Abstract
The NLRP3 (NOD−, LRR− and pyrin domain-containing protein 3) inflammasome plays a pivotal role in defending the host against infection as well as sterile inflammation. Activation of the NLRP3 inflammasome is critically regulated by a de-ubiquitination mechanism, but little is known about how ubiquitination restrains NLRP3 activity. Here, we showed that the membrane-bound E3 ubiquitin ligase gp78 mediated mixed ubiquitination of NLRP3, which inhibited NLRP3 inflammasome activation by suppressing the oligomerization and subcellular translocation of NLRP3. In addition, the endoplasmic reticulum membrane protein insulin-induced gene 1 (Insig-1) was required for this gp78–NLRP3 interaction and gp78-mediated NLRP3 ubiquitination. gp78 or Insig-1 deficiency in myeloid cells led to exacerbated NLRP3 inflammasome-dependent inflammation in vivo, including lipopolysaccharide-induced systemic inflammation and alum-induced peritonitis. Taken together, our study identifies gp78-mediated NLRP3 ubiquitination as a regulatory mechanism that restrains inflammasome activation and highlights NLRP3 ubiquitination as a potential therapeutic target for inflammatory diseases.
Similar content being viewed by others
Introduction
NLRP3 is an intracellular sensor that detects a broad range of microbial motifs, endogenous danger signals, and environmental irritants, resulting in the formation and activation of the NLRP3 inflammasome [1]. This inflammasome is composed of NLRP3, ASC, NEK7, and caspase-1. Upon activation, the NEK7–NLRP3 interaction increases, then NLRP3 oligomerizes and recruits ASC protein to nucleate helical ASC filament assembly, resulting in the formation of ASC speck. Assembled ASC recruits pro-caspase-1 and enables the auto-proteolytic activation of pro-caspase-1 into mature caspase-1, then caspase-1 processes inactive pro-IL-1β and pro-IL-18 into mature IL-1β and IL-18 [1]. Inappropriate activation of the NLRP3 inflammasome has been implicated in many inflammatory diseases, suggesting that this process needs to be tightly controlled.
A two-signal model, ‘priming’ signal and ‘activation’ signal, has been proposed for NLRP3 inflammasome activation. The priming signal serves for the expression of NLRP3 as well as pro-IL-1β, and the induction of posttranslational modifications (PTMs) of NLRP3, while the activation signal promotes inflammasome formation and full activation. Increasing evidence has shown that NLRP3 activation is tightly modulated by various PTMs, including phosphorylation and ubiquitination [1, 2]. For example, we previously reported that PKA kinase induces phosphorylation on Ser 291 and ubiquitination of NLRP3, which hinders activation of the inflammasome [3]. JNK1-mediated NLRP3 phosphorylation is critical for NLRP3 de-ubiquitination and facilitates its self-association and the subsequent inflammasome assembly [4].
Notably, the activation of NLRP3 inflammasome is tightly controlled by a de-ubiquitination process, during which the deubiquitinase BRCC3 promotes the de-ubiquitination of NLRP3 and is required for NLRP3 oligomerization and activation [5]. However, little is known about how NLRP3 acquires an ubiquitinated state in the resting stage to avoid unwanted inflammasome activation. Besides the diverse PTMs, NLRP3 activity is also tightly modulated by its subcellular translocation [6, 7], which ensures that the inflammasome is assembled and activated in the right place and at the right time. How NLRP3 executes this translocation and whether it is regulated by ubiquitination remain largely unknown.
gp78, a membrane-bound E3 ligase, usually in association with Insig-1, plays critical roles in diverse biological events, including cholesterol synthesis [8], antiviral immunity [9] etc., but its contribution to inflammation remain largely unknown. Here, we reveal a key role of gp78/Insig-1 in restraining NLRP3 inflammasome activation. Mechanistically, gp78 and Insig-1 mediate mixed ubiquitination of NLRP3, including K48- and K63-linked types, which inhibits NLRP3 oligomerization and subcellular translocation during inflammasome assembly.
This study suggests that the regulation of NLRP3 activity by ubiquitination might be a key step in avoiding unnecessary activation, indicating that NLRP3 ubiquitination is a promising target for treating NLRP3 inflammasome-dependent inflammatory diseases.
Results
Loss of gp78 enhances NLRP3 inflammasome activation in macrophages
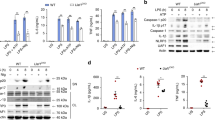
To investigate the role of gp78 in inflammasome activation, we first generated mice with myeloid cell-specific deletion of gp78 using the Lyz2-cre system (Lyz2-cre-gp78f/f). When treated with different NLRP3 agonists [ATP, nigericin, aluminum salts (alum), and monosodium urate crystals (MSU)], gp78-deficient peritoneal macrophages showed elevated IL-1β (Fig. 1A), significantly increased caspase-1 and IL-1β maturation (Fig. 1B) and increased IL-18 secretion (Fig. 1C) compared to control cells. Besides the release of IL-1β and IL-18, activation of the NLRP3 inflammasome also triggers pyroptotic cell death, which can be detected by the release of lactate dehydrogenase (LDH) [10] and we found increased LDH release in gp78-deficient macrophages (Fig. 1D). ASC nucleation-induced oligomerization is recognized as a common mechanism of NLRP3 inflammasome activation [11, 12]. Significantly increased ASC speck formation and ASC oligomerization were observed in the absence of gp78 (Fig. 1E and F). In contrast, activation of the NLRC4 and AIM2 inflammasomes, triggered by Salmonella typhimurium infection and poly (dA:dT) transfection, respectively, were not affected by gp78 depletion, suggesting the specific suppression of NLRP3 inflammasome activation by gp78 (Fig. 1G, H). Notably, the expression of NLRP3 inflammasome components and the secretion of tumor necrosis factor alpha (TNF-α) remained largely unaffected in response to gp78 depletion, indicating that gp78 deficiency does not affect the priming stage under these conditions (Figs. 1A, B, S1A). To further confirm the result, we checked the activation of the general signaling pathways mediated by TLR4, including the NF-κB and MAPK activation essential for TNF-α upon LPS stimulation and found gp78 deficiency in macrophages had little influence on LPS-induced NF-κB and MAPK activation (Fig. S1B). Consistent with the data from gp78 conditional knockout mouse, silencing of gp78 in mouse peritoneal macrophages by small-interfering RNAs (siRNAs) also showed similar results (Fig. S1C,S1D). These results indicate that gp78 specifically interferes with the activation step of the NLRP3 inflammasome without affecting the expression of its essential components.
A, B ELISA analysis of supernatant (SN) for IL-1β and TNF-α release (A, n = 4.), and immunoblots of SN (B) in lysates from peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice primed with LPS (500 ng/ml) for 4 h and treated with ATP (2 mM) or nigericin (10 μM) for 30 min and alum (300 μg/ml) or MSU (200 μg/ml) for 5 h. C, D ELISA analysis of IL-18 production (C) and LDH release (D) from SN of peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice primed with LPS (500 ng/ml) for 4 h and treated with ATP (2 mM) or nigericin (10 μM) for 30 min and alum (300 μg/ml) or MSU (200 μg/ml) for 5 h. N = 4. E Arrows showing ASC specks immunofluorescence in bone-marrow-derived macrophages from gp78f/f and Lyz2-cre-gp78f/f mice primed with LPS (500 ng/ml) for 4 h followed by treatment with 10 μM nigericin for 45 min (scale bar, 20 μm) (left). Quantification of ASC speck formations (right). F Immunoblots of ASC oligomerization in cross-linked cytosolic pellets from peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice primed with LPS (500 ng/ml) for 4 h followed by treatment with 10 μM nigericin for 45 min. G, H ELISA analysis of SN for IL-1β release (G, n = 3) and immunoblots of SN (H) in lysates of peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice primed with LPS (500 ng/ml) for 4 h followed by treatment with Salmonella or poly(dA:dT) transfection for 1 h. *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SD (A, C, D, G), mean ± SEM (E). Data are representative of three independent experiments.
As an E3 ubiquitin ligase, gp78 plays a sophisticated role in cholesterol biosynthesis and ablation of gp78 in the liver leads to decreased lipid biosynthesis [13]. To determine whether the enhanced NLRP3 activity in gp78-depleted macrophages is induced by a disturbance of cholesterol homeostasis, we supplemented wild-type and gp78-deficient macrophages with different doses of methyl-β-cyclodextrin (MCD)-cholesterol. Consistent with our previous study [14], MCD-cholesterol dose-dependently inhibited NLRP3 inflammasome activation (Fig. S1E). Notably, we found that MCD-cholesterol showed a similar kinetic pattern in both cells (Fig. S1E), suggesting that gp78 restrains NLRP3 activity directly, rather than through affecting cholesterol homeostasis.
Macrophage-specific gp78 transgenic mice show impaired NLRP3 inflammasome activation
To further confirm the role of gp78 in suppressing NLRP3 inflammasome activation, we adopted different gain-of-function strategies. First, lentiviral transduction of immortalized mouse macrophages (iBMDMs) with gp78 significantly inhibited IL-1β secretion (Fig. 2A), IL-1β and caspase-1 maturation (Fig. 2B) in response to LPS plus ATP stimulation. The TNF-α production remained largely unaffected (Fig. S2A). Furthermore, we generated gp78 transgenic mice (CD68-gp78 mice) with macrophage-specific transgenic expression of the mouse gp78 gene using CD68 promoter (Fig. S2B,S2C) and found that gp78 expression was markedly increased in the peritoneal macrophages of these mice (Fig. 2D). Peritoneal macrophages from CD68-gp78 mice showed impaired inflammasome activation compared with control mice, as evidenced by decreased IL-1β secretion (Fig. 2C), as well as suppressed IL-1β and caspase-1 maturation (Fig. 2D). gp78 transgenic macrophages also showed inhibited ASC oligomerization and ASC speck formation (Fig. 2E, F). Collectively, these results further confirmed the role of gp78 in restraining NLRP3 inflammasome activation.
A, B ELISA analysis of SN for IL-1β release (A, n = 4) and immunoblots of SN (B) in lysates of LPS-primed wild-type and gp78-overexpressing iBMDMs treated with ATP (2 mM) for 30 min. C, D ELISA analysis of SN for IL-1β and TNF-ɑ release (C, n = 4) and immunoblots of SN (D) in lysates of peritoneal macrophages from wild-type and CD68–gp78 mice primed with LPS (500 ng/ml) for 4 h followed by treatment with ATP (2 mM) or nigericin (10 μM) for 30 min. E Immunoblots of ASC oligomerization in cross-linked cytosolic pellets of peritoneal macrophages from wild-type and CD68-gp78 mice primed with LPS (500 ng/ml) for 4 h followed by treatment with nigericin (10 μM) for 30 min. F Arrows showing ASC specks immunofluorescence in BMDMs from wild-type and CD68-gp78 mice primed with LPS (500 ng/ml) for 4 h followed by treatment with 10 μM nigericin for 1.5 h (scale bar, 20 μm) (left). Quantification of ASC speck formations (right). *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SD (A, C), mean ± SEM (F). Data are representative of three independent experiments.
It was reported that gp78 is an essential E3 ligase in regulating Endoplasmic Reticulum Associated Degradation [15] and gp78 deficiency would lead to ER stress [16]. Moreover, previous study has shown that ER stress promotes the activation of NLRP3 inflammasome [17]. To check whether the elevated NLRP3 inflammasome activation in gp78-deficient macrophages is an indirect result of ER stress, we detected the markers of ER stress in WT and gp78-deficient macrophages. Consistent with previous reports [17,18,19], we found that compared with thapsigargin (TG, ER stress-inducing agent) treatment, LPS or LPS plus ATP stimulation minimally induced ER stress in WT macrophages (Figs. S2D–2F). Moreover, gp78-deficient macrophages did not exhibit significantly altered expression of ER stress-associated genes in response to LPS or LPS plus ATP treatment (Figs. S2D–2F). However, we noted that gp78 deficiency significantly promoted thapsigargin-induced expression of unfolded protein response-target genes, such as Dnajb9 (ERdj4), Hspa5 (Bip), Ddit3 (CHOP), and the splicing of Xbp1 (Figs. S2D–2F). These results indicate that in our experimental conditions, LPS or LPS plus ATP stimulation minimally induced ER stress, and that gp78 deficiency does not exhibit significantly altered expression levels of ER stress-associated genes in response to LPS or LPS plus ATP treatment. Taken together, the enhanced NLRP3 inflammasome activation in gp78-deficient macrophages is not an indirect result of ER stress.
gp78 interacts with NLRP3 via its CUE (coupling of ubiquitin to ER degradation) domain
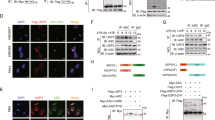
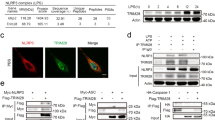
Next, we investigated the mechanism by which gp78 suppresses NLRP3 activity. By co-immunoprecipitation assays, we found that gp78 was associated with NLRP3 in LPS-primed peritoneal macrophages (Fig. 3A). Confocal microscopy also showed co-localization of gp78 and NLRP3 in LPS-primed peritoneal macrophages, LPS-primed THP-1 cells, and HEK293T cells overexpressing HA-tagged gp78 and Flag-tagged NLRP3 (Figs. 3B, S3A, S3B). Moreover, overexpression of gp78 and the components of the NLRP3 inflammasome in HEK293T cells showed that gp78 specifically interacted with NLRP3 (Fig. 3C, D).
A Immunoblots of lysates from wild-type peritoneal macrophages primed with LPS (500 ng/ml) for 4 h immunoprecipitated with anti-gp78 antibody or anti-rabbit IgG and then immunoblotted with the indicated antibodies. B Confocal microscopy images of BMDMs with or without LPS showing co-localization of NLRP3 (green) with gp78 (red) (scale bar, 5 μm). C Immunoblots of lysates immunoprecipitated with anti-Flag antibody and then with the indicated antibodies from HEK293T cells 24 h after transfection with HA-tagged NLRP3 and Flag-tagged gp78. D HA-tagged NLRP3 inflammasome components were individually transfected into HEK293T cells with Myc-tagged gp78. 24 h after transfection, cell lysates were immunoprecipitated with anti-HA antibody or anti-Mouse IgG and then immunoblotted with the indicated antibodies. E Schematic diagram of gp78 and its truncated mutants. F Myc-tagged gp78 or its truncated mutants were individually transfected into HEK293T cells with Flag-tagged NLRP3. 24 h after transfection, cell lysates were immunoprecipitated with anti-Myc antibody or anti-Mouse IgG and then immunoblotted with the indicated antibodies. Data are representative of three independent experiments.
To further delineate how gp78 interacts with NLRP3, we introduced a series of truncated mutants of both proteins according to their domain structures (Figs. 3E,S3C). We found that the CUE domain of gp78 was essential for its interaction with NLRP3 (Fig. 3F). Both NACHT and LRR domains of NLRP3 were involved in its interaction with gp78 (Fig. S3D). Altogether, these data suggest that both NACHT and LRR domains of NLRP3 and the CUE domain of gp78 are essential for the gp78–NLRP3 association.
gp78 mediates mixed ubiquitination of NLRP3
Recent studies have shown that the de-ubiquitination of NLRP3 is necessary for NLRP3 inflammasome activation [5, 20], suggesting that ubiquitination might be a key brake on NLRP3 activity. As E3 ubiquitin ligase, gp78 mediates the ubiquitination and degradation of target proteins such as HMG-CoA reductase [8] and the tumor metastasis suppressor KAI1 [21], etc. We next explored whether gp78 controls NLRP3 inflammasome activation via its E3 ligase activity. It has been reported that the Cys356 in the RING domain of gp78 is critical for its E3 ligase activity, so we generated two gp78 mutants, gp78 C356G (Cys356 mutation to Gly356) and gp78 ΔRING, both of which were deprived of E3 ubiquitin ligase activity [8]. Lentiviral transduction of Lyz2-cre-gp78f/f BMDMs with wild-type gp78 significantly inhibited caspase-1 activation and IL-1β production while the E3 ligase-dead mutant gp78 C356G failed (Fig. 4A, B), suggesting that gp78 restrains NLRP3 activation via its E3 ligase activity. In contrast, TNF-ɑ production was not affected in these experiments (Fig. S4A). We further investigated whether gp78 could affect the ubiquitination of NLRP3. LPS-induced NLRP3 ubiquitination was substantially decreased in gp78-depleted macrophages while it was increased in peritoneal macrophages from CD68-gp78 transgenic mice (Fig. 4C, D). Notably, the protein level of NLRP3 remained unchanged in peritoneal macrophages from gp78-deficient or transgenic mice (Fig. 4C, D). Consistent with these in vitro data, we found increased ubiquitination of NLRP3 in wild-type peritoneal macrophages upon LPS challenge in vivo (Fig. 4E), while this LPS-induced NLRP3 ubiquitination was strikingly downregulated in peritoneal macrophages from Lyz2-cre-gp78f/f mice (Fig. 4E). Moreover, overexpression of gp78 increased the ubiquitination of NLRP3 in a dose-dependent manner in HEK293T cells overexpressing Flag-tagged NLRP3 and HA-tagged ubiquitin (Fig. 4F), while the E3 ligase-dead mutants gp78 ΔRING and gp78 C356G failed (Fig. 4G). We also performed in vitro ubiquitination assays and confirmed that gp78 directly catalyzed the formation of ubiquitin chains attached to NLRP3, whereas gp78 C356G failed (Fig. 4H). In contrast, another ER-resident E3 ubiquitin ligase, Hrd1, which exhibits considerable homology to gp78 and plays critical roles in ER-associated degradation [22], did not catalyze the ubiquitination of NLRP3 (Fig. 4G).
A, B BMDMs from Lyz2-cre-gp78f/f mice were lentivirally transduced with Phage empty vector, Phage-FLAG gp78 or Phage-FLAG-gp78 C356G, then primed with 500 ng/ml LPS for 4 h followed by stimulating with ATP (2 mM) or nigericin (10 μM) for 30 min. Supernatants (SN) were analyzed by ELISA for IL-1β release (A, n = 4). SNs and cell extracts (Lysate) were also analyzed by immunoblotting (B). C Peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice were primed with or without LPS (500 ng/ml) for 4 h. Cell lysates were immunoprecipitated with anti-NLRP3 antibody or anti-mouse IgG and then immunoblotted with the indicated antibodies. D Peritoneal macrophages from wild-type and CD68-gp78 mice were primed with LPS (500 ng/ml) for 4 h. NLRP3 of peritoneal macrophages was immunoprecipitated for ubiquitination analysis. E gp78f/f and Lyz2-cre-gp78f/f mice were intraperitoneally injected with or without LPS (20 mg/kg body weight) for 4 h. NLRP3 protein of peritoneal cells was immunoprecipitated for ubiquitination analysis. F HEK293T cells were transfected with Flag-tagged NLRP3, HA-tagged Ub, along with different doses of Myc-tagged gp78. Cell lysates were immunoprecipitated with anti-Flag antibody and then immunoblotted with the indicated antibodies. G HEK293T cells were transfected with Flag-tagged NLRP3 and HA-tagged Ub, along with Myc-tagged gp78 and its catalytically inactive mutants (C356G) or Myc-tagged Hrd1. Cell lysates were immunoprecipitated with anti-Flag antibody and then immunoblotted with the indicated antibodies. H The in vitro ubiquitination reaction mixture contains E1, E2, Ub, GST-NLRP3 ΔLRR, and gp78 or gp78 C356G as indicated. After incubation at 30 °C for 30 min, the mixture was assessed by immunoblotting with the indicated antibodies. I HEK293T cells expressing Flag-tagged NLRP3, Myc-tagged gp78, and HA-tagged Ub, HA-tagged K48-Ub, or HA-tagged K63-Ub were immunoprecipitated with anti-Flag antibody for ubiquitination. J HEK293T cells expressing Flag-tagged NLRP3, HA-tagged Ub, and Myc-tagged gp78 or V5-tagged BRCC3 as indicated were immunoprecipitated with anti-Flag antibody for ubiquitination. K HEK293T cells expressing Flag-tagged NLRP3, Myc-tagged gp78, HA-tagged Ub, or HA-tagged K48/K63R Ub as indicated were immunoprecipitated with anti-Flag antibody for ubiquitination. *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SD (A). Data are representative of three independent experiments.
NLRP3 has been reported to be modified by a combination of K48- and K63-linked ubiquitination [5] and we found that both K48- and K63-linked ubiquitin chains were involved in gp78-mediated NLRP3 ubiquitination (Fig. 4I). Importantly, we found that BRCC3, a deubiquitinase essential for NLRP3 inflammasome activation [5], de-ubiquitinated this gp78-mediated ubiquitination of NLRP3 (Fig. 4J). Interestingly, when co-expressed with an ubiquitin mutant containing Lys48-to-Arg48 and Lys63-to-Arg63 mutation (Ub K48/K63R), gp78 also catalyzed evident NLRP3 ubiquitination (Fig. 4K), indicating that other types of ubiquitination may also be involved in this gp78-mediated suppression of NLRP3 activity. In line with this idea, we performed mass spectrometry and found that, besides K48- and K63-linked ubiquitination, K6- and K11-linked ubiquitination of NLRP3 also occurred in HEK293T cells overexpressing gp78 (Fig. S4B). In addition, we also found that the ubiquitination sites were mainly located on the NACHT and adjacent Linker1, Linker 2 domains (Fig. S4C). Altogether, these data indicated that gp78 mediates a mixed ubiquitination of NLRP3, which inhibits NLRP3 activity.
gp78 suppresses NLRP3 oligomerization and subcellular translocation during inflammasome activation
We next explored how the gp78-mediated NLRP3 ubiquitination inhibited NLRP3 inflammasome activation. NEK7–NLRP3 interaction, NLRP3 oligomerization, the recruitment of ASC to NLRP3 oligomers, and subsequent ASC speck formation are recognized to be critical steps for the formation of NLRP3 inflammasome complexes [23,24,25,26,27] (Fig. S5A). We found that ASC oligomerization and ASC specks were enhanced in gp78-depleted macrophages (Fig. 1E, F). To determine whether gp78 affected ASC self-association or NLRP3-dependent ASC oligomerization, we reconstituted the inflammasome in HEK293T cells by transfection of pro-IL-1β, pro-caspase-1, ASC, and NLRP3. Previous studies have shown that the expression of ASC leads to IL-1β secretion and the addition of NLRP3 enhances this secretion [28]. Our data showed that expression of gp78 inhibited NLRP3-dependent, rather than NLRP3-independent, ASC-dependent IL-1β release (Fig. 5A), suggesting that gp78 interferes with NLRP3-dependent ASC oligomerization. Consistent with these data, we found that the NLRP3–ASC association was markedly increased in gp78-deficient macrophages while it was decreased in CD68-gp78 macrophages (Fig. 5B, C). NEK7–NLRP3 interaction and NLRP3 oligomerization are upstream events during inflammasome assembly [1], so we next explored which step was affected by gp78 to inhibit NLRP3-dependent ASC oligomerization. Our data showed that gp78 had no influence on the NEK7–NLRP3 interaction (Fig. 5D). In contrast, NLRP3 oligomerization was significantly enhanced in gp78-deficient macrophages while it was dramatically impaired in CD68-gp78 macrophages (Fig. 5E, F), suggesting that gp78 inhibits NLRP3 oligomerization to interfere with subsequent NLRP3-dependent ASC oligomerization.
A Direct inhibition by gp78 of the reconstituted NLRP3 inflammasome. HEK293T cells were transfected with plasmids expressing the indicated components and IL-1β release was measured. Data are shown as the level of IL-1β measured for the fully-reconstituted NLRP3 inflammasome with or without gp78. N = 4. B Peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice were primed with LPS (500 ng/ml) for 4 h followed by ATP (2 mM) stimulation for 30 min. Cell lysates were immunoprecipitated with anti-ASC antibody or anti-Rabbit IgG and then immunoblotted with the indicated antibodies. C Peritoneal macrophages from wild-type and CD68-gp78 mice were primed with LPS (500 ng/ml) for 4 h followed by ATP (2 mM) stimulation for 30 min. Cell lysates were immunoprecipitated with anti- ASC antibody or anti-mouse IgG and then immunoblotted with the indicated antibodies. D Peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice were primed with LPS (500 ng/ml) for 4 h followed by nigericin (10 μM) stimulation for 30 min. Cell lysates were immunoprecipitated with anti-NEK7 antibody or anti-rabbit IgG and then immunoblotted with the indicated antibodies. E Peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice were primed with LPS (500 ng/ml) for 4 h followed by nigericin (10 μM) stimulation for 30 min, and then underwent immunoblot analysis of NLRP3 by SDD-AGE or SDS-PAGE assay. F Peritoneal macrophages from wild-type and CD68-gp78 mice were primed with LPS (500 ng/ml) for 4 h followed by nigericin (10 μM) stimulation for 30 min, and then underwent immunoblot analysis of NLRP3 by SDD-AGE or SDS-PAGE assay. G Peritoneal macrophages from gp78f/f and Lyz2-cre-gp78f/f mice were primed with LPS (500 ng/ml) for 4 h followed by nigericin (10 μM) stimulation for the indicated time. Then mitochondrial and cytosolic fractions were subjected to immunoblot. H Left panels, super-resolution and 3D-structured illumination microscopy (SIM) imaging in LPS-Primed BMDMs from gp78f/f and Lyz2-cre-gp78f/f mice stimulated with ATP (2 mM) for 30 min with staining for NLRP3 (green), and mitochondria (red) (scale bars, 1 μm). Right panel, quantification of Pearson’s coefficient for co-localization volume. *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SD (A), mean ± SEM (H). Data are representative of three independent experiments.
In addition to the requirement for interactions among NLRP3 inflammasome components, NLRP3 inflammasome assembly and activation also require the subcellular translocation of NLRP3 [6]. Upon activation, NLRP3 is translocated to other subcellular compartments, such as mitochondria [29], the Golgi complex [14, 30], and the microtubule-organizing center [31] for subsequent assembly and activation. However, little is known about the mechanisms that regulate this translocation. Modification of proteins with ubiquitin chains regulates a variety of signaling pathways through proteasomal/autophagic degradation or by altering protein activity and/or localization [5]. We found that gp78 deficiency led to the accumulation of NLRP3 in mitochondria and a decreased NLRP3 level in the cytosol upon Nigericin stimulation (Fig. 5G). This result indicated that gp78-mediated ubiquitination of NLRP3 might inhibit its translocation to mitochondria during inflammasome activation. Consistently, using super-resolution and 3D-structured illumination microscopy, we also found increased localization of NLRP3 with mitochondria in gp78-deficient cells upon inflammasome activation (Fig. 5H). Altogether, these data showed that gp78-mediated ubiquitination of NLRP3 might interfere with NLRP3 self-association and its translocation to mitochondria upon inflammasome activation, resulting in suppressed NLRP3 inflammasome assembly and activation.
Insig-1 is required for gp78-mediated NLRP3 ubiquitination
Under certain circumstances, gp78 mediates substrate ubiquitination in association with Insig-1 [32]. For example, gp78 interacts with Insig-1 to promote the sterol-triggered degradation of HMG-CoA reductase [8]. Insig-1 facilitates gp78 to associate with and ubiquitinate STING, thus potentiating STING signaling [9]. gp78-Insig mediates the ubiquitination and degradation of ACAT2, fine-tuning lipid homeostasis [33]. To explore whether Insig-1 is involved in the gp78-mediated suppression of NLRP3 activity, we generated Insig-1 conditional knockout mice with myeloid cell-specific deletion of Insig-1 (Lyz2-cre-Insig1f/f). The gene knockout efficiency was confirmed by RT-PCR analysis (Fig. S5B). We found that the association between gp78 and NLRP3 was evidently decreased in Insig-1-depleted macrophages (Fig. 6A). Moreover, the interaction between gp78 and NLRP3 was markedly enhanced in the presence of Insig-1 (Fig. 6B), suggesting that Insig-1 promotes the interaction between gp78 and NLRP3.
A Peritoneal macrophages from Insig1f/f or Lyz2-cre-Insig1f/f mice were primed with LPS (500 ng/ml) for 4 h and cell lysates were immunoprecipitated with anti-gp78 antibody or anti-rabbit IgG and then immunoblotted with the indicated antibodies. B HEK293T cells expressing Flag-tagged NLRP3 and Myc-tagged gp78 or HA-tagged Insig-1 were immunoprecipitated with anti-Myc antibody and then immunoblotted with the indicated antibodies. C Peritoneal macrophages from Insig1f/f or Lyz2-cre-Insig1f/f mice were primed with LPS (500 ng/ml) for 4 h. Cell lysates were immunoprecipitated with anti-NLRP3 antibody or anti-Mouse IgG for ubiquitination and then immunoblotted with the indicated antibodies. D, E ELISA analysis of SN for IL-1β and TNF-α production (D, n = 4) and immunoblots of SN (E) from lysates. Peritoneal macrophages from Insig1f/f or Lyz2-cre-Insig1f/f mice were primed with LPS (500 ng/ml) for 4 h followed by treatment with ATP (2 mM) or nigericin (10 μM) for 30 min. *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SD (D). Data are representative of three independent experiments.
Given that Insig-1 is required for the gp78–NLRP3 association, we explored whether Insig-1 deficiency affects the ubiquitination of NLRP3. Our data showed that this ubiquitination was significantly reduced in Insig-1-depleted macrophages, suggesting that Insig-1 is required in gp78-mediated NLRP3 ubiquitination (Fig. 6C). Moreover, activation of the NLRP3 inflammasome was markedly enhanced in Insig-1-deficient macrophages, as evidenced by elevated IL-1β secretion vs. control cells, without affecting TNF-α production (Fig. 6D) and the increased IL-1β and caspase-1 maturation (Fig. 6E). Collectively, these data suggest that Insig-1 suppresses inflammasome activation via facilitating the gp78-mediated ubiquitination of NLRP3.
gp78 and Insig-1 restrain NLRP3 inflammasome-dependent inflammatory diseases in vivo
We finally investigated whether gp78 and Insig-1 inhibit NLRP3 inflammasome activation in vivo. First, when challenged with intraperitoneally-injected LPS, Lyz2-cre-gp78f/f mice showed significantly increased serum IL-1β and IL-18 levels (Fig. 7A, B) while CD68-gp78 mice displayed significantly reduced serum IL-1β and IL-18 secretion (Fig. 7C, D). The increased TNF-α in Lyz2-cre-gp78f/f mice were probably due to the contribution of IL-1β to TNF-α production in vivo (Fig. 7A) while the TNF-α production were not affected in CD68-gp78 mice (Fig. 7C). To further confirm the inhibitory role of gp78 on NLRP3 inflammasome in vivo, we performed a MCC950 (a small molecule inhibitor of the NLRP3 inflammasome) [34] supplementation experiment and the data showed that the elevated serum IL-1β by gp78 deficiency was deminished by MCC950 treatment (Fig. S5C), indicating that the in vivo model is dependent on NLRP3 and that gp78 is involved in NLRP3-related pathways. Moreover, in an LPS-induced sepsis model, the data showed a decreased survival rate in Lyz2-cre-gp78f/f mice and an increased survival rate in CD68-gp78 mice (Fig. 7E, F). In addition, Lyz2-cre-Insig1f/f mice showed markedly upregulated serum IL-1β and barely affected TNF-α production upon LPS challenge, similar to those in Lyz2-cre-gp78f/f mice (Fig. 7G).
A ELISA of IL-1β and TNF-ɑ in serum from gp78f/f and Lyz2-cre-gp78f/f mice 4 h after intraperitoneal injection of LPS (25 mg/kg body weight). Data are the mean ± SEM (gp78f/f, n = 10; Lyz2-cre-gp78f/f, n = 10). B ELISA of IL-18 in serum from gp78f/f and Lyz2-cre-gp78f/f mice 4 h after intraperitoneal injection of LPS (25 mg/kg body weight). Data are the mean ± SEM (gp78f/f, n = 12; Lyz2-cre-gp78f/f, n = 13). C ELISA of IL-1β and TNF-ɑ in serum from wild-type and CD68–gp78 mice 4 h after intraperitoneal injection of LPS (25 mg/kg body weight). Data are the mean ± SEM (n = 16). D ELISA of IL-18 in serum from wild-type and CD68–gp78 mice 4 h after intraperitoneal injection of LPS (25 mg/kg body weight). Data are the mean ± SEM (wild-type, n = 8; CD68–gp78, n = 9). E Survival rates of gp78f/f and Lyz2-cre-gp78f/f mice injected with LPS (20 mg/kg body weight) (gp78f/f, n = 12; Lyz2-cre-gp78f/f, n = 11). F Survival rate of wild-type and CD68–gp78 mice injected with LPS (20 mg/kg body weight, n = 8). G ELISA of IL-1β and TNF-ɑ in serum from Insig1f/f and Lyz2–cre–Insig1f/f mice 4 h after intraperitoneal injection of LPS (25 mg/kg body weight). Data are the mean ± SEM (n = 13). H ELISA of IL-1β in the peritoneal cavity of gp78f/f and Lyz2-cre-gp78f/f mice intraperitoneally injected with alum (2 mg/mouse). Data are the mean ± SEM (n = 8). I ELISA of IL-1β in the peritoneal cavity of wild-type and CD68–gp78 mice intraperitoneally injected with alum (2 mg/mouse). Data are the mean ± SEM (n = 5). J FACS analysis of PECs (peritoneal exudate cells) and neutrophil numbers in the peritoneal cavity of gp78f/f and Lyz2–cre–gp78f/f mice intraperitoneally injected with alum (2 mg/mouse). Data are the mean ± SEM (gp78f/f, n = 6; Lyz2-cre-gp78f/f, n = 5). K FACS analysis of PECs and neutrophil numbers in the peritoneal cavity of wild-type and CD68–gp78 mice intraperitoneally injected with alum (2 mg/mouse). Data are the mean ± SEM (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001. Values are mean ± SEM.
We then addressed the role of gp78 in NLRP3 inflammasome activation in vivo using a model of alum-induced peritonitis. The data showed that IL-1β secretion in the lavage fluid was up-regulated in Lyz2-cre-gp78f/f mice (Fig. 7H) while it was downregulated in CD68-gp78 mice (Fig. 7I). Consistent with this, the peritoneal exudate cells and neutrophils recruited upon alum challenge were significantly increased in Lyz2-cre-gp78f/f mice (Fig. 7J) and decreased in CD68-gp78 mice (Fig. 7K). Collectively, these data suggest an immunosuppressive effect of gp78/Insig-1 during inflammatory responses in vivo, at least in the models of sepsis and peritonitis we explored.
Discussion
Due to the conserved biological function of the NLRP3 inflammasome in various physiological processes, fine-tuning of its activity is critical for the prevention of related diseases and the maintenance of immune homeostasis. The multi-level, fine-tuned regulation of NLPR3 inflammasome activation is ensured by different mechanisms at different stages. In the resting stage, low expression of NLRP3 ensures that NLRP3 inflammasome assembly is hardly induced. In the LPS-primed stage, NLRP3 expression is upregulated and its ubiquitination is used to limit its activity. The activation of NLRP3 inflammasome is restricted to an appropriate intensity and time course to avoid harmful effects.
Regarding the regulation of the inflammasome, one critical strategy is to control the protein level of NLRP3 during inflammasome activation. For example, E3 ligase TRIM31 directly binds to NLRP3, promoting the K48-linked ubiquitination and proteasomal degradation of NLRP3, which maintains the low expression of NLRP3 and prevents unwanted inflammasome activation in both resting and activation states [35]. Dopamine has been reported to inhibit NLRP3 inflammasome activation via the dopamine D1 receptor by promoting the MARCH7-dependent K48-linked ubiquitination and autophagic degradation of NLRP3 [36]. However, when encountering emergencies, the NLRP3 inflammasome needs to be assembled and activated instantly to respond to possible dangers. This time- and energy-consuming regulation of degradation has a fundamental effect on the activation of inflammasomes. Thus, fine-tuning NLRP3 activity for a quick response to potential threats may be needed.
Another way to regulate NLRP3 activity is to maintain its inactive state. Our study demonstrates that the E3 ubiquitin ligase gp78 restrains NLRP3 activity by suppressing its oligomerization and subcellular translocation. We found that, in HEK293T cells overexpressing gp78, the ubiquitinated sites are mainly located on the NACHT and its adjacent Linker1 and Linker2 domains (Fig. S4C), while interactions between NACHT domains have been reported to be necessary for NLRP3 oligomerization, suggesting that gp78 might inhibit NLRP3 oligomerization through interfering with the interactions between NACHT domains. In addition to the inhibition of NLRP3 oligomerization by gp78, we found that gp78 suppressed the subcellular translocation of NLRP3 during inflammasome activation. We speculate that when NLRP3 is ubiquitinated by gp78, it may undergo a conformational change or the ubiquitin chains on NLRP3 may serve as a platform to recruit some unknown proteins, which inhibit NLRP3 translocation to mitochondria resulting in decreased inflammasome activation. Compared with the regulation of degradation, this gp78-mediated NLRP3 ubiquitination keeps NLRP3 in check by restricting its activity without degrading the protein, enabling its rapid turnover for inflammasome assembly when required.
Besides the K48- and K63-linked ubiquitination, we found that gp78 mediated K6- and K11-linked ubiquitination of NLRP3. It is generally known that K48-linked ubiquitination regulates protein degradation, while K63-linked ubiquitination regulates protein trafficking and signaling [37]. Our data showed that gp78 restrains NLRP3 activity by inhibiting its self-association and subcellular translocation. A recent study showed that the E3 ligase HUWE1 mediates the K27-linked ubiquitination of NLRP3, which promotes NLRP3 oligomerization and subsequent inflammasome assembly [38]. Collectively, these findings suggest that the complex ubiquitin linkages on NLRP3 regulate the activation of the inflammasome, which may lead to distinct consequences in response to stimuli.
In conclusion, we revealed a previously unknown role for the E3 ubiquitin ligase gp78 in macrophages as an important suppressor of NLRP3 inflammasome activation. The NLRP3 inflammasome must be efficiently activated to defend the host from infection as well as sterile inflammation, but activation must be controlled to prevent hyperinflammation and its deleterious consequences.
Experimental model and subject details
Mice
C57BL/6 mice were purchased from the Model Animal Research Center of Nanjing University. gp78f/f and Insig1f/f mice were kindly provided by Prof. Bao-Liang Song of Wuhan University. Lyz2-cre mice were purchased from the Jackson Laboratory. All mice were on the C57BL/6 background. gp78f/f and Insig1f/f mice were separately crossed with Lyz2-cre mice to obtain Lyz2-cre-gp78f/f and Lyz2-cre-Insig1f/f mice. gp78 transgenic mice (CD68-gp78) were generated by GemPharmatech Co. Ltd. Fragments of Human CD68 promoter, IVS-1 enhancer, gp78 CDS, 3xFlag, and rabbit globin polyA were amplified by pfu DNA polymerase and ligated to the pMD18-T vector by SLIC after confirmation by PCR, enzyme digestion, and sequencing. The target region [the promoter of the gene encoding human CD68 and the macrophage-specific IVS-1 enhancer were upstream of the sequence encoding mouse gp78, and a Kozak sequence (GCCGCCACC) was added before the ATG; sequences encoding 3xFLAG epitope and poly A were added after gp78] on the final vector was released by Pvu I restriction enzyme digestion and confirmed by PCR, enzyme digestion, and sequencing before delivery into zygotes. Nine lines were initially analyzed, and subsequently a single line was used for most experiments.
Cells
Mouse BMDMs were generated as previously described. Briefly, cells were flushed from tibias and femurs with cold Dulbecco’s modified Eagle’s medium (DMEM) and were cultured in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 10 ng/ml macrophage colony-stimulating factor (M-CSF, PeproTech) to generate BMDMs.
Mouse peritoneal macrophages were collected 4 days after thioglycolate (Merck) injection. Immortalized mouse macrophages (iBMDMs) were kindly provided by Prof. Feng Shao (National Institute of Biological Sciences, Beijing). THP-1 cells were cultured in RPMI 1640 containing the same supplements. THP-1 cells were differentiated for 4 h with 100 nM phorbol myristate and re-plated.
For stimulation, 5 × 105 cells were plated in 12-well plates overnight. The medium was changed to opti-MEM the next day and then the cells were primed with LPS (500 ng/ml) for 4 h. After that, cells were stimulated with various NLRP3 activators (2 mM ATP for 30 min; 10 µM nigericin for 30 min; 200 μg/ml MSU for 5 h; 300 μg/ml alum for 5 h). For AIM2 inflammasome activation, poly(dA:dT) (1 µg/ml) was transfected using Lipofectamine 2000 following the manufacturer’s protocol (Invitrogen). For NLRC4 inflammasome activation, S. typhimurium was grown overnight in Luria-Bertani broth, and peritoneal macrophages were infected for 1 h with the Salmonella culture (1:100) and then incubated for another 1 h in the presence of gentamycin.
Reagents
ATP, LPS from Escherichia coli O111: B4, phorbol myristate, and MCD-cholesterol were from Sigma; nigericin, MSU, and poly(dA:dT) were from Invivogen; alum was from Thermo Fisher Scientific.
The antibody against pro-IL-1β was from R&D systems (AF-401-NA); anti-mouse caspase-1 (AG-20B-0042), anti-NLRP3 (AG-20B-0014), and anti-ASC (AG-25B-0006) were from AdipoGen; anti-gp78 (16675-1-AP), anti-NEK7 (EPR4900), anti-Insig-1 (55282-1-AP), anti-mouse IgG (B900620), anti-V5 (66007-1-Ig), anti-His (66005-1-Ig), and anti-Myc (60003-2-Ig) were from Proteintech; anti-Ub (sc-8017) was from Santa Cruz; anti-Flag antibodies were separately from Proteintech (20543-1-AP) and MBL (M185-3LL); anti-HA antibodies were separately from Thermo Fisher Scientific (26183) and MBL (M180-3); anti-β-actin (M1210-2), anti-β-tubulin (EM0103), anti-TOMM20 (ET1609-25), and anti-VDAC1 (ET1601-20) were from HuaAn Biotechnology Co., Ltd; anti-rabbit IgG (2729 P) was from Cell Signaling Technology; anti-Flag M2 magnetic beads (M8823) were from Sigma; Protein A/G (HY-K0202) beads were from MCE.
Method details
Transfection and co-immunoprecipitation
Briefly, plasmids were transfected into HEK293T cells using polyethyleneimine. After 24 h, the cells were collected and re-suspended in lysis buffer (50 mM Tris-HCl, 5 mM EDTA, 150 mM NaCl, 0.5% (vol/vol) Nonidet-P40, and 10% (vol/vol) glycerol, pH 7.4) supplemented with 1 mM PMSF and complete protease inhibitor cocktail. Cell lysates were immunoprecipitated with the indicated antibody and beads overnight at 4 °C with rotation. Immunocomplexes were washed three times in lysis buffer the following day, resolved by SDS-PAGE, and analyzed by western blotting.
LPS-primed peritoneal macrophages were re-suspended in SDS lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% (vol/vol) Nonidet-P40, 2% SDS, and 10% (vol/vol) glycerol) and then heated at 100 °C for 10 min followed by 10× dilution with lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% (vol/vol) Nonidet-P40, and 10% (vol/vol) glycerol). Proteins were immunoprecipitated from cell lysates with anti-NLRP3 antibody and then the ubiquitination of NLRP3 was assessed by western blot analysis.
Immunofluorescence staining and confocal microscopy
Cells were primed with LPS for 4 h, then fixed in 4% fixative solution (Solarbio) for 20 min and permeabilized with saponin (Beyotime) for 5 min. After blocking with 5% bovine serum albumin (BSA; Solarbio) for 1 h at room temperature, cells were incubated overnight with anti-gp78 and anti-NLRP3 antibodies [1:200 in phosphate-buffered saline (PBS) containing 5% BSA] followed by staining with DyLight 488-labeled (Multisciences) and Alexa Fluor 555-labeled secondary antibodies (Abcam). Nuclei were co-stained with DAPI (Roche). Stained cells were viewed under a confocal fluorescence microscope (LSM 880 with AiryScan, Nikon A1R).
ASC oligomerization and ASC speck formation
Peritoneal macrophages were plated in 6-cm dishes (4–5 × 106 cells per dish). Cells were primed with 500 ng/ml LPS for 4 h and treated with 10 µM nigericin for 45 min. After that, the supernatants were removed, the cells were rinsed in ice-cold PBS, and 500 µl ice-cold lysis buffer (50 mM Tris-HCl pH 7.5, 1% Triton X-100, 150 mM NaCl, 0.1 mM PMSF, and protease inhibitor cocktail) was added and incubated at 4 °C for 10 min. Cells were scraped and lysed by shearing ten times through a 21-gauge needle. The lysates were centrifuged at 6000 g for 15 min at 4 °C and then 50 µl of supernatant was removed for western blot analysis. The remaining pellets were washed twice in ice-cold PBS and re-suspended in 500 µl room-temperature PBS. Disuccinimidyl suberate (2 mM) was added to the re-suspended pellets and incubated at room temperature for 30 min with rotation. Samples were then centrifuged at 6000 g for 15 min, and the cross-linked pellets were re-suspended in 40 µl SDS loading buffer. Samples were boiled for 10 min at 100 °C and analyzed by western blotting.
For ASC speck formation, BMDMs were seeded at 5 × 105/ml on chamber slides and allowed to attach overnight. The following day, the cells were primed with 500 ng/ml LPS for 4 h and treated with 10 µM nigericin for the indicated time. The cells were fixed in 4% fixative solution followed by ASC and DAPI staining.
Cellular fractionation
Cytosol and mitochondria were isolated from peritoneal macrophages using a Mitochondria Isolation Kit (89874, Thermo Fisher Scientific) according to the manufacturer’s guidelines.
Super-resolution microscopy and structured illumination microscopy
Sample preparation was as described above. For three-dimensional reconstruction, images of BMDMs were captured using a Nikon structured illumination microscope equipped with an ECLIPSE Ti and the objective lens was a CFI Apochromat TIRF 100×H. Images were recorded as vertical z stacks and processed using NIS analysis and Imaris 8.4 software to generate three-dimensional images.
The Pearson coefficient of co-localized volume ranges from 1 to −1 and indicates the level of correlation between the channels analyzed. The closer a value approaches 1, the stronger the correlation or overlap between the two channels selected; a value closer to –1 corresponds to a lower correlation between the two channels of interest.
Lentivirus transduction of immortalized bone marrow-derived macrophages (iBMDMs) and gp78-deficient BMDMs
Cells were spin-infected at 32 °C with lentivirus encoding Flag-gp78 or gp78 C356G as indicated for 120 min at 1500 g. Forty-eight hours after infection, cells were selected by culture with 2 μg/ml puromycin (Sigma).
ELISA
Supernatants from cell culture and serum were collected and the concentrations of IL-1β, IL-18, and TNF-ɑ were determined according to the manufacturer’s instructions (Thermo Fisher Scientific).
In vivo LPS challenge
Mice were injected intraperitoneally with LPS (25 mg/kg body weight). After 4 h, they were sacrificed and the serum concentrations of IL-1β, IL-18, and TNF-ɑ were measured by ELISA (Thermo Fisher). For survival model, mice were injected with 20 mg/kg LPS.
Alum-induced peritonitis
To establish a peritonitis model, mice were injected intraperitoneally with 2 mg alum dissolved in 0.2 ml sterile PBS. After 6 h, the mice were sacrificed and the peritoneal cavity was flushed with cold PBS. The peritoneal lavage fluid was analyzed using FACS Calibur for the recruitment of polymorphonuclear neutrophils with the neutrophil markers FITC-Ly6G (RB6-8C5, Multisciences) and APC-CD11b (M1/70, Biolegend), and the IL-1β concentration in the lavage fluid was determined by ELISA.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
The oligomerization of NLRP3 was analyzed as previously reported. Peritoneal macrophages were lysed with Triton X-100 lysis buffer and then re-suspended in 1× sample buffer and loaded onto a vertical 1.5% agarose gel. After electrophoresis in running buffer (1× TBE and 0.1% SDS) for 1 h at a constant voltage of 80 V at 4 °C, the proteins were transferred to nitrocellulose filter membrane (Pall, #28637358) for immunoblotting.
The buffers used were as follows: 1×TBE buffer: 89 mM Tris pH 8.3, 89 mM boric acid, 2 mM EDTA; Triton X-100 lysis buffer: 0.5% Triton X-100, 50 mM Tris–HCl, 150 mM NaCl, 10% glycerol, 1 mM PMSF, and protease inhibitor cocktail; 1× sample buffer: 0.5× TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue.
Lactate dehydrogenase assay
The release of LDH into the culture medium was determined using an LDH Cytotoxicity Assay Kit (Promega) according to the manufacturer’s instructions.
Reconstitution of the NLRP3 inflammasome in HEK293T cells
The HEK293T cells were seeded into six-well plates (5 × 105 cells per well) in complete culture medium. Then cells were transfected with plasmids expressing HA-pro-IL-1β (1 μg), Myc-pro-caspase-1 (60 ng), Flag-ASC (150 ng), and Flag-tagged NLRP3 (200 ng) with or without Myc-gp78 using polyethyleneimine. The medium was replaced 6 h after transfection and supernatants were collected 34 h after medium change. The IL-1β maturation was determined by ELISA and cell lysates were analyzed by western blot.
Small-interfering RNA transfection
Peritoneal macrophages were plated in 12-well plates (3 × 105 cells per well) and then transfected with 100 nM siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. The siRNA for mouse gp78 (CGAAAGCGGUUCUUAAACAAATT) and the scrambled siRNA (UUCUCCGAACGUGUCACGUTT) were chemically synthesized by Genepharma Co., Shanghai, China.
Quantitative PCR
RNA was extracted using the RNA-Quick purification kit (ES Science). Complementary DNA was synthesized using the SuperScript First-Strand cDNA synthesis kit (Vazyme Biotech) according to the manufacturer’s protocols. Quantitative PCR was performed using SYBR Green (Vazyme Biotech) on a Light Cycler 480 (Roche Diagnostics). The samples were individually normalized to gapdh and the primers used were as follows.
Il1b-F | ATCAACCAACAAGTGATATTCTCCAT |
Il1b-R | GGGTGTGCCGTCTTTCATTAC |
Tnf-F | CCTGTAGCCCACGTCGTAG |
Tnf-R | GGGAGTAGACAAGGTACAACCC |
gapdh-F | AGGTCGGTGTGAACGGATTTG |
gapdh-R | TGTAGACCATGTAGTTGAGGTCA |
nlrp3-F | AGCCAGAGTGGAATGACACG |
nlrp3-R | CGTGTAGCGACTGTTGAGGT |
Il18-F | GACTCTTGCGTCAACTTCAAGG |
Il18-R | CAGGCTGTCTTTTGTCAACGA |
casp1-F | CACAGCTCTGGAGATGGTGA |
casp1-R | TCTTTCAAGCTTGGGCACTT |
Hspa5-F | GTGTGTGAGACCAGAACCGT |
Hspa5-R | GTTCTTGAACACACCGACGC |
Ddit3-F | GGAACCTGAGGAGAGAGTGTTC |
Ddit3-R | CTTCCTCTTCGTTTCCTGGGG |
Dnajb9-F | GAGGCTACTCGGCGTTCG |
Dnajb9-R | GCAGACTTTGGCACACCTAA |
Xbp1s-F | CTGAGTCCGCAGCAGGTG |
Xbp1s-R | GACCTCTGGGAGTTCCTCCA |
Xbp1-F | GAACCAGGAGTTAAGAACACG |
Xbp1-R | AGGCAACAGTGTCAGAGTCC |
Xbp-1 splicing detection
Total RNA was isolated from gp78f/f or Lyz2-cre-gp78f/f derived peritoneal macrophages, either untreated, treated with LPS (500 ng/ml, 4 h), LPS (500 ng/ml, 4 h) plus ATP (2 mM, 30 min), or thapsigargin (0.5 μM, 6 h), and RT-PCR analysis of total RNA was performed to simultaneously detect both spliced and unspliced XBP1 mRNA and GAPDH. RT-PCR products were separated by electrophoresis (2% agarose gel). The primers used are Xbp1-F and Xbp1-R.
In vitro ubiquitination assay
For the synthesis of polyUb chains, purified GST-NLRP3 ΔLRR, Flag-gp78 was mixed with E1 (80 nM), Ube2g2 (1 μM), and ubiquitin (50 µM) (kindly provided by Prof. Zong-Ping Xia of Zhengzhou University) in a reaction buffer containing 20 mM HEPES-K, pH 7.4, 5 mM MgCl2, 2 mM ATP, and 10% glycerol. The reaction was carried out at 30 °C for 30 min and then resolved by SDS-PAGE. Ubiquitinated products were detected by immunoblotting with the indicated antibodies.
Mass spectrometry
In-gel digestion and mass spectrometry analysis
The silver-stained gel band was excised, cut into small pieces, and washed with water followed by a 1:1 mixture of 30 mM K4Fe(CN)6 and 100 mM Na2S2O3. The protein was reduced with 10 mM Tris (2-carboxyethyl) phosphine (Thermo Scientific) in 100 mM NH4HCO3 at room temperature for 30 min and alkylated with 55 mM iodoacetamide or N-ethylmaleimide (Sigma) in 100 mM NH4HCO3 in the dark for 30 min. After that, the gel pieces were washed with 100 mM NH4HCO3 and 100% acetonitrile, then dried using a SpeedVac. Finally, they were digested with 12.5 ng/μL trypsin (Promega) in 50 mM NH4HCO3 for 16 h at 37°C, and the tryptic peptides were extracted twice with 50% acetonitrile/5% formic acid and dried using a SpeedVac. The sample was reconstituted with 0.1% formic acid, desalted using a MonoSpinTM C18 column (GL Science, Tokyo, Japan), and then dried with a SpeedVac.
LC/tandem MS (MS/MS) analysis of peptides
The peptide mixture was analyzed using a home-made 30 cm pulled-tip analytical column (75 μm ID packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch GmbH), which was then placed in-line with an Easy-nLC 1200 nano HPLC (Thermo Scientific, San Jose, CA) for mass spectrometry. Peptides eluted from the LC column were directly electro-sprayed into the mass spectrometer with a distal 2.5 kV spray voltage. Data-dependent tandem mass spectrometry (MS/MS) was performed with a Q Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA). A cycle of one full-scan MS spectrum (m/z 300–1800) was acquired followed by the top 20 MS/MS events, sequentially generated on the first to the twentieth most intense ions selected from the full MS spectrum at a 30% normalized collision energy. Full scan resolution was set to 70,000 with an automated gain control (AGC) target of 3e6. The MS/MS scan resolution was set to 17,500 with an isolation window of 1.8 m/z and an AGC target of 1e5. The number of microscans was one for both MS and MS/MS scans and the maximum ion injection times were 50 and 100 ms, respectively. The dynamic exclusion settings used were as follows: charge exclusion, 1 and >8; exclude isotopes, on; and exclusion duration, 30 s. MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific). The analytical column temperature was set at 55 °C during the experiments. The mobile phase and elution gradients used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 5–10% B; 1–96 min, 10–40% B; 96–104 min, 40–60% B; 104–105 min, 60–100% B; 105–120 min, 100% B. The flow rate was set at 300 nL/min.
Data analysis
The acquired MS/MS data were analyzed against a homemade database (including all target proteins) using PEAKS Studio 8.5. Cysteine alkylation by iodoacetamide or N-ethylmaleimide was specified as fixed modification with mass shift 57.02146 and methionine oxidation and protein n-terminal acetylation as variables. In addition, lysine ubiquitination was set as dynamic modification with mass shift 114.0429. In order to accurately estimate peptide probabilities and false discovery rates, we used a decoy database containing the reversed sequences of all the proteins appended to the target database.
Quantification and statistical analysis
For cell experiments, none of the samples was excluded. For animal experiments, none of the animals was excluded from the analysis except for animals that were dead or no enough sample were collected. The results for ELISA and q-PCR are expressed as the mean ± SD. For animal experiments, genotyping was performed before experiments. The mouse model data are expressed as the mean ± SEM and analyzed using two-tailed Student’s t test for two groups. For mouse survival rate analysis, GraphPad Prism7 was used to plot Kaplan–Meier survival curves and compared using log-rank tests. Differences were considered significant when *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–89.
Shim DW, Lee KH. Posttranslational Regulation of the NLR Family Pyrin Domain-Containing 3 Inflammasome. Front Immunol. 2018;9:1054.
Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016;45:802–16.
Song N, Liu ZS, Xue W, Bai ZF, Wang QY, Dai J, et al. NLRP3 Phosphorylation Is an Essential Priming Event for Inflammasome Activation. Mol Cell. 2017;68:185–97 e6.
Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–8.
Hamilton C, Anand PK. Right place, right time: localisation and assembly of the NLRP3 inflammasome. F1000Res. 2019;8:676.
Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 2013;153:348–61.
Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–40.
Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014;41:919–33.
Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–604.
Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014;156:1193–206.
Yu JW, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–49.
Liu TF, Tang JJ, Li PS, Shen Y, Li JG, Miao HH, et al. Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab. 2012;16:213–25.
Guo C, Chi Z, Jiang D, Xu T, Yu W, Wang Z, et al. Cholesterol Homeostatic Regulator SCAP-SREBP2 Integrates NLRP3 Inflammasome Activation and Cholesterol Biosynthetic Signaling in Macrophages. Immunity 2018;49:842–56 e7.
Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–7.
Ying Z, Wang H, Fan H, Zhu X, Zhou J, Fei E, et al. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum Mol Genet. 2009;18:4268–81.
Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nunez G, He Y, et al. Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3- and Caspase-2-Driven Mitochondrial Damage. Immunity 2015;43:451–62.
Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–8.
Lu Y, Qiu Y, Chen P, Chang H, Guo L, Zhang F, et al. ER-localized Hrd1 ubiquitinates and inactivates Usp15 to promote TLR4-induced inflammation during bacterial infection. Nat Microbiol. 2019;4:2331–46.
Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem. 2012;287:36617–22.
Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–9.
Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–34.
Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–35.
He Y, Zeng MY, Yang D, Motro B, Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016;530:354–7.
Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–8.
Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, et al. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem. 2016;291:103–9.
Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65.
Carty M, Kearney J, Shanahan KA, Hams E, Sugisawa R, Connolly D, et al. Cell Survival and Cytokine Release after Inflammasome Activation Is Regulated by the Toll-IL-1R Protein SARM. Immunity 2019;50:1412–24 e6.
Yang CS, Kim JJ, Kim TS, Lee PY, Kim SY, Lee HM, et al. Small heterodimer partner interacts with NLRP3 and negatively regulates activation of the NLRP3 inflammasome. Nat Commun. 2015;6:6115.
Chen J, Chen ZJ. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 2018;564:71–6.
Magupalli VG, Negro R, Tian Y, Hauenstein AV, Di Caprio G, Skillern W, et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science. 2020;369:eaas8995.
Joshi V, Upadhyay A, Kumar A, Mishra A. Gp78 E3 ubiquitin ligase: essential functions and contributions in proteostasis. Front Cell Neurosci. 2017;11:259.
Wang YJ, Bian Y, Luo J, Lu M, Xiong Y, Guo SY, et al. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat Cell Biol. 2017;19:808–19.
Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–55.
Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727.
Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 2015;160:62–73.
Zinngrebe J, Montinaro A, Peltzer N, Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014;15:28–45.
Guo Y, Li L, Xu T, Guo X, Wang C, Li Y, et al. HUWE1 mediates inflammasome activation and promotes host defense against bacterial infection. J Clin Investig. 2020;130:6301–16.
Acknowledgements
We thank Prof Bao-Liang Song (Wuhan University) for kindly providing gp78f/f and Insig1f/f mice, Dr. Tao Li (National Center of Biomedical Analysis) for providing plasmid of GST-NLRP3 ΔLRR, and Prof Zong-Ping Xia (Zhengzhou University) for kindly providing E1, E2, and ubiquitin proteins for the in vitro ubiquitination assays. We thank Dr I.C Bruce for reading the paper. We thank Dr Chao Peng and Su Chen of the Mass Spectrometry System at the National Facility for Protein Science in Shanghai (NFPS), the Zhangjiang Lab, Shanghai Advanced Research Institute, and the Chinese Academy of Science, China for data collection and analysis. We are grateful for technical support from the Core Facilities, Zhejiang University School of Medicine. This work was also supported by the Key Laboratory of Immunity and Inflammatory Diseases of Zhejiang Province.
Funding
This work was supported by the National Natural Science Foundation of China (81930042, 81730047, 82025017, 31800759, and 32000630) and the Fundamental Research Funds for the Central Universities. The work was supported by China National Postdoctoral Program for Innovative Talents (BX20200296) and China Postdoctoral Science Foundation (2021M692780). This work was also supported by the Key Laboratory of Immunity and Inflammatory Diseases of Zhejiang Province and Innovative Institute of Basic Medical Sciences of Zhejiang University.
Author information
Authors and Affiliations
Contributions
TX, WY, HF, ZC, YG, XG, DJ, ZW, KZ, SC, ML, JZ, DY, and QY performed the experiments; DW, and XZ designed the research; TX, WY, HF, DW, and XZ wrote the paper; DW and XZ supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All mice were housed in a specific pathogen-free facility in the Laboratory Animal Center of Zhejiang University. Animal experimental protocols were approved by the Review Committee of Zhejiang University School of Medicine and were in compliance with institutional guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by: R Johnstone
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, T., Yu, W., Fang, H. et al. Ubiquitination of NLRP3 by gp78/Insig-1 restrains NLRP3 inflammasome activation. Cell Death Differ 29, 1582–1595 (2022). https://doi.org/10.1038/s41418-022-00947-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00947-8
This article is cited by
-
HECTD3 inhibits NLRP3 inflammasome assembly and activation by blocking NLRP3-NEK7 interaction
Cell Death & Disease (2024)
-
Targeting NLRP3 inflammasome for neurodegenerative disorders
Molecular Psychiatry (2023)
-
Gp78 deficiency in hepatocytes alleviates hepatic ischemia-reperfusion injury via suppressing ACSL4-mediated ferroptosis
Cell Death & Disease (2023)
-
Deubiquitinase OTUD6A in macrophages promotes intestinal inflammation and colitis via deubiquitination of NLRP3
Cell Death & Differentiation (2023)
-
NEDD4 Regulated Pyroptosis Occurred from Co-infection between Influenza A Virus and Streptococcus pneumoniae
Journal of Microbiology (2023)