Abstract
The DNA damage response (DDR) is critical for maintaining cellular homeostasis and genome integrity. Mounting evidence has shown that posttranslational protein modifications play vital roles in the DDR. In this study, we showed that deubiquitinase OTUD6A is involved in the DDR and is important for maintaining genomic stability. Mechanistically, in response to DNA damage, the abundance of OTUD6A was increased; meanwhile, PP2A interacted with OTUD6A and dephosphorylated OTUD6A at sites S70/71/74, which promoted nuclear localization of OTUD6A. Subsequently, OTUD6A was recruited to the damage site, where it interacted with TopBP1 and blocked the interaction between TopBP1 and its ubiquitin E3 ligase UBR5, decreasing K48-linked polyubiquitination and increasing the stability of TopBP1. OTUD6A depletion impaired CHK1 S345 phosphorylation and blocked cell cycle progression under DNA replication stress. Consistently, knockout of OTUD6A rendered mice hypersensitive to irradiation, shortened survival, and inhibited tumor growth by regulating TopBP1 in xenografted nude mice. Moreover, OTUD6A is expressed at high levels in breast cancer, and OTUD6A overexpression promotes cell proliferation, migration and invasion, indicating that dysregulation of OTUD6A expression contributes to genomic instability and is associated with tumor development. In summary, this study demonstrates that OTUD6A plays a critical role in promoting tumor cell resistance to chemoradiotherapy by deubiquitinating and stabilizing TopBP1.
Similar content being viewed by others
Introduction
In response to genotoxic pressure from both endogenous and exogenous sources, cells must correctly repair DNA damage to maintain the integrity of their genomes [1, 2]. Proper functioning of the DDR is critical for cellular homeostasis because unscheduled execution of the DDR may result in chromosomal aberrations or deleterious mutations, which lead to multiple diseases such as cancer [3, 4].
The DDR is regulated by various posttranslational protein modifications (PTMs) in cells [5,6,7,8]. In addition to E3 ubiquitin ligases [9,10,11,12,13,14], deubiquitinases (DUBs) are also involved in the DDR via direct recruitment to DNA damage sites or catalytic removal of ubiquitin from key proteins in the DDR [15,16,17,18,19,20,21,22], and regulate apoptosis, cell cycle, and signaling pathways linked to the development of cancer [23,24,25]. OTU domain-containing protein 6A (OTUD6A), a member of the ovarian tumor domain protease (OTU) subfamily of deubiquitinases (DUBs), is up-regulated in patients with colorectal cancer, in which it promotes cancer cell proliferation by regulating Drp1 stability and mitochondrial fission [26]. However, it remains unclear whether OTUD6A functions in the DDR pathway or plays a role in the maintenance of genome stability.
Topoisomerase IIβ-binding protein 1 (TopBP1) contains multiple BRCA1 carboxyl-terminal (BRCT) domains that act as scaffolds to regulate DNA replication, cell cycle checkpoint signaling, and DNA damage repair [27,28,29,30,31,32]. In normal cells, TopBP1 expression and activity are finely regulated by multiple PTMs [29, 30, 33, 34]. Among these PTMs, ubiquitination plays a particularly important role in regulating protein degradation. Previous studies showed that TopBP1 is ubiquitinated by the HECT domain-containing ligase hHYD (UBR5 homologue in Drosophila) in unstressed cells, while X ray-irradiation reduces the levels of ubiquitinated TopBP1 [35]. Moreover, high levels of TopBP1 expression have been found in many types of cancer cells; for example, a higher TopBP1 protein level in breast cancer samples was shown to be associated with a higher tumor grade and shorter patient survival time [36,37,38]. Therefore, exploration of the mechanisms underlying up-regulation of the protein level of TopBP1 in cancer cells is of great clinical importance.
In this study, we identified an important role of OTUD6A in maintaining genome integrity by regulating TopBP1 ubiquitination and stability in response to DNA damage and replication stress. We showed that OTUD6A depletion renders cells and Otud6a−/− mice more sensitive to genotoxic insults, and ectopic expression of OTUD6A in breast cancer promotes cell proliferation and invasion. Our study suggests that OTUD6A may be a potential target for cancer therapy.
Materials and Methods
Animal models
Animals were housed in specific pathogen-free barrier facilities and handled following the ‘Principles for the Utilization and Care of Vertebrate Animals’ and the ‘Guide for the Care and Use of Laboratory Animals’. Animal studies were approved by the IACUC of the Center for Experimental Animal Research (China) and Peking University Laboratory Animal Center (IACUC No. LSC ZhengX-2-1). Female BALB/c nude mice (6 weeks of age) were purchased from Beijing Vital River Laboratory Animal Technology. MDA-MB-231 cells (stably expressing shOTUD6A-1, shOTUD6A-2, or the control plasmid) were injected subcutaneously into the armpits of BALB/c nude mice (mice were divided into 3 groups randomly, 6 female mice in each group, 2 × 106 cells per mouse). Tumor size was measured every 2 days using a caliper beginning from the 7th day, and tumor volume was calculated using the following formula: volume = length × width2 /2. Mice were sacrificed on the 13th day after injection of MDA-MB-231 cells. Tumor tissue samples were subjected to immunohistochemistry for Ki67 staining. Meanwhile, MDA-MB-231 cells (stably expressing OTUD6A, shTopBP1, OTUD6A/shTopBP1 or the control plasmid) were injected subcutaneously into the armpits of BALB/c nude mice (mice were divided into 4 groups randomly, 6 female mice in each group, 1.7 × 106 cells per mouse). Tumor volume and size were measured as described above. Investigators undertaking the animal monitoring were blinded to the genotype of the mice.
Antibodies and reagents
Rabbit polyclonal TopBP1 antibody (A300-111A-M) was purchased from Bethyl Laboratories. Mouse monoclonal Flag antibody (F3165), cycloheximide (CHX, C7698), puromycin (P8833), thymidine (CAS 50-89-5), nocodozole (CAS 31430-18-9) and etoposide (E1383) were purchased from Sigma-Aldrich. Mouse monoclonal anti-Myc (M047-3, RRID: AB_591112) and mouse monoclonal His (D291-3, RRID: AB_10597733) antibodies were purchased from MBL. Rabbit monoclonal phospho-Chk1 (Ser345) (133D3, #2348) antibody, rabbit monoclonal K48-linkage specific polyubiquitin (D9D5, RRID: AB_10859893) antibody, and rabbit monoclonal K63-linkage specific polyubiquitin (D7A11, RRID: AB_10827985) antibody were purchased from Cell Signaling Technology. Mouse monoclonal anti-γH2AX (05–636, RRID: AB_309864) antibodies were purchased from Millipore. Histone H2A.X (phospho-S139) rabbit polyclonal antibody (BS4760) was purchased from Bioworld. Histone H3 mouse monoclonal antibody (BE3015) was purchased from EASYBIO. Rabbit polyclonal OTUD6A antibody (24486-1-AP) and rabbit polyclonal CHK1 antibody (25887-1-AP) were purchased from Proteintech. Rabbit polyclonal lamin B1 antibody (HX1983) was purchased from Huaxingbio. Rabbit polyclonal Ki67 antibody (A11390), rabbit PPP2R1A antibody (A5799), rabbit PP2AA antibody (A6702), mouse monoclonal α-tubulin antibody (AC012) and rabbit polyclonal anti-β-actin antibody (AC026) were purchased from ABclonal Technology. MG132 (T2154) was purchased from TargetMol. Mouse monoclonal TopBP1 (sc-271043) antibody, mouse monoclonal EDD (UBR5, sc-515494) antibody, aphidicolin (sc-201535) and polybrene (sc-134220) were purchased from Santa Cruz Biotechnology. Rabbit polyclonal OTUD6A antibody (NBP1-91498) was purchased from Novus Biologicals. Ubiquitin monoclonal antibody (P4D1) was purchased from Enzo Life Sciences. Phosphoserine antibody (ab9332) was purchased form Abcam. RPA2 monoclonal antibody (9H8) was purchased from Invitrogen. LB-100 (SF1082-10 mM) was purchased from Beyotime Biotechnology. Rabbit PP2AB antibody (SRP07090) was purchased from Tianjin Saierbio.
Plasmids
OTUD6A cDNA was kindly provided by Prof. Lingqiang Zhang at the Beijing Institute of Radiation Medicine. OTUD6A WT, OTUD6A C152A, OTUD6A D149A/C152A/H268A (OTUD6A 3A), and OTUD6A phosphosite mutants were constructed and cloned into the pcDNA3 Flag vector. Myc-TopBP1 was kindly provided by Prof. Jiadong Wang at Peking University School of Basic Medical Sciences. Myc-UBR5 HECT WT/C2768A was kindly provided by Prof. Shimin Zhao at the Institutes of Biomedical Sciences at Fudan University. The cDNA of PP2AA was inserted into pCMV-HA vector. His-OTUD6A was amplified by PCR and cloned into the pET-28a vector. All plasmids were verified by DNA sequencing.
Cell culture
HEK293T, HeLa, MDA-MB-231, HCT116, MCF-7 and U2OS cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco, USA). MDA-MB-436, HCC1954, HCC1937, T47D, Hs578T, ZR-75-1, SKBR3 and BT20 cell lines were obtained from the American Type Culture Collection (ATCC) and maintained in DMEM media. BT-549 cells was obtained from the ATCC andmaintained in RPMI 1640 (Gibco, USA). The 4T1 murine cell line was a kind gift from Dr. Jiadong Wang (Peking University). The identities of cell lines were authenticated by short tandem repeat analysis. Cell lines were confirmed to have no mycoplasma contamination by PCR analysis. MEF cells were separated from mice embryos at E13.5D following a standard procedure and cultured in DMEM supplemented with 12% fetal bovine serum and 100 μM nonessential amino acids (Solarbio).
Generation of cell lines with knocked down OTUD6A or TopBP1 or overexpressed OTUD6A
The knockdown cell lines were constructed as described previously [22]. ShCTRL, shOTUD6A and shTopBP1 oligos were cloned into the pLKO.1 plasmid to obtain a lentiviral particle, and the target cell lines were infected by the indicated lentivirus. Next, 1 μg/mL puromycin was added to the media of the infected cells until the OTUD6A or TopBP1 protein level was stably knocked down. Protein knockdown was confirmed by immunoblotting. The sequences for shOTUD6A and shTopBP1 were as follows:
shCTRL: TTCTCCGAACGTGTCACGT
shOTUD6A-1: CAGGCTGAGATGTCGGAGCAC
shOTUD6A-2: CGACAGTAGCATTGAATCTGT
shTopBP1: GCTGCAAAGAAGTGGAATTTA
OTUD6A was inserted into the pLVX-IRES-ZsGreen1 vector and subjected to lentiviral particle packaging. The target cell lines were infected with the OTUD6A-containing lentiviral particle to obtain cells with stable OTUD6A overexpression.
Laser microirradiation
Laser microirradiation was carried out following procedures described previously [39]. HeLa cells were grown on thin glass-bottom plates and irradiated with an ultraviolet laser (16 Hz pulse, 60% laser output). Images were taken using a Dragonfly (Andor) confocal imaging system for the indicated period of time.
Cell fractionation assay
The cell fractionation assay was performed as described previously [40] with modifications. Briefly, cells were lysed in buffer A for 1 h on ice. The supernatant was ultracentrifuged and collected as the cytosolic fraction. Cell pellets were washed in buffer A and resuspended in NP-40 lysis buffer as the nuclear fraction.
Chromatin extraction assay
The chromatin extraction assay was performed as described previously [21, 40] with modifications. Cells were lysed in buffer A for 1 h on ice. The supernatant was ultracentrifuged and collected as the soluble fraction. The pellet was washed twice in buffer A and lysed for 30 min in buffer B on ice. After centrifugation, the insoluble pellet was washed twice in buffer B. The final chromatin pellet was resuspended in buffer C as the chromatin fraction and denatured with 2× SDS loading buffer.
In vitro dephosphorylation assay
The proteins of substrate OTUD6A and phosphatase PP2A were enriched by IP assay from HEK293T cell lysates transfected with the indicated plasmids, and then the indicated proteins were incubated in phosphatase assay buffer (20 mM Tris-HCl, 1 mM EGTA, 5 mM MgCl2, 0.02% 2-mercaptoethanol, BSA (0.2 mg/mL)) at 30 °C for 4 h, followed by immunoblotting.
Electrophoretic Mobility Shift Assays (EMSA)
Electrophoretic mobility shift assays were performed as previously described [41] with the following modification. Fork DNA substrate containing 20-bp dsDNA and two 19-bp ssDNA arms was generated by annealing oligonucleotides A and B, and a ss-dsDNA hybrid containing 20-bp dsDNA and one 19-bp ssDNA arm was generated by annealing oligonucleotides A and C. Oligonucleotides B and C were radiolabeled at the 5’-end by [γ32P]-ATP (PerkinElmer) using T4 polynucleotide kinase (New England Biolabs Inc.), and then annealed with oligonucleotide A. After that, the fork DNA or ss-dsDNA hybrid and purified OTUD6A were incubated in binding buffer at 4 °C for 30 min. The mixture was separated by 5% Native-PAGE (polyacrylamide gel electrophoresis) (Tris borate-EDTA buffer) for 1 h at 110 V and 4 °C. The gel was dried by Gel Drying Equipment (Bio-Rad) and [32 P] DNA was detected by autoradiography. The sequences of oligonucleotides A, B and C were as follows:
A: 5′-CCAGTGAATTGTTGCTCGGTACCTGCTAACGGTAATCGG-3′
B: 5′-CAGCTATGGGACATTCGATACCGAGCAACAATTCACTGG-3′
C: 5′-TACCGAGCAACAATTCACTGG-3′
Detection of the genotype of Otud6a −/− C57BL/6 mice and sensitivity of mice to irradiation
Otud6a−/− C57BL/6 mice were purchased from Cyagen. The genotype of the mice was confirmed by PCR with the following primers:
Forward primer (F1): 5′-AAGTTCAGTGGTCAAGAGACCTG-3′
Reverse primer (R1): 5′-TCCCTGCCACATGAAAGGAGC-3′
Forward primer (F2): 5′-CTCACCCACCATTGTCATCGG-3′
17 Otud6aWT, Otud6a+/− and Otud6a−/− littermates were irradiated with 7 Gy of whole body IR, and survival rates were calculated every day after irradiation for 7 weeks.
Metaphase spread of MEFs
Otud6aWT, Otud6a+/− and Otud6a−/− MEF cells were generated and cultured as described above. Metaphasic spread of MEFs was performed as described previously [22] with modifications. Briefly, cells were treated with nocodozole (1 μM) for 12 h and harvested in 15 mL tubes (200 g per tube). For each sample, at least 30 metaphase spreads were analyzed. Chromosomes were stained with DAPI, and images were obtained using a confocal microscope (Zeiss LSM-710) with a 63 oil objective lens. Quantification analysis was performed using Imaris 7.6 software (Bitplane).
Duolink proximity ligation assay (PLA)
PLAs were performed using the Duolink® In Situ PLA® kit (DUO92101, Sigma-Aldrich) as described previously [42]. ShCTRL, shOTUD6A-1 and shOTUD6A-2 MDA-MB-231 cells were used in this assay.
Tissue arrays and immunohistochemistry (IHC)
Human breast cancer tissue arrays containing 45 cancer specimens were purchased from Shanghai Biochip Company Ltd. (Shanghai, China). Immunohistochemistry staining was performed by Servicebio (Wuhan, China). OTUD6A and TopBP1 expression levels were assessed by ImageJ software. The intensity of staining was scored from 1 to 4, and the extent of staining was scored from 0 to 100%. The final staining score was obtained by multiplying the two scores. The slides were analyzed by two independent pathologists.
Cell invasion assay
Control cells (1.8 × 104 cells), OTUD6A-overexpressing MDA-MB-231 cells (1.8 × 104 cells), shCTRL cells (2 × 104 cells) and shOTUD6A MDA-MB-231 cells (2 × 104 cells) were seeded in Transwell inserts (Corning) containing Matrigel (BD Biosciences)-coated porous membranes. After 24 h of incubation, the cells remaining in the insert were removed with cotton swabs and the cells that were adherent to the lower side of the membrane were fixed with methanol, subjected to crystal violet staining, and counted.
Cell scratch wound healing assay
MDA-MB-231 cells were grown to confluence in 6-well plates, and monolayer cells were wounded with a sterile plastic tip. After the adherent cells were washed three times with PBS, the wounded monolayers were cultured in DMEM. After incubation for the indicated number of hours, repopulation of the wounded areas was observed by microscopy (Olympus) and quantified using ImageJ software.
Cell synchronization and fluorescence-activated cell sorting
HeLa cells were treated with 2 mM thymidine for 19 h and released in fresh medium for 10 h. Thymidine (2 mM) was added again, and cells were incubated for another 17 h in order to arrest cells in the G1 phase before releasing them again in fresh medium. Cells were collected at the indicated time points for cell-cycle analysis. The synchronized cells were washed twice with PBS, resuspended in 1 mL 70% ethanol and stored at 4 °C overnight. Next, the fixed cells were washed and incubated in PBS containing 100 μg/mL propidium iodide (Beyotime) and 100 μg/mL RNase A (Yuanyebio) at 37 °C for 30 min. Finally, FACS analysis was performed using a FACSVerse cytometer (BD Biosciences) and the percentages of cells in different phases were quantified using FlowJo software v10 (BD Biosciences).
Apoptosis assays
Apoptosis assays were performed using the AnnexinV-FITC Apoptosis Detection Kit (Beyotime Biotechnology). Cells were collected and stained with AnnexinV and propidium iodide (PI). Apoptosis was determined by FACS analysis. Wild-type cells shown in the Q4 quadrant are negative for both stains. The Q3 quadrant shows Annexin V-positive cells, which are in the early stage of apoptosis. The Q2 quadrant shows cells that are both Annexin V- and PI-positive, which are in the late stage of apoptosis. Quantitative apoptotic measurement was performed for quadrants Q2 and Q3, and 104 cells were counted for each group.
Statistical analyses
The values reported in each graph represent the averages of at least three independent experiments, and ‘n’ describes the number of biological replicates in each figure legend. The statistical significance of comparisons of means was assessed by Student’s t-test (variance is similar between the groups). Statistical details and error bars ( ± SEM) are defined in each figure legend: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. For each figure, statistical tests are justified as appropriate. Analyses and graphical presentation were performed using the GraphPad Prism 8.0 software.
Full-length original western blots of these results are provided in Supplementary File 1.
Results
OTUD6A is recruited to DNA damage sites
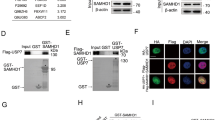
To investigate the function of OTUD6A in the DDR, we performed laser microirradiation, which revealed that GFP-OTUD6A was recruited to DNA damage sites after microirradiation (Fig. 1A). Moreover, the protein level of OTUD6A gradually increased after DNA damage (Fig. 1B, and supplementary Fig. S1A, B). Interestingly, OTUB2 promoted OTUD6A abundance and bleomycin treatment enhanced the OTUB2-OTUD6A interaction (supplementary Fig. S1C, D). Moreover, OTUB2 deubiquitinated K48-linked polyubiquitination of OTUD6A (supplementary Fig. S1E, F). Cell fractionation and chromatin extraction assays showed that in response to DNA damage, OTUD6A translocated from the cytosol to the nucleus (Fig. 1C), where more OTUD6A was recruited to the chromatin (Fig. 1D). These results indicate that OTUD6A is involved in the DDR.
A The dynamic accumulation of GFP-OTUD6A at DSB sites in U2OS cells was detected by laser microirradiation assay. The red line indicates the positions for laser microirradiation. Scale bar, 5 μm. B The change of OTUD6A protein level was measured in HeLa cells treated with 3 μΜ bleomycin. C Cell fractionation assays were performed to measure the OTUD6A cellular distribution in OTUD6A-overexpressed HeLa cells after bleomycin treatment (3 μΜ). D The recruitment of OTUD6A to chromatin in MCF7 cells treated with bleomycin (3 μΜ) was measured by chromatin extraction assay. E The effect of OTUD6A depletion on genome stability in HeLa cells treated with or without 10 Gy IR was measured by neutral comet assays. Images and quantified data are shown. The graphs show the mean ± SEM. About 100 cells were counted in each group. Statistical analysis was performed using Student’s t-test (**P < 0.01; ****P < 0.0001). n.s., not significant. F OTUD6A S3A (S70/71/74 A) mutant translocated to the nuclear fraction. Sequence alignment of OTUD6A from different organisms revealed that S70/71/74 are conserved in different species. The subcellular distributions of different OTUD6A mutants in HeLa cells were analyzed by IF and cell fractionation assays. Representative images of the Flag-OTUD6A WT/ S3A/ S3D distributions are shown. G The dynamic accumulation of GFP-OTUD6A WT and S3A at DSB sites was monitored by laser microirradiation assay. Representative images are shown, and 6 cells were counted for each group. Scale bar, 5 μm. H The chromatin recruitment of different OTUD6A mutants in HeLa cells treated with bleomycin (3 μΜ) was examined by chromatin extraction assay. I Co-IP assays were performed to measure the interaction between OTUD6A and PP2A subunits in HEK293T cells treated with or without bleomycin (3 μΜ). J Denatured IP assays were performed to detect the dephosphorylation of OTUD6A or OTUD6A S3A by PP2AA in HEK293T cells with or without DNA damage induced by bleomycin (3 μΜ). K Dephosphorylation of OTUD6A by PP2A was confirmed by in vitro dephosphorylation assay. L PP2AA promoted the nuclear translocation of OTUD6A. HeLa cells expressing the indicated plasmids were treated with or without 2 μΜ LB-100 for 6 h, after which cell fractionation assays were performed. DNA damage was induced by 3 μΜ bleomycin for 2 h in I, J, L.
Neutral comet assays were then conducted to explore the role of OTUD6A in maintaining genome stability. At 1 h after IR treatment, both OTUD6A WT and knockdown cells showed an increased comet tail length. Strikingly, OTUD6A depletion significantly increased the comet tail length at 24 h after IR treatment (Fig. 1E and supplementary Fig. S1G), suggesting that depletion of OTUD6A resulted in impaired DNA damage repair and decreased genome stability.
Dephosphorylation of OTUD6A by PP2A promotes its response to DNA damage by increasing its nuclear localization and chromatin recruitment
Because phosphorylation/dephosphorylation regulate protein subcellular localization [43,44,45], and OTUD6A, which is distributed in the cytoplasm and the nucleus (supplementary Fig. S2A), could translocate from the cytoplasm to the nucleus following DNA damage (Fig. 1C), we then assessed whether OTUD6A was phosphorylated and whether this modification affected OTUD6A subcellular localization. IP-MS assays identified some potential phosphorylated sites of OTUD6A (supplementary Fig. S2B, C). Among them, sites Ser70/71/74 were conserved in many species (Fig. 1F). We therefore constructed mutants with multiple phosphorylated sites and found that the phosphorylation-deficient triple mutant OTUD6A S3A (S70/71/74 A) had the greatest effect on protein nuclear localization (Fig. 1F, and supplementary Fig. S2D–F). In response to DNA damage, more OTUD6A S3A proteins were recruited to the damage site than OTUD6A WT (Fig. 1G). Consistently, more OTUD6A S3A protein, but not S3D protein, was recruited to the chromatin in comparison with OTUD6A WT (Fig. 1H). These results indicate that dephosphorylation of OTUD6A at S70/71/74 promotes its nuclear localization following DNA damage.
Next, we performed IP-MS assay to identify the phosphatase that dephosphorylated OTUD6A in response to DNA damage. The subunits of PP2A, PPP2R1A and PP2AB, were identified as potential partners of OTUD6A (supplementary Fig. S2G). Therefore, we examined the interaction between OTUD6A and the subunits of PP2A, including PPP2R1A, PP2AA, and PP2AB. Indeed, the interaction between PP2A and OTUD6A was confirmed, which was enhanced in cells after bleomycin treatment (Fig. 1I). Consistently, PP2A dephosphorylated OTUD6A, and the effect of PP2A on OTUD6A dephosphorylation was enhanced after DNA damage (Fig. 1J). Moreover, in comparison to its effect on OTUD6A WT phosphorylation, PP2A only slightly reduced the phosphorylation level of the OTUD6A S3A mutant (Fig. 1J right panel, lane 3 vs. lane 2), and, in cells treated with bleomycin, almost no change in the phosphorylation level of OTUD6A S3A was observed in comparison with that of cells that were not treated with bleomycin (Fig. 1J right panel, lane 4 vs. lane 3). These data indicate that, in response to DNA damage, PP2A mainly dephosphorylated OTUD6A at serines 70, 71 and 74. Furthermore, in vitro dephosphorylation assays confirmed the dephosphorylation of OTUD6A by PP2A, and inhibition of PP2A by LB-100, a specific inhibitor of PP2A, blocked its effect on the dephosphorylation of OTUD6A (Fig. 1K). In addition, OTUD6A abundance in the nucleus increased by PP2AA overexpression but diminished by LB-100 (Fig. 1L). Taken together, these results indicate that in response to DNA damage, PP2A dephosphorylates OTUD6A and promotes the nuclear translocation of OTUD6A.
OTUD6A interacts with TopBP1 in vivo and in vitro
Identification of the interacting proteins of OTUD6A by IP-MS analysis showed that both E3 ligase UBR5 and TopBP1 were potential partners of OTUD6A (Fig. 2A). Enriched biological pathway analysis revealed that functionally related proteins are involved in DNA repair and cell cycle regulation pathways (Fig. 2B). As TopBP1 ubiquitination decreased after DNA damage [35], and the deubiquitinase of TopBP1 remains unclear, we wondered whether OTUD6A functioned as a DUB of TopBP1. The interactions between OTUD6A and TopBP1 (Fig. 2C–E) or UBR5 (supplementary Fig. S3A) were confirmed, and the OTUD6A-TopBP1 interaction was promoted by DNA damage (Fig. 2C, D). To elucidate the mechanism underlying the OTUD6A-TopBP1 interaction, we constructed the indicated OTUD6A mutants and performed co-IP assays. The C terminal catalytic domain of OTUD6A was critical for its interaction with TopBP1 (Fig. 2F). Surprisingly, the deubiquitinase-defective OTUD6A mutant (Flag-OTUD6A 3A) in which the catalytic triad D149/C152/H268 was mutated also interacted with TopBP1 (Fig. 2F). In addition, the N and C termini of TopBP1 interacted with OTUD6A (supplementary Fig. S3B, C). Notably, consistent with its increased nuclear translocation, the OTUD6A S3A mutant showed a stronger interaction with TopBP1 in comparison with that of OTUD6A WT (Fig. 2G and supplementary Fig. S3D). Taken together, these results indicate that DNA damage promotes the interaction between TopBP1 and OTUD6A.
A The interactors of OTUD6A were identified by IP-Mass-spectrometry using HEK293T cells transfected with Flag-OTUD6A. The silver staining result is shown, and major hits from the mass spectrometry analysis are shown in the table. B Enriched biological pathway analysis of OTUD6A potential interacting targets identified by mass spectrometry. Bar graph of enriched terms across OTUD6A targets was provided by Metascape. C, D The interaction between OTUD6A and TopBP1 was promoted after DNA damage. Co-IP assays were performed using cellular extracts from HEK293T cells treated with or without 10 Gy IR (C) or 3 μM bleomycin (D). E The direct interaction between TopBP1 and OTUD6A proteins was examined by in vitro interaction assay. Myc-TopBP1 was immunoprecipitated from Myc-TopBP1-transfected HEK293T cells; His-OTUD6A protein was purified from E. coli. F Mapping of OTUD6A domains critical for TopBP1 binding by co-IP assay in HEK293T cells. Schematic representation of various OTUD6A truncations is shown. G Co-IP assays were performed to compare the interaction between OTUD6A WT or S3A mutant and TopBP1 in HEK293T cells treated with or without bleomycin (3 μM).
OTUD6A stabilizes TopBP1 by inhibiting K48-linked polyubiquitination of TopBP1
Considering TopBP1 interacts with OTUD6A and it is ubiquitinated by UBR5 (supplementary Fig. S4A, B), we examined whether OTUD6A increased the protein level of TopBP1 by regulating its ubiquitination. As expected, OTUD6A significantly deubiquitinated TopBP1 and OTUD6A depletion increased TopBP1 ubiquitination (Fig. 3A, B). In addition, the protein level of TopBP1 was elevated along with gradual overexpression of OTUD6A (Fig. 3C). Consistently, chase experiments showed that the half-life of TopBP1 was prolonged in OTUD6A-overexpressed cells (Fig. 3D). Moreover, OTUD6A specifically reduced TopBP1 K48-linked but not K63-linked polyubiquitination (Fig. 3E and supplementary Fig. S4C). These results suggest that OTUD6A stabilizes TopBP1 through inhibiting K48-linked polyubiquitination of TopBP1.
A The effect of OTUD6A on TopBP1 ubiquitination in HEK293T cells was measured by His-ubiquitin pull down assay. B The influence of OTUD6A depletion on endogenous TopBP1 ubiquitination was detected by IP assay in OTUD6A knockdown HCT116 cells. C The effect of OTUD6A on TopBP1 abundance was analyzed by immunoblotting using HEK293T cells transfected with different amounts of Flag-OTUD6A. D Half-life analysis of TopBP1 in OTUD6A-overexpressing HeLa cells treated with 50 µg/mL cycloheximide (CHX) for the indicated times. E The effect of OTUD6A on K48-linked polyubiquitination of Myc-TopBP1 was analyzed by denatured IP assay using anti-K48-ubiquitin linkage-specific antibodies. F The effects of OTUD6A truncations on TopBP1 ubiquitination were detected by His-ubiquitin pull down assays. G The influence of catalytic mutant OTUD6A 3A on TopBP1 abundance was detected in HEK293T cells transfected with different amounts of Flag-OTUD6A 3A. H Co-IP assays were performed to detect the effects of OTUD6A and catalytic mutant OTUD6A 3A on the interaction between TopBP1 and UBR5 in HEK293T cells. I The effect of OTUD6A depletion on the endogenous interaction between TopBP1 and UBR5 was detected in MDA-MB-231 cells by PLAs. Representative images are shown and about 30 cells were counted to analyze the number of TopBP1-UBR5 PLA dots. OTUD6A knockdown efficiency was detected by immunoblotting. Statistical analysis was performed using two-tailed Student’s t-tests (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). The graphs show the mean ± SEM.
Next, we performed His-ubiquitin pulldown assays to determine whether deubiquitination of TopBP1 by OTUD6A is dependent on its catalytic activity. And we found that in addition to OTUD6A WT, the C terminus of OTUD6A and catalytic mutant OTUD6A 3A that interacted with TopBP1 also down-regulated TopBP1 ubiquitination (Fig. 3F). Because C152 in the catalytic triad is critical for its DUB activity [26, 46, 47], we also constructed a OTUD6A C152A mutant and found that C152A mutant also inhibited TopBP1 K48-linked but not K63-linked polyubiquitination (supplementary Fig. S4D, E). Furthermore, TopBP1 protein level was also elevated in catalytic mutant OTUD6A 3A-overexpressing cells (Fig. 3G), suggesting that OTUD6A inhibits TopBP1 ubiquitination in a deubiquitinase activity-independent manner.
To further explore the mechanism underlying the inhibition of TopBP1 ubiquitination by OTUD6A, we investigated the effect of OTUD6A on TopBP1-UBR5 interaction. The interaction between TopBP1 and UBR5 was significantly attenuated by both OTUD6A WT and OTUD6A 3A mutant (Fig. 3H). PLAs showed that OTUD6A depletion increased the in situ TopBP1-UBR5 interaction (Fig. 3I). Taken together, these results indicate that OTUD6A stabilizes TopBP1 by blocking the interaction between TopBP1 and its E3 ligase UBR5 and thus inhibiting K48-linked polyubiquitination of TopBP1.
OTUD6A promotes CHK1 S345 phosphorylation and OTUD6A depletion leads to S phase cell cycle arrest
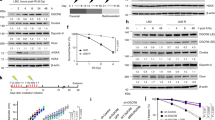
Under replication stress, TopBP1 is recruited to the replication fork and activates the ATR pathway. Subsequently, ATR phosphorylates substrates, including RPA and CHK1, regulates the cell cycle and DDR [28, 48, 49]. Considering that OTUD6A interacted with TopBP1 (Fig. 2C–E), we applied EMSA and chromatin extraction assays to examine whether OTUD6A is recruited to replication forks. Indeed, OTUD6A bound to fork DNA and a ss-dsDNA hybrid directly (Fig. 4A), and the chromatin-bound OTUD6A increased upon replication stress (supplementary Fig. S5A), suggesting that OTUD6A could be recruited to the replication fork.
A The direct binding of OTUD6A to fork DNA and a ss-dsDNA hybrid was confirmed by EMSA assay. His-OTUD6A protein was purified from E. coli. B The effect of OTUD6A depletion on the stability of TopBP1 at the replication fork was investigated by IF assays in shCTRL- and shOTUD6A-MDA-MB-231 cells treated with 5 mM HU for 4 h. OTUD6A knockdown efficiency was detected by immunoblotting. Representative images of 10 cells were counted to analyze the colocalization percentage of TopBP1 with RPA2. Scale bar, 10 μm. C, D The influence of OTUD6A on CHK1 S345 phosphorylation was assessed by immunoblotting. OTUD6A WT or OTUD6A-overexpressing HeLa cells treated with or without 3 μΜ bleomycin for 2 h (C) or 10 Gy IR (D) were collected at different time points as indicated and then subjected to immunoblotting. E, F The effects of OTUD6A on the cell cycle were measured in HeLa cells under normal conditions (E) and DNA replication stress induced by cisplatin treatment (2 μM,12 h) (F) using a FACSVerse cytometer. OTUD6A was reintroduced in OTUD6A-depleted HeLa cells in E. G OTUD6A depletion prolonged the S phase. ShCTRL- and shOTUD6A- HeLa cells were synchronized at the G1/S phase transition, after which the cells were released and collected at different time points for FACS analysis to assess their ability to pass the S phase. H The effect of OTUD6A knockdown on apoptosis was assessed by FCAS analysis in HeLa cells treated with 2 μM cisplatin for 12 h. I, J Clonogenic survival assays were performed to assess the effect of OTUD6A on cell survival in HeLa cells treated with 0 to 0.9 μM bleomycin (I) or 0 to 250 μM HU (J) for 24 h. GFP, GFP-OTUD6A, GFP-OTUD6A S3A, and GFP-OTUD6A S3D were reintroduced in OTUD6A-knockdown HeLa cells in I. K The OTUD6A knockdown efficiency of the HeLa cells used in F–H, J was confirmed by immunoblotting. Statistical analyses were performed for the results shown in B, E, F, H–J using two-tailed Student’s t-tests (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). n.s., not significant. The graphs show the mean ± SEM (n = 10 in B; n = 3 in E, F, H–J).
Next, we explored whether TopBP1 affected recruitment of OTUD6A to replication forks upon replication stress. TopBP1 depletion had no effect on the recruitment of OTUD6A to chromatin (supplementary Fig. S5B), suggesting that TopBP1 does not affect the recruitment of OTUD6A to replication forks. As TopBP1 is accumulated to stalled replication forks induced by the replication inhibitor HU [48, 50], we also conducted chromatin extraction assays to examine whether replication stress-induced recruitment of TopBP1 to the replication forks is influenced by OTUD6A. The results showed that OTUD6A depletion decreased TopBP1 abundance on chromatin after HU treatment, but this effect was prevented by the presence of proteasome inhibitor MG132 which inhibited TopBP1 degradation (supplementary Fig. S5C). Consistently, IF assays showed that OTUD6A depletion reduced the colocalization of TopBP1 and RPA2 (a replication fork marker) (Fig. 4B). These observations suggest that OTUD6A stabilizes TopBP1 at replication forks upon replication stress, but it does not affect recruitment of TopBP1 to replication forks.
Given that TopBP1 expression increased at the G1/S transition and S phase [37, 51], we assessed the effect of OTUD6A on TopBP1 foci in early S phase. Knockdown of OTUD6A obviously reduced the number of TopBP1 foci in response to DNA damage (supplementary Fig. S5D). Furthermore, CHK1 activation was enhanced in OTUD6A-overexpressed cells (Fig. 4C, D). Depletion of OTUD6A led to a much higher percentage of cells in S phase in comparison with that of the control group, and this phenotype was rescued by reintroducing OTUD6A (Fig. 4E and supplementary Fig. S5E). Consistently, OTUD6A depletion also led to S phase arrest under cisplatin-induced DNA replication stress (Fig. 4F). In addition, at 12 h after UV treatment, control cells recovered from the damage and entered into the normal cell cycle, while depletion of OTUD6A still caused cell cycle arrest at S phase (supplementary Fig. S5F), indicating that OTUD6A deletion leads to a prolonged S phase and renders cells more sensitive to replication stress. Moreover, when cells were synchronized in the G1/S phase and then released, OTUD6A-depleted cells were arrested in the S phase (Fig. 4G). We also observed that OTUD6A depletion promoted apoptosis after cisplatin treatment (Fig. 4H and supplementary Fig. S5G) and reduced cell survival rate after bleomycin or HU treatment (Fig. 4I–K and supplementary Fig. S5H). The reduced cell survival rate in OTUD6A-depleted cells was compensated obviously by reintroducing OTUD6A-WT, and in comparison with OTUD6A-WT, introducing the OTUD6A S3A mutant showed a higher rescue efficacy, while the OTUD6A S3D mutant displayed lower efficacy (Fig. 4I and supplementary Fig. S5H). These results demonstrate that OTUD6A overexpression promotes CHK1 activation, and depletion of OTUD6A results in cell cycle arrest and renders cancer cells more sensitive to chemotherapeutic drugs.
Deletion of OTUD6A renders mice hypersensitive to IR
Given that increased radiation sensitivity is a hallmark of a defective DDR, we then investigated the role of OTUD6A in maintaining genome stability using Otud6a−/− mice. The survival rates of Otud6a−/−, Otud6a+/− and Otud6aWT littermates (n = 17) were monitored after irradiation (7 Gy) (Fig. 5A and supplementary Fig. S6A). All Otud6a−/− and Otud6a+/− mice died within 14 and 16 days, respectively, while 17% of Otud6aWT mice were alive 1 month after irradiation (Fig. 5B), suggesting that deletion of Otud6a renders mice hypersensitive to IR. Moreover, the spleens of Otud6a−/− mice displayed abnormal splenic cysts after irradiation (supplementary Fig. S6B). IHC assays revealed that cells from the livers and spleens of Otud6a−/− mice exhibited more γH2AX staining than those from Otud6a+/− and Otud6aWT mice (Fig. 5C and supplementary Fig. S6C). In addition, we examined chromosome aberrations by chromosome separation assays using MEF cells from different genotype mice. MEF cells from Otud6a−/− mice had the highest frequency of chromosomal breaks and fusion (Fig. 5D and supplementary Fig. S6D). Consistently, neutral comet assays also confirmed that Otud6a deletion resulted in genome instability in MEF cells (Fig. 5E, F and supplementary Fig. S6D). These results indicate that OTUD6A plays an important role in maintaining genome integrity and its deletion renders mice more sensitive to genotoxic insults.
A, B Otud6aWT, Otud6a+/− and Otud6a−/− female littermates (n = 17 for each genotype) were exposed to a whole body dose of 7 Gy (A) and the survival rate was monitored for 20 days (B). C IHC staining of γH2AX from the spleen and liver tissues of Otud6aWT, Otud6a+/− and Otud6a−/− female littermates after exposure to 5 Gy IR for 7 days. Representative γH2AX staining is shown. D The effect of OTUD6A depletion on chromosome aberrations was visualized by performing metaphase spread assays. MEFs isolated from Otud6aWT, Otud6a+/− and Otud6a−/− mice were treated with nocodozole (1 μM) for 12 h and subjected to the metaphase spread assays. Chromosome aberrations such as break and fusion were analyzed in about 30 metaphase spreads for each genotype. Representative aberrations and the aberration percentage of Otud6aWT, Otud6a+/− and Otud6a−/− MEFs are shown. The experiments were performed three times. E The influence of OTUD6A depletion on genome integrity was detected by neutral comet assay. Otud6aWT, Otud6a+/− and Otud6a−/− MEFs were treated with 3 μM bleomycin for 3 h and then restored for 24 h, after which the cells were subjected to the neutral comet assay. Images and quantified data are shown. More than 200 cells were counted in each group. F The genotypes of the MEFs used in D, E were confirmed by PCR. The statistical analyses in D, E were performed using two-tailed Student’s t-tests (*P < 0.05; **P < 0.01; ****P < 0.0001). n.s., not significant. The graphs show the mean ± SEM.
OTUD6A promotes cell proliferation and invasion in breast carcinoma
TopBP1 is highly expressed in the basal-like breast cancer subtype (triple-negative) [52]. Because OTUD6A regulated TopBP1 abundance, we then determined whether OTUD6A modulated the proliferation and invasion of breast carcinoma through regulation of TopBP1. IHC assays showed that breast cancer tissue samples displayed higher levels of both TopBP1 and OTUD6A than that of adjacent normal tissue (Fig. 6A, B). In addition, the protein level of OTUD6A was positively correlated with that of TopBP1 in a panel of human breast cancer cell lines (Fig. 6C and supplementary Fig. S7A). Next, we explored the regulatory influence of OTUD6A on TopBP1 using MDA-MB-231 cells, a triple-negative breast cancer cell line. The endogenous interaction between TopBP1 and OTUD6A was confirmed by co-IP assay (Fig. 6D). In response to DNA damage, the protein level of OTUD6A gradually increased (Fig. 6E), which was accompanied by its increased recruitment to chromatin (Fig. 6F). Moreover, the effects of OTUD6A on the endogenous ubiquitinated-TopBP1 level, TopBP1 stability, CHK1 S345 phosphorylation, and cell cycle were confirmed in MDA-MB-231 cells (Fig. 6G–K). PLAs revealed that OTUD6A depletion increased endogenous ubiquitinated-TopBP1 abundance (Fig. 6G, H). Consistently, OTUD6A depletion decreased the protein level of TopBP1, which was restored after MG132 treatment (Fig. 6I). Moreover, OTUD6A knockdown impaired phosphorylation at CHK1 S345 (Fig. 6J) and resulted in S phase cell cycle arrest under replication stress (Fig. 6K). Similar effects of OTUD6A on the cell cycle were also observed in OTUD6A-depleted MCF7 cells (supplementary Fig. S7B).
A, B IHC staining of TopBP1 (A) and OTUD6A (B) in human breast cancer specimens (n = 45). Representative images of IHC staining of breast cancer tissue and paired adjacent normal tissue from the same patient are shown. The protein expression scores of TopBP1 (A) and OTUD6A (B) in adjacent normal tissue and paired breast cancer tissue are shown. Statistical analysis was performed using paired Student’s t-tests. The staining shown in A, B was quantified by ImageJ software. Scale bars, 20 µm. C Immunoblotting of TopBP1 and OTUD6A expression in the indicated human breast cancer cell lines. D The interaction of endogenous TopBP1 with OTUD6A in MDA-MB-231 cells stably expressing OTUD6A was measured by co-IP assay using the indicated antibodies. E The OTUD6A protein levels gradually increased in MDA-MB-231 cells after bleomycin (3 μΜ) treatment. F OTUD6A was recruited to chromatin in MDA-MB-231 cells after 3 μΜ bleomycin treatment. G, H The effect of OTUD6A knockdown on the level of endogenous ubiquitinated-TopBP1 was detected by PLAs. Representative images are shown (G), and about 50 MDA-MB-231 cells were counted to analyze the number of TopBP1-ubiquitin PLA dots (H). I MDA-MB-231 cells with depleted OTUD6A were treated with or without 10 μΜ MG132 for 10 h and then used to assess TopBP1 expression. J The effect of OTUD6A depletion on CHK1 S345 phosphorylation was examined by immunoblotting with MDA-MB-231 cells harboring shCTRL and shOTUD6A, which were treated with or without 3 μΜ bleomycin for 2 h and collected at the indicated time points. K, L FCAS analysis of the effects of OTUD6A depletion on the cell cycle (K) and apoptosis (L) in OTUD6A-depleted MDA-MB-231 cells that were treated with 2 μM cisplatin for 12 h. M, N CCK8 analysis was performed to detect the effects of OTUD6A and TopBP1 on cell proliferation in different MDA-MB-231 cell lines with abnormally expressed OTUD6A or depleted TopBP1. O, P Analyses of cell invasion ability in MDA-MB-231 cells with overexpressed (O) and depleted OTUD6A (P) by Transwell assays. Representative Images and quantified data are shown. Q Analyses of the cell migration ability of MDA-MB-231 cells harboring shCTRL, shOTUD6A, and shOTUD6A & OTUD6A by cell scratch wound healing assays. The OTUD6A protein level was examined by immunoblotting. Representative images and quantified data are shown. The statistical analyses in A, B, H, K–Q were performed using two-tailed Student’s t-tests (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). n.s., not significant. The graphs show the mean ± SEM (n = 3 in K–N, Q, n = 5 in O, P).
The results described above drove us to investigate the role of OTUD6A in regulating breast cancer progression. In comparison to wild-type MDA-MB-231 cells, cells with depleted OTUD6A had a higher apoptosis rate after cisplatin or paclitaxel treatment (Fig. 6L and supplementary Fig. S7C). In addition, CCK8 assay showed that cell proliferation was inhibited by OTUD6A depletion (Fig. 6M) but promoted by OTUD6A overexpression (Fig. 6N). More importantly, when TopBP1 was depleted, cell proliferation rate was reduced significantly whether OTUD6A was overexpressed or not (Fig. 6N and supplementary Fig. S7D), suggesting that OTUD6A promotes cell proliferation in a TopBP1-dependent manner. Furthermore, cell invasion was significantly promoted by OTUD6A overexpression but inhibited by OTUD6A depletion (Fig. 6O, P and supplementary Fig. S7E, F). Wound healing assays showed that OTUD6A depletion suppressed cell migration, which could be compensated by OTUD6A re-introduction (Fig. 6Q). These results indicate that OTUD6A promotes cell proliferation and invasion in breast carcinoma.
OTUD6A depletion inhibits breast tumor growth in a xenograft mouse model
To confirm the role of OTUD6A in regulation of tumor growth, we established MDA-MB-231 xenografts in female BALB/c nude mice. As expected, OTUD6A depletion significantly reduced the size and weight of breast cancer xenografts (Fig. 7A–D). In addition, the levels of Ki67 and TopBP1 were greatly reduced in tumors of MDA-MB-231 xenografts with depleted OTUD6A (Fig. 7E). In contrast, OTUD6A overexpression significantly increased tumor growth, whereas depletion of endogenous TopBP1 dramatically reduced tumor size and weight in vivo no matter OTUD6A was overexpressed (Fig. 7F–H). Furthermore, the number of Ki67-positive cells increased significantly in tumor specimens with ectopically expressed OTUD6A, but decreased when TopBP1 was knocked down (Fig. 7I, J). We also assessed the effects of OTUD6A depletion on tumor development by injection of 4T1, a mouse breast cancer cell line, into Otud6a+/− and Otud6a−/− female C57BL/6 N mice. Otud6a−/− mice displayed smaller tumors in comparison with those of Otud6a+/− mice (supplementary Fig. S8A–C). These results indicate that OTUD6A promotes the growth of breast cancer xenografts.
A 2 × 106 shCTRL- or shOTUD6A- MDA-MB-231 cells were injected into the armpit of 6 week-old female BALB/c nude mice to induce tumorigenesis. Photographs of the tumors of each group (n = 6) at the end of the experiment are shown. (B, C) The graph shows the tumor growth (B) and tumor weight (C) for each group (n = 6). D The OTUD6A protein level of the MDA-MB-231 cells used in A–C was confirmed by immunoblotting. E IHC staining of Ki67 and TopBP1 (scale bar, 50 µm) in tumor sections (n = 4 for each group). The immunostainings of TopBP1 were evaluated blindly and the intensity of staining was scored from 1 to 4, and the extent of staining was scored from 0 to 100%. The images and quantified data are shown. F Four different MDA-MB-231 cell types (CTRL, OTUD6A, shTopBP1 and OTUD6A/shTopBP1) were constructed for the in vivo tumorigenesis assay. 1.7 × 106 MDA-MB-231 cells from each group were injected into the armpit of 6-week-old female BALB/c nude mice to induce tumorigenesis. Photographs of the tumors from each group (n = 6) at the end of the experiment are shown. G, H The graph shows the tumor growth (G) and tumor weight (H) for each group (n = 6). I IHC staining of Ki67 and TopBP1 (scale bar, 50 µm) in tumor sections (n = 3 for each group). The images and quantified data are shown. J The protein levels of OTUD6A and TopBP1 were confirmed by immunoblotting. Statistical analyses in B, C, E, G–I were performed using two-tailed Student’s t-tests (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). n.s., not significant. The graphs show the mean ± SEM.
Discussion
Here, we found that OTUD6A functions in the DDR by down-regulating ubiquitination of TopBP1. In response to DNA damage, OTUD6A is dephosphorylated by PP2A at sites S70/71/74 and rapidly recruited to DNA damage sites, where it promotes TopBP1 stability by inhibiting K48-linked polyubiquitination, and subsequently enhances CHK1 S345 phosphorylation, which is important for cell cycle progression (Fig. 8). We showed that OTUD6A is highly expressed in breast cancer and promotes cancer cell proliferation and invasion. Using xenograft nude mice models, we demonstrated that OTUD6A enhances tumor growth in a TopBP1-dependent manner. Moreover, OTUD6A deletion rendered mice hypersensitive to irradiation and shortened survival.
Under DNA damage, OTUD6A is dephosphorylated at serine 70, 71, and 74 by PP2A, and OTUD6A subsequently translocates to the nucleus, where it is recruited to DNA damage sites and inhibits ubiquitination of TopBP1 by blocking the interaction between TopBP1 and its E3 ligase UBR5. Consequently, the protein stability of TopBP1 is promoted and CHK1 is activated, which subsequently promotes cell cycle progression, genome stability and breast cancer development.
Phosphorylation/dephosphorylation regulate the catalytic activity, cellular distribution or function of deubiquitinases For example, the phosphorylation of OTUB1 at S16 is important for its nuclear accumulation [43], whereas dephosphorylation of YB1 and HDAC4 could promote their nuclear localization [44, 45]. In this study, we found that PP2A-modulated dephosphorylation of OTUD6A at S70/71/74 facilitated its nuclear localization and chromatin recruitment, as well as interaction with TopBP1. We attempted to generate specific antibodies targeting phosphorylated OTUD6A at S70/71/74 to assess endogenous phosphorylation at these three sites, but we failed to obtain them. Therefore, specific pS70/71/74 antibodies should be generated to allow further exploration of changes in the phosphorylation levels at sites S70/71/74 in OTUD6A, as well as the mechanisms through which OTUD6A phosphorylation is regulated in response to DNA damage. In addition, it is worth mentioning that although the OTUD6A S240A mutant had no effect on the nuclear localization of OTUD6A, because the PTM score of OTUD6A S240 increased obviously after bleomycin treatment (supplementary Fig. S2C), a specific phosphorylation antibody targeting OTUD6A S240 should be generated to verify whether Ser 240 is phosphorylated and regulated by bleomycin-induced DNA damage in the future.
Recently, Kim et al. showed that USP13 regulates the replication stress response by deubiquitinating and stabilizing TopBP1 [34]. In our study, we identified OTUD6A as a DUB of TopBP1. In comparison to the other OTUs we screened (supplementary Fig. S9), OTUD6A showed the strongest inhibitory effect on TopBP1 ubiquitination. Notably, in contrast with USP13, which regulates TopBP1 ubiquitination in a DUB activity-dependent manner, OTUD6A inhibits TopBP1 ubiquitination in a DUB activity-independent manner by disrupting the interaction between TopBP1 and its E3 ligase UBR5 (Fig. 3H), suggesting that different deubiquitinases target the same substrate through different mechanisms. Moreover, previous studies demonstrated that TopBP1 is recruited to the replication forks and activates the ATR pathway under replication stress [28, 48, 49], and our study showed that OTUD6A is recruited to the replication forks where it stabilizes TopBP1. However, the recruitment of OTUD6A to the replication forks does not affect recruitment of TopBP1, and vice versa, suggesting that other factors might affect the recruitment of OTUD6A to the replication forks upon replication stress. The mechanism of OTUD6A recruitment and the relationship between the recruitment of OTUD6A and TopBP1 to the replication fork merit exploration in future experiments.
Taken together, our results provide novel insight into the role of OTUD6A in the DDR pathway and identify TopBP1 as a substrate of OTUD6A when the latter is activated in response to DNA damage. More importantly, we demonstrate that OTUD6A promotes breast cancer proliferation, invasion and breast cancer growth, and these effects are dependent on TopBP1. In conclusion, OTUD6A is critical for maintaining genomic stability and is a promising clinical target for preventing breast cancer progression.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary data files and all original data are available from the corresponding authors upon request.
References
Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl J Med. 2009;361:1475–85.
Pilie PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071–8.
Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204.
Sarangi P, Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci. 2015;40:233–42.
Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell. 2013;49:897–907.
Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26.
Kim JJ, Lee SY, Miller KM. Preserving genome integrity and function: the DNA damage response and histone modifications. Crit Rev Biochem Mol Biol. 2019;54:208–41.
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009;136:435–46.
Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, et al. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015;10:226–38.
Krais JJ, Wang Y, Bernhardy AJ, Clausen E, Miller JA, Cai KQ, et al. RNF168-mediated ubiquitin signaling inhibits the viability of BRCA1 null cancers. Cancer Res. 2020;80:2848–60.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007;131:887–900.
Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–78.
Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007;316:1198–202.
Cheng YC, Shieh SY. Deubiquitinating enzyme USP3 controls CHK1 chromatin association and activation. Proc Natl Acad Sci USA 2018;115:5546–51.
Li F, Sun Q, Liu K, Han H, Lin N, Cheng Z, et al. The deubiquitinase OTUD5 regulates Ku80 stability and non-homologous end joining. Cell Mol Life Sci. 2019;76:3861–73.
Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, et al. The Deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 2015;13:93–107.
Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9.
Peng Y, Liao Q, Tan W, Peng C, Hu Z, Chen Y, et al. The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat Commun. 2019;10:1224.
Wijnhoven P, Konietzny R, Blackford AN, Travers J, Kessler BM, Nishi R, et al. USP4 Auto-Deubiquitylation promotes homologous recombination. Mol Cell. 2015;60:362–73.
Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, et al. A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. 2018;78:1069–82.
Yang Y, Yang C, Li T, Yu S, Gan T, Hu J, et al. The Deubiquitinase USP38 promotes NHEJ repair through regulation of HDAC1 activity and regulates cancer cell response to genotoxic insults. Cancer Res. 2020;80:719–31.
Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012;31:2373–88.
Komander D, Clague MJ, Urbe S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63.
Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in Deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers (Basel) 2020;12:1579.
Shi L, Liu J, Peng Y, Zhang J, Dai X, Zhang S, et al. Deubiquitinase OTUD6A promotes proliferation of cancer cells via regulating Drp1 stability and mitochondrial fission. Mol Oncol. 2020;14:3169–83.
Wardlaw CP, Carr AM, Oliver AW. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst). 2014;22:165–74.
Sokka M, Parkkinen S, Pospiech H, Syvaoja JE. Function of TopBP1 in genome stability. Subcell Biochem. 2010;50:119–41.
Ma S, Cao C, Che SY, Wang YJ, Su DX, Liu S, et al. PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability. Sci Adv 2021;7:eabf7684.no.19
Liu T, Lin YH, Leng W, Jung SY, Zhang H, Deng M, et al. A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint. Mol Cell. 2014;56:681–95.
Leimbacher PA, Jones SE, Shorrocks AK, de Marco Zompit M, Day M, Blaauwendraad J, et al. MDC1 Interacts with TOPBP1 to Maintain Chromosomal Stability during Mitosis. Mol Cell. 2019;74:571–83 e8.
Bigot N, Day M, Baldock RA, Watts FZ, Oliver AW, Pearl LH. Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint. Elife 2019;8:e44353.
Feng H, Lu J, Song X, Thongkum A, Zhang F, Lou L, et al. CK2 kinase-mediated PHF8 phosphorylation controls TopBP1 stability to regulate DNA replication. Nucleic Acids Res. 2021;49:2400–1.
Kim W, Zhao F, Gao H, Qin S, Hou J, Deng M, et al. USP13 regulates the replication stress response by deubiquitinating TopBP1. DNA Repair (Amst). 2021;100:103063.
Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, et al. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem. 2002;277:3599–605.
Forma E, Krzeslak A, Bernaciak M, Romanowicz-Makowska H, Brys M. Expression of TopBP1 in hereditary breast cancer. Mol Biol Rep. 2012;39:7795–804.
Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, et al. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol Cell Biol. 2009;29:2673–93.
Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–81.
Peng B, Wang J, Hu Y, Zhao H, Hou W, Zhao H, et al. Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res. 2015;43:5936–47.
Hu B, Li S, Zhang X, Zheng X. HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res. 2014;42:5582–93.
Lim KS, Li H, Roberts EA, Gaudiano EF, Clairmont C, Sambel LA, et al. USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol Cell. 2018;72:925–41.e4.
Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–89.
Herhaus L, Perez-Oliva AB, Cozza G, Gourlay R, Weidlich S, Campbell DG, et al. Casein kinase 2 (CK2) phosphorylates the deubiquitylase OTUB1 at Ser16 to trigger its nuclear localization. Sci Signal. 2015;8:ra35.
Chen C, Wei X, Wang S, Jiao Q, Zhang Y, Du G, et al. Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation. Biochim Biophys Acta. 2016;1863(7 Pt A):1633–42.
Mehta S, McKinney C, Algie M, Verma CS, Kannan S, Harfoot R, et al. Dephosphorylation of YB-1 is required for nuclear localisation during G2 phase of the cell cycle. Cancers (Basel). 2020;12:315.
Peng Y, Liu J, Wang Z, Cui C, Zhang T, Zhang S, et al. Prostate-specific oncogene OTUD6A promotes prostatic tumorigenesis via deubiquitinating and stabilizing c-Myc. Cell Death Differ. 2022. https://doi.org/10.1038/s41418-022-00960-x.
Fu X, Zhao J, Yu G, Zhang X, Sun J, Li L, et al. OTUD6A promotes prostate tumorigenesis via deubiquitinating Brg1 and AR. Commun Biol. 2022;5:182.
Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006;124:943–55.
Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37.
Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol. 2011;193:267–73.
Yoshida K, Inoue I. Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene 2004;23:6250–60.
Liu K, Graves JD, Lee YJ, Lin FT, Lin WC. Cell Cycle-Dependent Switch of TopBP1 Functions by Cdk2 and Akt. Mol Cell Biol. 2020;40:e00599–19.
Acknowledgements
We sincerely thank Prof. Lingqiang Zhang for providing OTUD6A cDNA, Prof. Jiadong Wang for providing the Myc-TopBP1 plasmid, Prof. Shimin Zhao for providing Myc-UBR5 plasmids, and Prof. Daochun Kong for providing materials used in EMSA experiment. We thank the National Center for Protein Sciences at Peking University, particularly Liying Du, Huan Yang, Dong Liu, Qi Zhang, Hongxia Lv, LiQin Fu, and Guilan Li, for technical help. We also appreciate the assistance of Xiaochen Li and Siying Qin from the Core Facilities of Life Sciences at Peking University for their assistance with microscopic imaging. This work was supported by the National Natural Science Foundation of China (82130081 and 81730080) and the Natural Science Foundation of Beijing Municipality (5212008).
Author information
Authors and Affiliations
Contributions
YZ designed and performed the experiments, analyzed the data, and wrote the manuscript. XH, DZ, and JL performed the experiments. MW analyzed the pathway data and performed experiments. SY provided technical support for metaphase spread assays. YT performed the cancer cell injection into mice. XZ designed and supervised this study and wrote the manuscript.
Corresponding author
Ethics declarations
COMPETING INTERESTS
The authors declare no competing interests.
Ethical approval
All animal procedures were performed under the guidelines of the Ethics Committee of Peking University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by K. Engeland
Rights and permissions
About this article
Cite this article
Zhao, Y., Huang, X., Zhu, D. et al. Deubiquitinase OTUD6A promotes breast cancer progression by increasing TopBP1 stability and rendering tumor cells resistant to DNA-damaging therapy. Cell Death Differ 29, 2531–2544 (2022). https://doi.org/10.1038/s41418-022-01036-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01036-6
This article is cited by
-
Genomic and transcriptomic profiling of hepatocellular carcinoma reveals a rare molecular subtype
Discover Oncology (2024)
-
Deubiquitylase YOD1 regulates CDK1 stability and drives triple-negative breast cancer tumorigenesis
Journal of Experimental & Clinical Cancer Research (2023)
-
Deubiquitinase OTUD6A in macrophages promotes intestinal inflammation and colitis via deubiquitination of NLRP3
Cell Death & Differentiation (2023)
-
A BRCA2 germline mutation and high expression of immune checkpoints in a TNBC patient
Cell Death Discovery (2023)
-
A primary luminal/HER2 negative breast cancer patient with mismatch repair deficiency
Cell Death Discovery (2023)