/Ptch1fl/fl mice, transduced with GFP (control) or PDLIM3-GFP, were detected by immunofluorescence (ARL13B in red). Boxed regions were magnified at the right. The percentage of cells with GFP + cilia in ciliated cells was quantified in D. Scale bars: 5 μm. E, F A schematic diagram showing generated constructs encoding GFP-tagged full length of PDLIM3, as well as GFP-tagged PDZ domain, ZM domain or LIM domain (E). GFP-tagged protein expression in NIH3T3 cells transduced with the above constructs were examined by western blotting using an antibody against GFP (F). G Cilia of NIH3T3 cells transduced with GFP or designated GFP-tagged constructs, were examined by immunofluorescene (ARL13B in red). Boxed regions were magnified at the below. Scale bars: 2 μm.
PDLIM3 contains at least three conserved domains: a PDZ domain, a ZM domain and a LIM domain [27, 28]. To determine which domain of PDLIM3 is responsible for its cilia localization, we generated constructs encoding a GFP fusion protein with the full-length construct, PDZ domain, ZM domain or LIM domain of PDLIM3 (Fig. 4E, F), which were then introduced into NIH3T3 cells by transfection. Based on the GFP signal, both the full-length and PDZ domain of PDLIM3 were found in the cilia of transfected NIH3T3 cells (Fig. 4G). However, the isolated ZM or LIM domains were absent in the cilia (Fig. 4G). These data indicate that the PDZ domain predominantly mediates the cilia localization of PDLIM3 in NIH3T3 cells.
PDLIM3 deletion impairs cilia formation in NIH3T3 cells
To investigate possible functions of PDLIM3 in cilia, we examined the cilia formation in NIH3T3 cells after deletion of PDLIM3 by CRISPR (Fig. 5A). As shown in Fig. 5A, cilia were readily detected in the control NIH3T3 cells. However, the percentage of ciliated cells was significantly decreased after PDLIM3 deletion (Fig. 5B). Moreover, the cilia in PDLIM3-null cells were significantly shorter than in control cells (Fig. 5C, D). These data suggest that PDLIM3 deletion interferes with cilia formation in NIH3T3 cells.
A, B Cilia of control NIH3T3 cells (WT) and PDLIM3 null cells were detected by Immunofluorescence (ARL13B in red). Cells were counterstained with DAPI (A). The percentage of ciliated cells in total cells was quantified in B. Scale bars: 10 μm. C, D Ciliar length was shown in 3 randomly selected control NIH3T3 cells (WT) and PDLIM3 null cells after immunofluorescence of ARL13B in red and counterstained with DAPI (C). The length of cilia from control NIH3T3 cells (WT, n = 67) and PDLIM3 null cells (KO, n = 33) was quantified in D. Scale bars: 5 μm. E–H PDLIM3 null NIH3T3 cells were transduced with a GFP-tagged plasmid carrying an empty vector (GFP) or full-length PDLIM3 (PDLIM3), PDZ domain only (PDZ) or full-length PDLIM3 lacking PDZ domain (FL▵PDZ). Cilia of NIH3T3 cells were detected by immunofluorescence ((ARL13B in red, E). The percentage of ciliated cells in transduced cells (GFP+), and the length of cilia in transduced cells were quantified in F and G (GFP, n = 53; PDLIM3, n = 70, PDZ, n = 42, and FL▵PDZ, n = 37), respectively. GLI1 protein in PDLIM3 null NIH3T3 cells transduced with GFP, PDZ or FL▵PDZ, in the presence/absence of SHH-CM, was examined by western blotting (H). Scale bars: 10 μm.
We next restored PDLIM3 expression in PDLIM3-null cells by transfection with a construct of full-length PDLIM3, PDZ domain only, a mutant form of PDLIM3 lacking the PDZ domain (FL▵PDZ), or an empty GFP construct as a control (Fig. 5E). As expected, the percentage of ciliated cells was significantly increased in PDLIM3-null cells upon transfection with the full-length PDLIM3 or the PDZ domain compared with the control transfection (Fig. 5F). Moreover, the cilia were significantly elongated in PDLIM3-null cells after restoration of the full-length PDLIM3 expression or the PDZ domain of PDLIM3 (Fig. 5G). However, no significant alterations in the number of cilia or cilia length were observed in PDLIM3-null cells transduced with the mutant PDLIM3 lacking the PDZ domain (Fig. 5E–G). These data confirm that compromised cilia formation in PDLIM3-null cells is indeed due to lack of PDLIM3, and the critical function of PDLIM3 in cilia formation is predominately mediated by the PDZ domain. In addition, we examined GLI1 protein in PDLIM3-null NIH3T3 cells transduced with the PDZ domain, FL▵PDZ or an empty GFP vector, in the presence/absence of SHH-CM. The GLI1 protein level was significantly increased in PDLIM3-null cells after transfection with the PDZ domain, but not with the GFP vector or the FL▵PDZ construct (Fig. 5H). These data confirm that the PDZ domain of PDLIM3 is critical for SHH signaling transduction.
PDLIM3 mediates ciliary delivery of cholesterol in NIH3T3 cells and MB cells
It was previously reported that many PDZ domain-containing scaffold proteins can modulate cell signaling through interaction with cholesterol [13]. Moreover, cholesterol is essential for hedgehog signaling transduction [29,30,31,32]. In order to test whether PDLIM3 can interact with cholesterol as well, we performed pull-down experiments using beads covalently coupled with cholesterol. Cholesterol beads were incubated with lysates from NIH3T3 cells transduced with GFP or GFP fused to the full length (GFP-PDLIM3), PDZ domain (GFP-PDZ) or LIM domain (GFP-LIM) of PDLIM3 (Fig. 6A). As shown in Fig. 6B, both the full-length PDLIM3 protein and the PDZ domain were pulled down by the cholesterol beads. However, we found no evidence for cholesterol binding in the case of lysates of NIH3T3 cells transfected with GFP alone or LIM domain of the PDLIM3. These data suggest that PDLIM3 can bind cholesterol, and that such interaction is mediated by the PDZ domain. To further test the physical interaction between cholesterol and PDLIM3 or its PDZ domain of PDLIM3, we expressed full-length PDLIM3 or the PDZ domain in E. coli (supplementary fig. S3). Harvested full-length PDLIM3 and the PDZ domain were captured by cholesterol beads, but not by control beads (Fig. 6C, D). We next performed cholesterol competition experiments by incubating PDLIM3 or the PDZ domain with cholesterol beads in the presence of water-soluble (WS) cholesterol. As shown in Fig. 6E, F, the addition of cholesterol significantly reduced the amount of PDLIM3 or PDZ domain captured by the cholesterol beads. These data demonstrated that PDLIM3 can directly bind with cholesterol through the PDZ domain. In an effort to obtain more quantitative information on binding affinity and stoichiometry, we monitored the changes in intrinsic fluorescence of PDLIM3 and its isolated PDZ domain upon addition of WS cholesterol. As shown in Fig. 6G, titration of cholesterol against a 5 µM solution of full-length PDLIM3 resulted in a sharp decrease in fluorescence at low cholesterol concentrations followed by a plateau at concentrations above 5 µM. This behavior is fully consistent with a 1:1 binding equilibrium with dissociation constant, Kd « 5 µM, as documented by the quality of the non-linear least-squares fit of this model (solid line in Fig. 6G). When we repeated this experiment for the isolated PDZ domain, we observed a similar decrease in tryptophan fluorescence. This observation confirms that cholesterol binds to a specific site on PDZ, giving rise to a significant decrease in fluorescence emission of tryptophan, either due to a direct contact or a secondary conformational change resulting in enhanced fluorescence quenching.
A, B Lysate of NIH3T3 cells transduced with the PDLIM3-GFP, PDZ-GFP or LIM-GFP construct, were used for cholesterol binding assays using beads coated with cholesterol. Input (A) and captured proteins (B) were examined by western blotting using an antibody against GFP. C–F Synthesized proteins of full-length PDLIM3 (C) or PDZ domain (D) were used for cholesterol binding assays using beads coated with cholesterol or control beads (C, D). In E and F, synthesized proteins were incubated with cholesterol-coated beads in the absence/presence of 50 μM WS cholesterol. Captured proteins were examined by western blotting using anti-His antibody. G Tryptophan (Trp) fluorescence quenching assay to examine the direct interaction between PDLIM3 and cholesterol. Trp fluorescence levels of full length PDLIM3 (left image) or PDZ domain (right image) in the presence of WS cholesterol. H–J Immunocytochemistry analyses of primary cilia in NIH3T3 cells transfected with GFP control, PDLIM3 knockout (KO) cells with or without PDLIM3 overexpression, or PDLIM3 KO cells treated with WS cholesterol. Primary cilia were immunostained with the ARL13B antibody (red), and cholesterol was labeled with filipin (blue). Boxed regions in left images are magnified at the right. (KO, n = 26; KO/Cholesterol, n = 34). Scale bars: 3 μm. Percentage of ciliated cells (I, n = 200) and the length of cilia (J; KO cells: n = 26; KO cells with cholesterol treatment: n = 34) in PDLIM3-KO cells with/without cholesterol treatment.
Having observed the physical interaction between cholesterol and PDLIM3, we next tested whether the disrupted ciliogenesis in PDLIM3-null cells is due to lack of cholesterol in the cilia. As shown in Fig. 6H, cholesterol was readily detected in the cilium of NIH3T3 cells by the filipin staining, which is consistent with previous findings that cholesterol is enriched in primary cilia [33, 34]. However, in PDLIM3-null cells, the intensity of filipin signal was markedly reduced in the cilia (Fig. 6H). Moreover, ciliary cholesterol was restored in the PDLIM3-null cells by forced expression of PDLIM3 (Fig. 6H). These data suggest that the presence of ciliary cholesterol relies on PDLIM3 expression in NIH3T3 cells. We then treated PDLIM3 null NIH3T3 cells with WS cholesterol. As expected, cholesterol was found in the cilia of PDLIM3-null cells in the presence of WS cholesterol. More importantly, the percentage of ciliated cells was significantly increased (Fig. 6I), and cilia length was increased in PDLIM3 null NIH3T3 cells after treatment with WS cholesterol (Fig. 6J), suggesting that WS cholesterol treatment rescued the deficiency of cilia formation in PDLIM3 null NIH3T3 cells. These data suggest that PDLIM3 may support ciliogenesis by recruiting cholesterol to cilia.
PDLIM3 is necessary for MB cells proliferation and tumor growth
To test the possible role of PDLIM3 in MB progression, we decided to examine the growth of MB cells after deletion of PDLIM3. For this purpose, we crossed conditional Ptch1 knockout mice (Math1-Cre/Ptch1fl/fl mice) with Cas9 mice that constitutively express CRISPR associated protein (Cas9) endonuclease [35]. MB cells isolated from Math1-Cre/Ptch1fl/fl /Cas9 mice, were infected with a lentivirus carrying mCherry-tagged guide RNA specific for PDLIM3, or an empty mCherry vector (Fig. 7A). The percentage of proliferative cells (Ki67+) was significantly reduced in MB cells infected with PDLIM3 guide RNA, compared to infection with control virus (Fig. 7B, C). These data suggest that PDLIM3 deletion in MB cells represses their proliferation. Expression levels of GLI1 and PTCH2 mRNAs were significantly reduced in MB cells infected with the PDLIM3 gRNA, compared with control virus (Fig. 7D), suggesting that PDLIM3 deletion inhibited SHH signaling in MB cells. We then intracranially transplanted MB cells infected with virus containing mCherry-tagged PDLIM3 gRNA or an empty vector, into CB17/SCID mice as in our previous studies [20]. At 2 weeks after the transplantation, majority of control tumor cells (tranduced with the empty vector) were found positive for Ki67 (Fig. 7E), suggesting that these cells were extensively proliferative. However, almost no Ki67+ cells were found in tumor cells transduced with PDLIM3 gRNA (Fig. 7E), and most of those PDLIM3 null tumor cells expressed NeuN, suggesting that PDLIM3 null tumor cells have differentiated after the transplantation. No significant alterations in the apoptosis were detected between PDLIM3 null tumor cells and control tumor cells (data not shown). GLI1 mRNA expression levels were significantly reduced in recipient cerebella transplanted with PDLIM3 null cells, compared with those in the cerebella transplanted with control tumor cells (Fig. 7F). These data further confirm that PDLIM3 deletion represses MB cell proliferation. Finally, tumor cells infected with the empty virus developed tumors in recipient mice with 100% penetrance (Fig. 7G). However, the tumor latency of PDLIM3 null MB cells was significantly prolonged (p < 0.01). These data indicate that PDLIM3 deletion significantly inhibits tumor growth. The above results demonstrate the important functions of PDLIM3 in the proliferation and tumorigenicity of MB cells.
A–D MB cells were infected by control virus (with an empty mCherry vector) or a virus carrying PDLIM3 gRNA for 48 h. Western blot analysis of PDLIM3 protein in infected MB cells (A); and (B) Immunofluorescence analyses of cell proliferation (Ki67+) in infected MB cells (mCherry+). Scale bars: 20 μm. The percentage of proliferative cells in infected MB cells was quantified in C. mRNA expressions of Gli1 and Ptch2 in infected MB cells were examined by q-PCR (D). E–G Cerebellar sections were prepared from recipient mice after two weeks following the transplantation (E). Injected tumor cells were identified based on the mCherry signal (circled by dotted lines). DAPI was used to counterstain cell nuclei. Cerebellar sections were immunostained for Ki67 and NeuN. Magnified view of tumor cells are shown in the right panels. Scale bars: 40 μm. Gli1 mRNA expression in the recipient cerebella was examined by q-PCR (F). Survival curves of CB17/SCID mice transplanted with tumor cells virally transduced with PDLIM3-gRNA or an empty vector (control) (G). Median survival: control, 50 days; PDLIM3-gRNA, undefined.
Discussion
Previous studies of PDLIM3 have been limited to muscle cells. PDLIM3 knockout in mice causes ventricular chamber dilation and dysfunction during embryonic development, which was due to destabilization of actin anchorage sites in cardiac muscle [36]. Although PDLIM3 is considered a diagnostic marker for SHH-group MB, its functions in the tumorigenesis of MB are not well understood. Here we have thoroughly analyzed the PDLIM3 expression during the development of cerebellar GNPs (the putative origin for SHH-MB) and the differentiation of MB cells, and found that PDLIM3 expression in GNPs and MB cells closely correlates with the levels of SHH pathway activation in these cells. These observations suggest that PDLIM3 plays an important role in regulating SHH signaling transduction. Deletion of PDLIM3 significantly repressed SHH signaling and inhibited the proliferation of MB cells, further confirming that PDLIM3 is required for SHH signaling transduction and proliferation in MB cells. These studies demonstrate the critical functions of PDLIM3 in the tumorigenesis of SHH-MB, further supporting the use of PDLIM3 as a biomarker for subgrouping SHH-MB in the clinic.
PDLIM3 deficiency compromised Hh pathway activation in MEFs upon SHH treatment, Ptch1 deletion, Smo activation or Sufu deletion, but failed to interfere with SHH signaling in MEFs after forced expression of Gli1. These results suggest that PDLIM3 impinges on hedgehog signaling transduction at a level above the Gli1, presumably during the formation of cilia, where Ptch1, Smo or Sufu regulates hedgehog signaling. Indeed, deletion of PDLIM3 by CRISPR significantly inhibited cilia formation and shorten the cilia length in NIH3T3 cells and MB cells, which was rescued by the restoration of PDLIM3 expression. These findings reveal that PDLIM3 is required for cilia formation. It is well established that cholesterol is essential for the formation and stability of cilia [31, 33, 37]. Using beads covalently coated with cholesterol, we further demonstrate that both endogenous and synthesized PDLIM3 can bind cholesterol. Moreover, exogenous cholesterol significantly enhanced cilia formation and elongation in PDLIM3 null cells. These data suggest that PDLIM3 promotes ciliogenesis through cholesterol delivery. We cannot completely exclude the possibility that PDLIM3 may affect the ciliogenesis and/or cilia functions through interaction with other cilia associated proteins, such as intraflagellar transport proteins and kinesin superfamily proteins [38, 39].
It is well established that cholesterol is indispensable for hedgehog signaling transduction [29, 30, 32, 40, 41]. Both Smo and Ptch proteins are found to physically interact with cholesterol, which regulate the activities of these two proteins through conformational changes. However, it is still unknown how the highly hydrophobic cholesterol can access Ptch1 or Smo to regulate the hedgehog pathway activation in cilia. It was previously postulated that an unidentified lipid transfer protein may deliver the cholesterol to Smo [41]. Our studies suggest that PDLIM3 may serve this role, based on its cilia localization and ability to bind cholesterol through its PDZ domain. In agreement with our findings, many PDZ domain-containing proteins including NHERF1/EBP50, 3 syntrophin isoforms (α, β2 and γ2), tamalin, PDZK2, PSD95 and SAP97, were found to interact with cholesterol and have cholesterol binding sites [13]. Interestingly, Chong et al. reported that Dlg5 (encoded by Discs large homolog 5) can physically interact with Smo through the PDZ domain [42]. These reports support the hypothesis that PDLIM3 may regulate the activity of Smo and Ptch1 by cholesterol delivery.
Disrupted ciliogenesis in PDLIM3 null cells was rescued by treatment with WS cholesterol, suggesting that impaired cilia formation in these cells is predominantly due to lack of ciliary cholesterol. These findings are consistent with recent studies that disruption in cilia transportation of cholesterol resulted in impaired ciliogenesis in hereditary Zellweger syndrome [34]. In addition, pharmacological cholesterol depletion or disruption of cholesterol metabolism disturbs the formation of primary cilia [37, 43]. Although cholesterol is enriched in the cilia, the source of cholesterol in cilia is still not clear. In our studies, PDLIM3 was found to reside both in cilia and cytoskeletal fibers that are responsible for transferring cholesterol from cytoplasm to cilia [34, 44]. Therefore, it is likely that PDLIM3 is involved in the cilia transportation of cholesterol from the cytoplasm into cilia. However, recent studies revealed that membrane cholesterol is the main source of ciliary cholesterol [31, 33]. It remains possible that PDLIM3 tethers membrane cholesterol to facilitate cilia formation and regulate Hh signaling. Future studies are warranted to explore these possibilities.
The critical role of SHH signaling in MB tumorigenesis makes this pathway an attractive therapeutic target for MB treatment. Currently most hedgehog pathway inhibitors such as vismodegib and sonidegib target Smo, appear to be ineffective in MB cells with mutations downstream of Smo [45]. Moreover, these Smo antagonists often cause drug resistance in MB cells due to the conformational changes of Smo [46,47,48]. Therefore, alternative targets are still needed to inhibit the SHH pathway in MB cells. Here our studies revealed that PDLIM3 is critical for SHH signal transduction in MB cells, and deletion of PDLIM3 significantly represses tumor growth in vivo. Therefore, PDLIM3 is a promising therapeutic target for MB treatment. Because PDLIM3 deficiency represses Hh signaling by interfering with cilia formation, approaches for targeting PDLIM3 protein or inhibiting PDLIM3 expression could provide a valuable tool to overcome anti-Smo resistance (intrinsic and acquired) of MB cells.
Data availability
All constructs generated in our studies are available to the scientific community upon request.
References
Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72.
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754 e736.
Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–14.
Dahmane N. Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100.
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–34.
Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–45.
Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87.
Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–72.
Bangs F, Anderson KV. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9:a028175.
Liu X, Fuentes EJ. Emerging themes in PDZ domain signaling: structure, function, and inhibition. Int Rev Cell Mol Biol. 2019;343:129–218.
Ponting CP, Phillips C, Davies KE, Blake DJ. PDZ domains: targeting signalling molecules to sub-membranous sites. Bioessays. 1997;19:469–79.
Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, et al. Syndecan recycling [corrected] is controlled by syntenin-PIP2 interaction and Arf6. Dev Cell. 2005;9:377–88.
Sheng R, Chen Y, Yung Gee H, Stec E, Melowic HR, Blatner NR, et al. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun. 2012;3:1249.
Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17.
Ohsawa N, Koebis M, Suo S, Nishino I, Ishiura S. Alternative splicing of PDLIM3/ALP, for alpha-actinin-associated LIM protein 3, is aberrant in persons with myotonic dystrophy. Biochem Biophys Res Commun. 2011;409:64–69.
Pomies P, Macalma T, Beckerle MC. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem. 1999;274:29242–50.
Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–15.
Shou Y, Robinson DM, Amakye DD, Rose KL, Cho YJ, Ligon KL, et al. A five-gene hedgehog signature developed as a patient preselection tool for hedgehog inhibitor therapy in medulloblastoma. Clin Cancer Res. 2015;21:585–93.
Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–26.
Gordon RE, Zhang L, Peri S, Kuo YM, Du F, Egleston BL, et al. Statins synergize with hedgehog pathway inhibitors for treatment of medulloblastoma. Clin Cancer Res. 2018;24:1375–88.
Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci USA. 2007;104:2973–8.
Cheng Y, Franco-Barraza J, Wang Y, Zheng C, Zhang L, Qu Y, et al. Sustained hedgehog signaling in medulloblastoma tumoroids is attributed to stromal astrocytes and astrocyte-derived extracellular matrix. Lab Invest. 2020;100:1208–22.
Cheng Y, Liao S, Xu G, Hu J, Guo D, Du F, et al. NeuroD1 dictates tumor cell differentiation in medulloblastoma. Cell Rep. 2020;31:107782.
Ellis T, Smyth I, Riley E, Graham S, Elliot K, Narang M, et al. Patched 1 conditional null allele in mice. Genesis. 2003;36:158–61.
Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–51.
Krcmery J, Camarata T, Kulisz A, Simon HG. Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. Bioessays. 2010;32:100–8.
Klaavuniemi T, Alho N, Hotulainen P, Kelloniemi A, Havukainen H, Permi P, et al. Characterization of the interaction between Actinin-Associated LIM Protein (ALP) and the rod domain of alpha-actinin. BMC Cell Biol. 2009;10:22.
McKeown CR, Han HF, Beckerle MC. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev Dyn. 2006;235:530–8.
Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, et al. Cellular cholesterol directly activates smoothened in hedgehog signaling. Cell. 2016;166:1176–1187 e1114.
Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, et al. Cholesterol activates the G-protein coupled receptor smoothened to promote hedgehog signaling. Elife. 2016;5:e20304.
Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA. Rapid, direct activity assays for smoothened reveal hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci USA. 2017;114:E11141–E11150.
Kinnebrew M, Luchetti G, Sircar R, Frigui S, Viti LV, Naito T, et al. Patched 1 reduces the accessibility of cholesterol in the outer leaflet of membranes. Elife. 2021;10:e70504.
Kinnebrew M, Iverson EJ, Patel BB, Pusapati GV, Kong JH, Johnson KA, et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife. 2019;8:e20304.
Miyamoto T, Hosoba K, Itabashi T, Iwane AH, Akutsu SN, Ochiai H, et al. Insufficiency of ciliary cholesterol in hereditary Zellweger syndrome. EMBO J. 2020;39:e103499.
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55.
Pashmforoush M, Pomies P, Peterson KL, Kubalak S, Ross J Jr, Hefti A, et al. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med. 2001;7:591–7.
Maerz LD, Burkhalter MD, Schilpp C, Wittekindt OH, Frick M, Philipp M. Pharmacological cholesterol depletion disturbs ciliogenesis and ciliary function in developing zebrafish. Commun Biol. 2019;2:31.
Reilly ML, Benmerah A. Ciliary kinesins beyond IFT: cilium length, disassembly, cargo transport and signalling. Biol Cell. 2019;111:79–94.
Ishikawa H, Marshall WF. Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol. 2017;9:a021998.
Bidet M, Joubert O, Lacombe B, Ciantar M, Nehme R, Mollat P, et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS One. 2011;6:e23834.
Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, et al. Structural basis for the recognition of sonic hedgehog by human Patched1. Science. 2018;361:eaas8935.
Chong YC, Mann RK, Zhao C, Kato M, Beachy PA. Bifurcating action of smoothened in hedgehog signaling is mediated by Dlg5. Genes Dev. 2015;29:262–76.
Suzuki A, Ogata K, Yoshioka H, Shim J, Wassif CA, Porter FD, et al. Disruption of Dhcr7 and Insig1/2 in cholesterol metabolism causes defects in bone formation and homeostasis through primary cilium formation. Bone Res. 2020;8:1.
Maharjan Y, Lee JN, Kwak SA, Dutta RK, Park C, Choe SK, et al. TMEM135 regulates primary ciliogenesis through modulation of intracellular cholesterol distribution. EMBO Rep. 2020;21:e48901.
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405.
Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–8.
Gajjar A, Stewart CF, Ellison DW, Kaste S, Kun LE, Packer RJ, et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clin Cancer Res. 2013;19:6305–12.
Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–4.
Acknowledgements
We would like to thank Dr. James Chen at Stanford University for providing Sufu−/− cells, Dr. Pascal Pomies at the University Montpellier in France for providing PDLIM3 antibody. We also appreciate Drs. Y Eugene Chinn and Jiawei Wu for critical discussion. This research is supported by the Department of Health in Pennsylvania (to ZY), and by National Natural Science Foundation of China (82072798 to LZ, 82073873 to YW).
Author information
Authors and Affiliations
Contributions
ZY and LZ conceived and supervised the project. JZ; YY; XL; GL and TM performed experiments and analyzed data. YL; YW; GX; HR and LZ designed the experiments and analyzed data. JZ; LZ and ZY wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All experiments with mice were conducted under approval of and following the guideline of the Institutional Animal Care and Use committee at Fox Chase Cancer Center.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Yang, Y., Li, X. et al. PDLIM3 supports hedgehog signaling in medulloblastoma by facilitating cilia formation. Cell Death Differ 30, 1198–1210 (2023). https://doi.org/10.1038/s41418-023-01131-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01131-2
This article is cited by
-
PDLIM3 knockdown promotes ferroptosis in endometriosis progression via inducing Gli1 degradation and blocking Hedgehog signaling pathway
Journal of Assisted Reproduction and Genetics (2024)