Abstract
Ciliogenesis proteins orchestrate vesicular trafficking pathways that regulate immune synapse (IS) assembly in the non-ciliated T-cells. We hypothesized that ciliogenesis-related genes might be disease candidates for common variable immunodeficiency with impaired T-cell function (T-CVID). We identified a heterozygous, predicted pathogenic variant in the ciliogenesis protein CCDC28B present with increased frequency in a large CVID cohort. We show that CCDC28B participates in IS assembly by regulating polarized T-cell antigen receptor (TCR) recycling. This involves the CCDC28B-dependent, FAM21-mediated recruitment of the actin regulator WASH to retromer at early endosomes to promote actin polymerization. The CVID-associated CCDC28BR25W variant failed to interact with FAM21, leading to impaired synaptic TCR recycling. CVID T cells carrying the ccdc28b 211 C > T allele displayed IS defects mapping to this pathway that were corrected by overexpression of the wild-type allele. These results identify a new disease gene in T-CVID and pinpoint CCDC28B as a new player in IS assembly.
Similar content being viewed by others
Introduction
Antibody deficiency is the hallmark of CVID, the most common inheritable immunodeficiency. This defect is associated with highly heterogeneous clinical presentations that include, beside infections, chronic lung disease, granulomatous disease, autoimmunity, lymphomas, and gastrointestinal disease [1]. Based on this heterogeneity, CVID is believed to group different immune disorders unified by hypogammaglobulinemia. Consistently, a substantial proportion of CVID patients displays defects not only in B-cell maturation and antibody class switching, but also in T-cell activation and helper function [2]. To date 68 disease genes have been identified, the majority of which encode proteins directly implicated in B-cell costimulation or immunoglobulin class switching [3, 4]. Gene products not directly implicated in these processes include the T-cell costimulatory receptor ICOS [5], the tyrosine kinase Itk [4], and the co-inhibitory receptor CTLA-4 [4, 6]. We and others have reported defects in TCR signaling in CVID patients with impaired T-cell responses (T-CVID) [7,8,9], which we have circumscribed in a subgroup of patients to a deficiency in the guanine nucleotide exchanger Vav1 [10, 11].
TCR signaling is orchestrated at the immune synapse (IS), a specialized interface formed by T cells upon contact with cognate APC [12]. Dysfunctional synapses have been observed in immune-related disorders, highlighting IS assembly as a potential new disease process in T-cell related immunodeficiencies. Our finding that components of the intraflagellar transport (IFT) system that controls ciliogenesis regulate IS assembly in the non-ciliated T cell [13] has pinpointed the ciliogenesis machinery as a source of candidate genes. Indeed, we and others have implicated a variety of ciliogenesis regulators in IS assembly and T-cell activation [14,15,16]. Here we have tested the hypothesis that mutations in ciliopathy-related genes may underlie T-CVID of unknown etiology. Starting with the identification of an increased frequency in a cohort of 101 CVID patients of a potentially pathogenic SNP in the gene encoding the ciliary protein Coiled-Coil Domain Containing 28B (CCDC28B) [17, 18], we addressed the function of CCDC28B in IS assembly. We show that CCDC28B promotes actin polymerization at endosomal TCRs by recruiting the actin regulator WASH and its partner FAM21, allowing for their recycling to the IS, a process essential to sustain signaling during T-cell activation [19, 20]. Additionally, we provide evidence that the CVID-associated SNP impairs the ability of CCDC28B to recruit the FAM21-WASH complex to endosomal TCRs and its association with the retromer complex, resulting in their failure to undergo polarized recycling. The data identify a new pathogenic mutation in T-CVID and highlight the ciliary protein CCDC28B as a new player in TCR trafficking and IS assembly.
Results
A pathogenic ccdc28b variant is present with increased frequency in CVID
To investigate the potential implication of ciliogenesis proteins in the T-cell defects associated with T-CVID we performed an exome analysis of the known ciliopathy-related genes (Table S1) on seven CVID patients classified as T-CVID based on their impaired T-cell proliferative response to TCR engagement. High coverage exon sequencing revealed the presence of known or novel predicted pathogenic variants in five ciliopathy-related genes in individual patients, all heterozygous (Table S2).
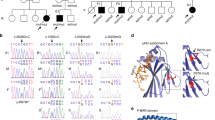
A cohort of 101 CVID patients was tested by Sanger sequencing for the presence of the exon variants in the ciliopathy-related genes identified by NGS. These variants were present only in the individual patients initially subjected to exon sequencing with the exception of one, mapping to ccdc28b encoding the ciliogenesis protein CCDC28B [17, 18], which was found in heterozygosity in 9/101 patients, hence with a frequency of 4.45%. This variant (Ensembl rs1407134) was also present in the healthy population, however with a frequency in the corresponding ethnic group (Central European) of 2%, based on allele frequencies in the 1000 Genomes Project Phase 3 and the Genome Aggregation database. This variant has a C > T transversion at position 211 of the transcript (NM_024296.5), which results in a R > W substitution at position 25 of the protein (Fig. 1A). The increased frequency of the R25W variant in CVID prompted us to investigate the role of CCDC28B in IS assembly and assess the potential pathogenic outcome of the R25W variant.
A Sanger sequencing chromatograms of the PCR amplification products of ccdc28b (NG_012178.1, nucleotide positions 6289-6510, corresponding to cDNA nucleotide positions 116-302, covering the entire exon 2) on genomic DNA from representative CVID patients carrying either the wild-type ccdc28b allele (left) or heterozygous for the 211 C > T allele (right). A scheme of the domain organization of CCDC28B is shown below. B Representative immunoblot of CCDC28B in lysates of hTERT-immortalized fibroblasts, Jurkat T-cells, and primary T-cells. C, D Immunofluorescence analysis and co-localization of CCDC28B and in Jurkat cells co-stained with antibodies to γ-tubulin (centrosome), Rab5 (early endosome), or GM130 (Golgi apparatus), and Jurkat cells transfected with CCDC28B-GFP co-stained for Rab11 (recycling endosome). Representative images (medial optical sections) are shown in C. The quantification (mean ± SD) using Mander’s coefficient of the weighted co-localization of CCDC28B with each marker is shown in D. 30 cells were analyzed for each marker (n = 3). E, F Left, Immunofluorescence analysis of CCDC28B (see Fig. S1E for antibody specificity control) and γ-tubulin in conjugates of Jurkat cells and SEE-pulsed Raji cells (APC) (E), or primary T cells and Raji cells pulsed with a mix of SEA, SEB, and SEE (SAg) (F). Conjugates formed in the absence of SEE/SAg were used as negative controls. Representative images (medial optical sections) are shown (n = 3). Size bar, 5 μm. Right, histograms showing the quantification (mean ± SD) using Mander’s coefficient of the weighted co-localization of CCDC28B with γ-tubulin (left) and the mean distance of CCDC28B from the T-cell:APC contact site (μm) (right). Measurements were taken on 10 conjugates (n ≥ 3). Error bars, SD. ***p < 0.001 (Student’s t test).
CCDC28B is required for IS assembly
CCDC28B is a coiled-coil (CC) domain-containing protein that interacts and colocalizes with the Bardet-Biedl syndrome (BBS) proteins at the basal body [17] (Fig. 1A). CCDC28B modulates cilia length in vitro and in vivo [17, 18], however the underlying mechanisms remain elusive. Jurkat and primary T-cells were found to express CCDC28B at levels comparable to the ciliated human hTERT-immortalized fibroblasts (Fig. 1B). Similar to ciliated cells, CCDC28B showed a centrosomal localization in T-cells (Fig. 1C, D) and additionally partially co-localized with early (Rab5+) and recycling (Rab11+) endosomes (Fig. 1C, D). The CCDC28B+ compartment polarized toward the T-cell:APC contact together with the centrosome in conjugates formed by Jurkat cells with SEE-loaded Raji cells used as APC (Fig. 1E). This result was validated in primary T-cells, stimulated with Raji cells pulsed with a combination of SEA, SEB, and SEE to maximize coverage of the TCR-Vβ repertoire (Fig. 1F).
To address the role of CCDC28B in IS assembly Jurkat cells were depleted of CCDC28B by siRNA-mediated silencing (Fig. S1A). Control and CCDC28B-KD cells, which expressed similar levels of surface CD3 (Fig. S1B), were compared for their ability to form functional mature synapses (15-min conjugation). Accumulation of the TCR:CD3 complex and phosphotyrosine signaling at the T-cell:APC contact, and centrosome polarization toward the APC, were used as readouts. IS defects were observed in CCDC28B-KD cells, as shown by impaired TCR and tyrosine-phosphoprotein accumulation at the IS (Fig. 2A, B). Defective phosphoprotein signaling was confirmed by flow cytometry (Fig. S2A). Synaptic TCR and tyrosine-phosphoprotein accumulation was compromised also at longer times points (Fig. S2B). Similar to the plasma membrane pool, the endosomal TCR pool (eCD3) failed to polarize toward the IS in CCDC28B-KD cells (Figs. 2A, B and S2C). The results were validated on primary CCDC28B-KD T-cells (Figs. 2C and S1B, C). The defects associated with CCDC28B deficiency were rescued by restoring CCDC28B expression (Fig. 2D and S1D), confirming its implication in IS assembly.
A Immunofluorescence analysis of CD3ζ (top), tyrosine phosphoproteins (PTyr) (middle), or γ-tubulin (bottom) in 15-min conjugates of control or CCDC28B KD Jurkat cells and SEE-pulsed Raji cells (APC). The histograms show the quantification (%, mean±SD) of conjugates with CD3ζ, PTyr or γ-tubulin staining at the T-cell:APC contact site 5 min and 15 min after conjugate formation. At least 100 conjugates were analyzed for each marker (n ≥ 3). B Left, Histograms showing the relative fluorescence of CD3ζ (top; includes membrane and endosomal CD3ζ) or PTyr (bottom) at the T-cell:APC contact site compared to the remaining T-cell area (recruitment index). Right, histogram showing the mean distance of CD3ζ+ endosomes (eCD3, top) or of the centrosome (γ-tub, bottom) from the T-cell:APC contact site (μm). Measurements were taken on 50 conjugates (n ≥ 3). C Immunofluorescence analysis of CD3ζ (top), tyrosine phosphoproteins (PTyr) (middle) or γ-tubulin (bottom) in conjugates of control or CCDC28B KD primary T-cells cells and Raji cells (APC) pulsed with a mix of SEA, SEB, and SEE. The histograms show the quantification (%, mean±SD) of conjugates with CD3ζ, PTyr or γ-tubulin staining at the IS 15 min after conjugate formation. At least 100 conjugates were analyzed for each marker (n ≥ 3). D Histograms showing the quantification (%, mean ± SD) of 15-min conjugates with CD3ζ, PTyr or γ-tubulin staining at the IS in conjugates of control or CCDC28B KD Jurkat cells, transfected with either empty vector (ctr, KD) or the same vector encoding wild-type CCDC28B (KD + CCDC28B), and SEE-pulsed Raji cells. At least 50 conjugates were analyzed for each marker (n ≥ 3). Size bar, 5 μm. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
CCDC28B is required for polarized TCR recycling to the IS
Centrosome repositioning at the IS is triggered by TCR signaling [12]. Centrosome polarization occurs normally in CCDC28B-deficient T-cells (Fig. 2A, B), suggesting that the initial steps in IS assembly do not require this protein. Consistently, the initial TCR and tyrosine-phosphoprotein accumulation at the nascent IS (5-min conjugation) was not affected by CCDC28B deficiency (Fig. 2A). Since TCR signaling is sustained by the recycling endosome-associated pool [21], we hypothesized that the defect in TCR accumulation at the mature IS in CCDC28B-deficient T cells could result from impaired recycling. To test this hypothesis surface TCRs were labeled with anti-CD3 mAb and incubated at 37 °C to allow for internalization of TCR:mAb complexes. Internalized TCRs were tracked using fluorochrome-labeled secondary antibodies in SEE-specific conjugates, either non-permeabilized to visualize TCRs that had recycled to the plasma membrane, or permeabilized to detect their intracellular localization.
Imaging of non-permeabilized antigen-specific conjugates showed that internalized antibody-tagged TCRs (tCD3) failed to effectively concentrate at the IS in CCDC28B-deficient cells (Fig. 3A, B and S2D, E), indicating a defect in polarized recycling. Analysis of permeabilized conjugates showed that tCD3+ dots, identified as endosomes by staining for Rab5 and Rab11 (Fig. S3A), remained dispersed away from the T-cell:APC contact (Fig. 3A, B) and from the polarized centrosome (Fig. S2E). Hence CCDC28B is required for the polarized transport of endosomal TCRs to the IS.
A, B Immunofluorescence analysis of recycling TCRs (tCD3) in control or CCDC28B KD Jurkat cells conjugated for 15 min with SEE-pulsed Raji cells (APC) (A) or primary T-cells conjugated with SEA/SEB/SEE-pulsed Raji cells (B). Before conjugation, cells were added with anti-CD3ε mAb (OKT3) and incubated at 37 °C for 2 h to allow for internalization of CD3-Ab complexes. Following acid-stripping to remove residual anti-CD3 mAb bound at the cell surface. Conjugates were stained with secondary fluorescently labeled antibodies, after fixing either under non-permeabilizing conditions (top) or after cell permeabilization (bottom). The histograms show the quantification (%, mean ± SD) of conjugates with tCD3 at the IS (left). The histograms on the right show either the relative fluorescence of tCD3 at the T-cell: APC contact site compared to the remaining T-cell area (recruitment index, mean±SD) for non-permeabilized conjugates, or the mean distance of tCD3+ endosomes from the T-cell:APC contact site (μm) for permeabilized conjugates. At least 20 conjugates were analyzed in each experiment (n ≥ 3). Size bar, 5 μm. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
CCDC28B is required for TCR-dependent F-actin polymerization
Endosome recycling is coordinated by the actin and tubulin cytoskeletons. F-actin polymerization at endosomes generates force for the abscission of vesicles enriched in recycling cargo [22, 23]. The actin phenotype observed in ccdc28b-deficient zebrafish, with disorganized F-actin bundles in muscle fibers [18] suggest that CCDC28B may participate in actin polymerization pathways in T-cells. TCR-dependent F-actin polymerization was impaired in CCDC28B-deficient T-cells, as measured by flow cytometry (Fig. 4A). Additionally, CCDC28B-deficient cells failed to efficiently spread and formed an irregular F-actin ring when plated on immobilized anti-CD3 mAb, as assessed by TIRF microscopy (Fig. 4B). Consistently, a reduction in F-actin accumulation at the synaptic membrane was observed in the absence of CCDC28B (Fig. 4C, D), which could be accounted for by the defect in sustained signaling caused by impaired TCR recycling. These results map CCDC28B to the pathway that couples TCR signaling to actin polymerization.
A Flow cytometric analysis of F-actin polymerization in response to TCR engagement in phalloidin-stained control or CCDC28B KD Jurkat cells. Cells were stimulated with soluble anti-CD3ε mAb (UCHT-1) for 15 min. The data are expressed as fold increase (mean ± SD) of F-actin in stimulated vs non-stimulated cells (n = 3). A representative FACS profile of phalloidin staining is shown on the right. Unlabeled cells were used as negative control. B TIRF microscopy-based analysis of F-actin in phalloidin-stained control or CCDC28B KD Jurkat cells activated by plating on anti-CD3ε mAb (UCHT-1)-coated coverslips for 15 min. The histogram shows the cell area (μm2) as a measure of cell spreading (n cells = 20, n = 3). C, D Immunofluorescence analysis of F-actin in 15 min-conjugates of control or CCDC28B KD Jurkat cells and SEE-pulsed Raji cells (APC) (C), or control or CCDC28B KD primary T cells and SEA/SEB/SEE-pulsed Raji cells (D) stained with fluorochrome-tagged phalloidin. The histograms show the quantification (%, mean ± SD) of conjugates with F-actin staining at the IS (at least 100 conjugates were analyzed, n ≥ 3) (left) or the relative fluorescence of F-actin at the T-cell: APC contact site compared to the remaining T-cell area (recruitment index) (n = 25 conjugates). E, F Immunofluorescence analysis of recycling TCRs (tCD3) and F-actin in control or CCDC28B KD Jurkat cells conjugated for 15 min with SEE-pulsed Raji cells (APC) (E) or primary T-cells conjugated with SEA/SEB/SEE- pulsed Raji cells (F). Before conjugation, cells were added with anti-CD3ε mAb (OKT3) and incubated at 37 °C for 2 h to allow for internalization of CD3-Ab complexes. Following acid-stripping to remove residual anti-CD3 mAb bound at the cell surface. Conjugates were permeabilized/fixed and stained with secondary fluorescently labeled antibodies and fluorochrome-labeled phalloidin. The histograms show the mean distance of F-actin+ dots from the T-cell:APC contact site (top, μm); the co-localization of tCD3 with F-actin on individual dots (bottom left, Mander’s coefficient); and the quantification (mean±SD) of tCD3+ dots positive for F-actin (bottom right). Measurements were taken on 50 conjugates from at least three independent experiments and a mean of 10 dots per cell were analyzed. Size bar, 5 μm. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
As expected, F-actin was also found at Rab5+ endosomes and, to a lesser extent, Rab11+ endosomes (Fig. S3B). The ability of F-actin+ endosomes to polarize toward the IS was reduced in CCDC28B-deficient Jurkat cells (Fig. 4E). Co-staining tCD3 with phalloidin in SEE-specific conjugates revealed a reduction in the proportion of tCD3+ endosomes that were positive for F-actin in CCDC28B-deficient cells (Fig. 4E). tCD3 co-localization with F-actin on individual endosomes was also reduced (Fig. 4E). These results suggest that the TCR recycling defects observed in CCDC28B-deficient T cells may result from a failure to polymerize F-actin on endosomes. These results were validated on primary T-cell blasts (generated as in Fig. S4A), which had a larger endosomal TCR pool compared to freshly purified T cells (Figs. 4F and S4B).
CCDC28B is required for recruitment of the actin regulator WASH and its adapter FAM21 to retromer-associated TCRs at early endosomes
WASH recruits the Arp2/3 nucleation factor to endosomes to promote F-actin polymerization through its interaction with FAM21 [24]. WASH+ endosomes failed to polarize toward the IS in CCDC28B-deficient cells, concomitant with a reduction in the proportion of WASH+ endosomes positive for F-actin (Figs. 5A and S5A), suggesting a role for CCDC28B in WASH recruitment to endosomes and local F-actin polymerization. Consistently, the proportion of tCD3+ endosomes positive for WASH was reduced in CCDC28B-deficient cells, as was tCD3 co-localization with WASH on individual endosomes (Figs. 5B and S5B). Similar results were obtained when cells were co-stained for tCD3 and FAM21 (Figs. 5C and S5C). Hence CCDC28B couples the FAM21/WASH complex to recycling TCRs to promote F-actin polymerization.
A Immunofluorescence analysis of WASH in 15 min-conjugates of control or CCDC28B KD Jurkat cells and SEE-pulsed Raji cells (APC) co-stained for F-actin. The histograms show the mean distance of WASH+ dots from the T-cell:APC contact site (top) and the quantification of WASH+ dots positive for actin (%, bottom). B, C Immunofluorescence analysis of recycling TCRs (tCD3; see legend to Fig. 4E for experimental setting) in control or CCDC28B KD Jurkat cells conjugated for 15 min with SEE-pulsed Raji cells (APC) and co-stained for WASH (B) or FAM21 (C). For each panel the histograms show the quantification of tCD3+ dots positive for WASH or FAM21 (top, %, mean ± SD); and the co-localization of tCD3 with WASH or FAM21 on individual dots (bottom, Mander’s coefficient, mean ± SD). Measurements were taken on 50 conjugates from at least three independent experiments and a mean of 10 dots per cell were analyzed. Size bar, 5 μm. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
WASH-dependent actin polymerization occurs at early, Rab5+ endosomes (EE) [24]. WASH localizes at EEs through FAM21, which interacts with the EE-associated retromer complex [25, 26]. A reduction in WASH and FAM21 co-localization with Rab5 on individual endosomes was observed in CCDC28B-deficient cells (Fig. 6A, B), suggesting that CCDC28B regulates TCR recycling by promoting the FAM21-dependent recruitment of WASH to EEs.
A–C Immunofluorescence analysis of WASH (A) and FAM21 (B) in 15 min-conjugates of control or CCDC28B KD Jurkat cells and SEE-pulsed Raji cells (APC) co-stained for Rab5. The histograms show the co-localization of WASH or FAM21 with Rab5 on individual dots (Mander’s coefficient, mean ± SD). Measurements were taken on 50 conjugates from at least three independent experiments and a mean of 10 dots per cell were analyzed. C Immunoblot analysis of tCD3-specific immunoprecipitates from post-nuclear supernatants of control or CCDC28B KD Jurkat cells. Cells were added with anti-CD3ε mAb (OKT3) and incubated at 37 °C for 2 h to allow for internalization of CD3-Ab complexes. Following acid-stripping to remove residual anti-CD3 mAb bound at the cell surface, tCD3s were immunoprecitated with secondary antibodies and protein A-Sepharose (tCD3 IP) (see scheme in Fig. S6A). Immunoprecipitates from lysates of cells treated similarly without the addition of anti-CD3ε mAb were used as negative controls (neg ctr). Lysates (lys) belong to the same immunoblot (different exposure). Immunoprecipitates were probed with the indicated antibodies. The results from two representative experiments are shown (n = 4). The quantifications (mean ± SD) of the relative intensities of the immunoreactive bands (KD vs ctr) are shown in the box. D Immunoblot analysis of GFP-specific immunoprecipitates from post-nuclear supernatants of Jurkat cells transiently transfected with a plasmid construct encoding GFP-tagged CCDC28B (CCDC-GFP IP). Immunoprecipitates carried out with non-immune Abs were used as negative control (neg ctr). Lysates (lys) belong to the same immunoblot (different exposure). E, F Immunoblot analysis of WASH-specific (E) or VPS35-specific (F) immunoprecipitates from post-nuclear supernatants of control or CCDC28B KD Jurkat cells. Immunoprecipitates carried out with non-immune Abs were used as negative control (neg ctr). Lysates (lys) belong to the same immunoblot (different exposure). For each panel the quantifications (mean ± SD) of the relative intensities of the immunoreactive bands (KD vs ctr) are shown in the box (n = 4). G, H Immunoblot analysis of GSH-Sepharose pull-down assays on post-nuclear supernatants of Jurkat cells using either GST fusion proteins encoding the N-terminal (GST-FAMn) and C-terminal (GST-FAMc) portions of the L-F-[D/E](3-10)-L-F repeat-rich tail of FAM21 (G), or a GST fusion protein encoding a portion of WASH spanning the WDH1, WDH2 and PR domains (H) (see schemes in Fig. S6B). Recombinant GST was used as negative control. Lysates (lys) belong to the same immunoblot (different exposure). For each filter the Ponceau staining is shown (n = 3). Total post-nuclear supernatants (lys) were included in all gels. The migration of molecular mass markers is shown for each filter. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
We used an immunoprecipitation approach to selectively pull-down tCD3s to ask whether CCDC28B interacts with recycling TCRs, and to address the outcome of CCDC28B deficiency on the local recruitment of FAM21/WASH and retromer (Fig. S6A). CCDC28B was found to associate with tCD3s (Fig. 6C). Consistent with the role of WASH in retromer-mediated TCR sorting, an association of VPS35 (a core retromer component) [27], FAM21 and WASH with tCD3s was also detected (Fig. 6C). The ability of tCD3s to recruit FAM21 and WASH, but not VPS35, was compromised in CCDC28B-deficient cells (Fig. 6C). Hence CCDC28B couples the FAM21/WASH complex to retromer-associated recycling TCRs.
To elucidate the interactions among CCDC28B, WASH/FAM21 and VPS35 we tested their association in Jurkat cells transfected with GFP-tagged CCDC28B. VPS35, FAM21 and WASH formed a complex with GFP-CCDC28B (Figs. 6D and S7A, B). CCDC28B deficiency impaired the ability of WASH to interact with VPS35 and FAM21 (Fig. 6E), as well as with its effectors α-tubulin and the Arp2/3 subunit ARPC3 (Fig. S7C). Similarly, the ability of VPS35 to interact with WASH and FAM21 was impaired in CCD28B-deficient cells (Fig. 6F).
To map CCDC28B on FAM21 and WASH we used a panel of GST-tagged deletion mutants (Fig. S6B). FAM21 associates with WASH through its N-terminal domain and with VPS35/retromer at EEs through multiple L-F-[D/E](3–10)-L-F repeats within its disordered tail (Fig. S6C) [25, 28, 29]. Pull-down experiments using the GST-tagged N- and C-terminal portions of the repeat-rich tail of FAM21 (FAMn and FAMc; Fig. S6B) revealed that CCDC28B interacts selectively with the C-terminal region of the FAM21 tail (Fig. 6G). This FAM21 region is also responsible for α-tubulin binding (Fig. S7D). No interaction with WASH was observed (Fig. 6G), consistent with the lack of the N-terminal domain of FAM21 where WASH binds [29] (Fig. S6B, C). Similar assays using a GST-tagged WASH construct spanning the FAM21 (WHD1) and α-tubulin (WHD2) binding domains (Fig. S6B) confirmed its ability to associate with CCDC28B (Fig. 6H). Hence CCDC28B couples FAM21/WASH to VPS35 through an interaction involving the C-terminal portion of the multiple repeat-domain of FAM21, where the molecular determinant for its interaction with retromer and EE association also maps [25].
T cells from CVID patients carrying the ccdc28b 211 C > T variant form dysfunctional immune synapses
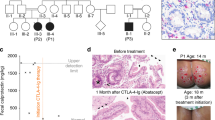
We next addressed the pathogenic potential of the CVID-associated ccdc28b 211 C > T SNP on 7 CVID patients carrying one 211 C > T ccdc28b allele (P1-P7) (Table 1) and 4 healthy donors (HD1-4). Expression of the wild-type and mutant allele was comparable in T-CVID T-cells, as assessed by allele-specific qRT-PCR (Figs. 7A and S8A). Proliferation assays of PBMCs in response to plate-bound anti-CD3 mAb showed defective T-cell responses in all patients carrying the R25W variant, who were on this basis classified as T-CVID (Fig. 7B). Surface CD3 and CCDC28B protein levels were comparable among individuals (Fig. S8B, C).
A Allele-specific real-time RT-PCR analysis of CCDC28B mRNA using specific primer pairs to amplify the wild-type transcript (wt) or the transcript carrying the 211 C > T mutation (mut) (see Table S4) on PBMCs from five healthy donors (HD) and seven T-CVID patients heterozygous for the 211 C > T ccdc28b allele (T-CVID). The relative gene transcript abundance was determined on triplicate samples using the ddCt method. B 3H-thymidine incorporation in triplicate samples of PBMC purified from four healthy donors (HD), and seven patients heterozygous for the 211 C > T ccdc28b allele (T-CVID) and activated on plate-bound anti-CD3ε mAb (OKT3) for 72 h. Data are expressed as counts per minute (cpm, mean ± SD). C–E Immunofluorescence analysis of CD3ζ (C), tyrosine phosphoproteins (D), or γ-tubulin (E) in 15-min conjugates of HD and T-CVID T-cells and Raji cells pulsed with a mix of SEA, SEB, and SEE. For each panel the scattered dot plot show, for each individual, the quantification (%) of conjugates with CD3ζ, PTyr, or γ-tubulin staining at the IS (n conjugates = 100) (C–E, left); for CD3ζ and PTyr the relative fluorescence at the T-cell: APC contact site compared to the remaining T-cell area (recruitment index) (C, D, right); for γ-tubulin the mean distance from the T-cell:APC contact site (μm) (n = 25 conjugates) (E, right). F, G Graphs showing, for each individual, the quantification (%) of conjugates with CD3ζ (F) or PTyr (G) accumulation at the IS in 15-min conjugates of HD and T-CVID T cells, nucleofected with either a GFP-encoding vector or the same vector encoding wild-type GFP-tagged CCDC28B and SEA/SEB/SEE-pulsed Raji cells (n conjugates = 30). The levels of mRNA encoding GFP-tagged wild-type CCDC28B were 3.39 ± 0.58 compared to endogenous ccdc28b mRNA in all samples (HD and T-CVID T cells), as assessed by allele-specific real-time RT-PCR. Expression of endogenous 211 C > T ccdc28b in T-CVID patients was not modified by overexpressed GFP-tagged wild-type CCDC28B (0.91 ± 0.21 compared to cells transfected with the empty vector). H, I Intracellular staining flow cytometry of HD and T-CVID T cells, nucleofected with either a GFP-encoding vector or the same vector encoding wild-type GFP-tagged CCDC28B, and stimulated for 6 h on immobilized anti-CD3ε (OKT3) and anti-CD28 mAb. Cells were permeabilized and stained with anti-IL-2 or anti-IFNγ mAb and the percentage of IL-2-positive or IFNγ-positive cells was quantified by flow cytometry. Statistical significance is shown only for activated sample. The differences of activated samples compared to the respective non-activated control were all statistically different. J Immunofluorescence analysis of F-actin in 15 min-conjugates of HD and T-CVID T-cells stained with fluorochrome-tagged phalloidin. The scattered dot plots show, for each individual, the quantification (%) of conjugates with F-actin staining at the IS (n conjugates = 100) (left) or the relative fluorescence of F-actin at the T-cell:APC contact site compared to the remaining T-cell area (recruitment index) (n = 25 conjugates) (right). K–M Immunofluorescence analysis of recycling TCRs (tCD3; see legend to Fig. 4E for experimental setting) in HD and T-CVID T-cells conjugated for 15 min with SEA/SEB/SEE-pulsed Raji cells and co-stained for F-actin (G), WASH (H), or FAM21 (I). For each panel the histograms show, for each individual, the co-localization of tCD3 with F-actin, WASH or FAM21 on individual dots (left, Mander’s coefficient), or the quantification of tCD3+ dots positive for the same markers (right, %). Measurements were taken on 50 conjugates and a mean of 10 dots per cell were analyzed. All graphs show the mean values ±SD of the HD and T-CVID groups. Error bars, SD. ****p ≤ 0.0001; ***p ≤ 0.001 (one-way analysis of variance (ANOVA), multiple comparisons).
Similar to CCDC28-KD T-cells, the synaptic accumulation of TCRs and tyrosine phosphoproteins was profoundly impaired in T-cells from all patients carrying the 211 C > T ccdc28b allele compared to healthy controls (Figs. 7C, D and S8D) despite normal centrosome polarization (Figs. 7E and S8D). To understand whether the IS defects were caused by 211 C > T ccdc28b heterozygosity, T-cells were transfected with a construct encoding GFP-tagged wild-type CCDC28B or GFP control. Overexpression of wild-type CCDC28B restored the ability of CCDC28BR25W-expressing T-CVID T-cells to form functional synapses (Figs. 7F, G and S8E). The IS defects in T-CVID T-cells led to impaired TCR-dependent cytokine production, which could be restored by overexpression of the wild-type allele (Fig. 7H, I), whereas healthy donor T-cells were unaffected (Fig. 7F–I). Hence 211 C > T ccdc28b heterozygosity impairs the ability of T-CVID T-cells to assemble functional synapses.
To address the underlying mechanism, we asked whether 211 C > T ccdc28b heterozygosity in T-CVID T-cells leads to the F-actin phenotypes observed in CCDC28 KD T-cells (Fig. 4). Defective synaptic F-actin accumulation was observed in these T-cells compared to healthy donors (Figs. 7J and S8F). Additionally, the proportion of tCD3+ endosomes positive for F-actin, WASH or FAM21, as well as tCD3 co-localization of with these molecules, was profoundly impaired (Figs. 7K–M and S9A–C). These data indicate that CCDC28BR25W expression in T-CVID T-cells leads to dysfunctional synapses by impairing the ability of wild-type CCDC28B to recruit FAM21/WASH to recycling TCRs, thereby inhibiting local F-actin polymerization.
The CCDC28B residue mutated in CVID is required for coupling FAM21/WASH to retromer
The investigate how CCDC28BR25W uncouples recycling TCRs from FAM21/WASH we transfected Jurkat cells, which carry the wild-type ccdc28b allele, with a GFP-tagged construct encoding CCDC28BR25W, reproducing the heterozygosity found in patients (Figs. 8A and S10A). CCDC28BR25W overexpression impaired IS assembly, as assessed by the defective synaptic accumulation of TCRs, phosphoproteins and F-actin (Figs. 8B–D and S10B, C). Similar to the CCDC28BR25W-expressing T-CVID T-cells, CCDC28BR25W expression in Jurkat cells impaired the association of FAM21 and WASH with tCD3+ endosomes and the accumulation of F-actin thereon (Fig. 8E–G), highlighting R25 as a critical determinant in CCDC28B-dependent FAM21/WASH recruitment to recycling TCRs. Consistently, GFP-specific immunoprecipitates from Jurkat cells expressing wild-type CCDC28B or CCDC28BR25W revealed that the R25W substitution compromised the interaction of CCDC28B with FAM21 and WASH, notably without affecting its interaction with VPS35 (Fig. 8H). These results were confirmed by pull-down assays using GST-tagged CCDC28BR25W (Fig. 8I and S6B). Hence CCDC28B couples FAM21/WASH to retromer through R25.
A Immunoblot analysis with anti-CCDC28B antibodies of Jurkat cells transiently transfected with plasmids encoding GFP-tagged wild-type CCDC28B (wt) or GFP-tagged CCDC28BR25W (mut). The recombinant and endogenous forms are indicated. Actin was used as loading control. The levels of mRNA encoding GFP-tagged mutant CCDC28B were 4.81 ± 0.56 compared to endogenous ccdc28b mRNA, as assessed by allele-specific real-time RT-PCR (n = 2), while the levels of the respective protein quantified on the immunoblots were 1.89 ± 0.35 (n = 3). B–D Immunofluorescence analysis of CD3ζ (B), tyrosine phosphoproteins (C) or F-actin (D) in 15-min conjugates of Jurkat cells expressing GFP-tagged wild-type CCDC28B (wt) or GFP-tagged CCDC28BR25W (mut) and SEE-pulsed Raji cells (APC). In each panel the histograms show the quantification (%, mean ± SD) of conjugates with CD3ζ, PTyr, or F-actin staining at the T-cell:APC contact site (at least 100 conjugates were analyzed for each marker) (n ≥ 3) (left) and the relative fluorescence of CD3ζ, PTyr, or F-actin at the T-cell:APC contact site compared to the remaining T-cell area (recruitment index, mean±SD) (n = 30 conjugates) (right). E–G Immunofluorescence analysis of recycling TCRs (tCD3; see legend to Fig. 4E for experimental setting) in 15-min conjugates of Jurkat cells expressing GFP-tagged wild-type CCDC28B (wt) or GFP-tagged CCDC28BR25W (mut) and SEE-pulsed Raji cells (APC), co-stained for F-actin (E), WASH (F), or FAM21 (G). For each panel the histograms show the quantification (mean ± SD) of tCD3+ dots positive for F-actin, WASH or FAM21 (left, %); and the co-localization of tCD3 with F-actin, WASH or FAM21 on individual dots (bottom, Mander’s coefficient, mean ± SD). Measurements were taken on 50 conjugates from at least three independent experiments and a mean of 10 dots per cell were analyzed. H Immunoblot analysis of GFP-specific immunoprecipitates (CCDC-GFP IP) from post-nuclear supernatants of Jurkat cells transiently transfected with plasmids encoding GFP-tagged wild-type CCDC28B (wt) or GFP-tagged CCDC28BR25W (mut). Immunoprecipitates carried out with non-immune Abs were used as negative control (neg ctr). Lysates (lys) belong to the same immunoblot (different exposure). The quantifications of the relative intensities of the immunoreactive bands (mut vs ctr) are shown in the box (n = 5). I Immunoblot analysis of GSH-Sepharose pull-down assays on post-nuclear supernatants of Jurkat cells using GST fusion proteins encoding wild-type CCDC28B (GST-wt), or CCDC28BR25W (GST-mut), or the CCDC28B CC domain (GST-CC), or a CCDC28 deletion mutant lacking the CC domain (ΔCC) (see schemes in Fig. S6B). Recombinant GST was used as negative control. Lysates (lys) belong to the same immunoblot (different exposure). For each filter the Ponceau staining is shown (n = 3). Total post-nuclear supernatants (lys) were included in all gels. The migration of molecular mass markers is shown for each filter. Error bars, SD. ***p < 0.001; **p < 0.01; *p < 0.05 (Student’s t test).
To test whether the CC domain mediates CCDC28B binding to retromer we generated GST-tagged constructs encoding either the isolated CC domain or lacking the CC domain (ΔCC-CCDC28B) (Fig. S6B). VPS35 was pulled down by the GST-tagged CC domain, while the interaction of CCDC28B with VPS35 was lost in the absence of its CC domain (Fig. 8I). Consistent with the finding that CCDC28B interacts with FAM21/WASH through R25, FAM21 and WASH were pulled down by the ΔCC-CCDC28B GST fusion but not by the isolated CC domain (Fig. 8I). Hence CCDC28B exploits its CC domain to interact with retromer, and the molecular determinant containing R25 to recruit FAM21/WASH (Fig. S6D).
Discussion
Polarized recycling is exploited by T-cells to sustain TCRs accumulation at the IS [19, 20]. Starting from the finding of an increased frequency of a potentially pathogenic SNP in the gene encoding the ciliopathy-related protein CCDC28B in CVID, we identify CCDC28B as a new player in this pathway acting at EEs, where it couples the actin regulator WASH and its interactor FAM21 to retromer to promote actin polymerization (Fig. S11). The ability of CCDC28B to recruit FAM21/WASH to retromer is dependent on R25, which is mutated in the CVID-associated variant. This interaction maps to the C-terminal portion of the multiple-repeat domain of FAM21, which mediates its association with retromer at EEs [25, 28, 29]. In turn, CCDC28B binds to VPS35/retromer through its C-terminal CC domain (Fig. S6D).
ccdc28b was identified in a screen for disease modifying-genes in BBS [17]. CCDC28B colocalizes with the core BBS protein complex, the BBSome, at the basal body of ciliated cells [17]. ccdc28b knockdown results in impaired ciliogenesis and developmental defects that recapitulate BBS phenotypes [18]. These observations, together with the epistatic effect of heterozygous ccdc28b mutations on BBS severity [17], indicate that CCDC28B may facilitate BBSome function. Interestingly, recycling has been implicated in targeting ciliary proteins to the basal body [30]. Our finding that CCDC28B promotes TCR recycling to the IS in the non-ciliated T-cell suggests a role in BBSome-dependent ciliary protein transport. The 211 C > T ccdc28b variant has been proposed as a disease gene in a family affected by another ciliopathy, Joubert syndrome [31]. Although its pathogenicity in Joubert syndrome remains to be confirmed [32], CCDC28BR25W expression in ciliated cells results in ciliogenesis defects (unpublished), indicating that the related processes of IS and primary cilium assembly are similarly affected by this protein variant. Abnormalities in immune cell development have been recently reported in BBS patients [33], suggesting the possibility that ccdc28b might act as a disease modifier also in the context of these immune-related defects.
The genetic etiology of CVID still poses a formidable challenge. The heterogeneity of clinical presentations and immune defects suggests a multigenic origin, which is supported by the growing number of disease genes that directly or indirectly affect the maturation of class-switched antibody-producing plasma cells [34]. A significant proportion of CVID patients has defects in T-cell activation and helper function [2, 5]. Here we show that T-cells from CVID patients heterozygous for 211 C > T ccdc28b display defective TCR-dependent proliferation and cytokine production, which can be accounted for by their inability to form functional immune synapses. This defect is causally linked to an impairment in actin polymerization at endosomal TCRs, caused by defective recruitment of FAM21/WASH to retromer through a determinant containing R25 and responsible for the FAM21 binding. That this interaction is impaired in T-CVID T-cells, which co-express wild-type and mutant CCDC28B, suggests that CCDC28BR25W functions as a dominant negative mutant. In support of this notion, introducing the mutant allele in Jurkat cells, which express only the wild-type allele, reproduced the phenotype observed in T-CVID T-cells, indicating that CCDC28BR25W interferes with the function of wild-type CCDC28B. The fact that CCDC28BR25W associates with VPS35 but not with FAM21/WASH, and hence does not promote endosomal F-actin polymerization, suggests that CCDC28BR25W limits the access of wild-type CCDC28B to VPS35, thereby acting in a dominant negative fashion. Increasing the amount of wild-type CCDC28B in T-CVID T-cells through forced expression can be expected to result in an effective competition with CCDC28BR25W for binding to VPS35, thereby correcting the defect.
The pathogenic role of the 211 C > T ccdc28b SNP in T-CVID is supported by the restoration of functional synapses in T-cells from these patients through forced expression of wild-type CCDC28B. While this highlights ccdc28b as a potential new disease gene in T-CVID, the 211 C > T SNP is also present in the healthy population, albeit at a lower frequency. This suggests that this ccdc28b allele might interact epistatically with other deleterious gene mutations to modify disease presentation or penetrance, similar to BBS [17]. While our exome sequencing of the ciliopathy-related genes in the initial 7 T-CVID patients ruled out the presence of mutations in the respective coding sequence, we cannot rule out expression defects resulting from mutations in non-coding parts of these genes. Additionally, sequencing of the ciliopathy-related genes has not been carried out yet on the T-CVID patients belonging to the enlarged cohort, who were only tested for the presence of 211 C > T ccdc28b SNP. We cannot exclude however the presence of deleterious mutations in genes not directly implicated in ciliopathies in these patients. In particular, based on the implication of CCDC28B in a key step in TCR recycling, which is regulated by a number of proteins that could have some degree of functional redundancy [20, 35], we can hypothesize epistatic interactions of ccdc28b with the genes encoding components of the TCR sorting and recycling machinery.
Ciliogenesis proteins have emerged as important regulators of IS assembly [36]. Ciliary proteins have been identified at different steps of this process, from actin clearance from the synaptic contact (Inpp5e [37]), to centrosome polarization (BBS1 [38]), to polarized TCR recycling (IFT20, IFT52, IFT57 [13, 39]). The identification of CCDC28B as component of the TCR trafficking machinery provides further evidence of the conservation in the pathways that control IS and primary cilium assembly. Taken together with the increased frequency of a pathogenic ccdc28b mutation in CVID, these data highlight ciliogenesis proteins as promising disease candidates for primary immunodeficiencies or other immune-related disorders associated with dysfunctional immune synapses.
Materials and methods
Patients and cells
Patients were classified as CVID according to using established criteria [40]. Hematologic and immunophenotypical characterization of patients is shown in Table 1. NGS analysis on these patients ruled out the presence of genes know to be associated with defective B cell function (unpublished results). Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of healthy donors and CVID patients by density centrifugation on Lympholyte-H Cell Separation Media (Euroclone, Milan, Italy). Experiments were performed after approval by the institutional review board of the Universities of Florence and Brescia for these studies. Peripheral blood was obtained after informed consent according to the Declaration of Helsinki, and sample size was kept small according to the guidelines of the ethics committee. No overt infectious disease was present at the time of blood sampling.
Cells lines included Jurkat T-cells, Raji B-cells and the BJ-5ta human hTERT-immortalized fibroblast line. Primary T-cells purified from peripheral blood of healthy donors using the Rosette Sep kit followed by density centrifugation on Lympholyte. The latter were expanded by stimulation with 0.1 µg/ml Staphylococcal enterotoxin E (SEE), B (SEB), and A (SEA) (Toxin Technology, Sarasota, FL, USA) and 130 U/ml human IL-2 (Miltenyi Biotec) for 7–10 days (Fig. S4A) in order to increase the number of antigen-specific T-cells.
Antibodies, reagents, plasmid, and GST fusions
IgG from OKT3 (anti-human CD3ɛ, IgG2) hybridoma supernatants was purified using Mabtrap (Amersham Biosciences, Inc., Piscataway, NJ, USA) and titrated by flow cytometry. All primary commercial antibodies used in this work are listed in Table S3, together with information about the dilutions used for immunoblotting, immunofluorescence and immunoprecipitation. Secondary peroxidase-labeled antibodies were from Amersham Biosciences, Alexa Fluor 488- and 555-labeled secondary Abs from Molecular Probes (Invitrogen, Eugene, OR, USA). Cell Tracker Blue was from Molecular Probes (Invitrogen) and poly-l-lysine from Sigma-Aldrich. 16% Formaldehyde was from Thermo Scientific.
GFP-tagged wild-type CCDC28B (wt) was obtained by cloning CCDC28B wt cDNA in pEGFP-N1 digested with XhoI and BamHI restriction enzymes (Thermo Fisher Scientific, Waltham, MA USA). GFP-tagged CCDC28BR25W (mut) was obtained using the QuikChange II site-directed mutagenesis kit (Agilent, Santa Clara, USA) and the mutated sequence was verified by DNA sequencing (Bio-Fab, Rome, Italy). Primers used for cloning are listed in Table S4.
The GST-tagged proteins used in this study are schematized in Fig. S6B. The constructs encoding the GST-tagged N- and C-terminal portions of the FAM21 multiple repeat domain of FAM21 (FAMn and FAMc, respectively) and GST-tagged WASH lacking the VCA domain were previously described [25, 28]. The cDNAs encoding CCDC28B wt, CCDC28B mut, CCDC28B depleted of the coiled-coil domain (∆CC) and the CCDC28B coiled-coil domain (CC) were inserted into the bacterial expression plasmid pGEX-6P-2, using EcoRI and XhoI restriction enzymes (Thermo Fisher Scientific, Waltham, MA USA). The recombinant fusion proteins, as well as control GST, were affinity purified on GSH-Sepharose (GE Healthcare) from bacterial cultures incubated for 4 h at 37 °C with 0.25 mM isopropyl-β-d-thiogalactopyranoside and lysed by sonication in PBS-1% Triton X-100.
DNA extraction, NGS analysis, and PCR sequencing
DNA was extracted from peripheral blood from CVID patients using QIAamp DNA Mini Kit or QIAmp Blood DNA Mini Kit (Qiagen GmbH, Hilden, Germany). The extraction was performed according to the manufacturer’s instructions for genomic DNA purification from blood. Quality control of DNA samples from 7 T-CVID patients and high-coverage NGS analysis of the exons of approximately 100 genes encoding proteins implicated in ciliogenesis, vesicular trafficking and TCR signaling were performed by Oxford Gene Technology-The Molecular Genetics Company™ (Begbroke, Oxfordshire, UK) (unpublished results). Variants were annotated with gene and gene function data from Ensembl [41]. Allele frequencies in the matched ethnic group were from 1000 Genomes Project Phase 3 [42] (Central European) and the Genome Aggregation database (gnomAD; Broad Institute) [43]. Variants of five ciliopathy-related gene variants predicted as likely to cause significant functional problems were analyzed by classical Polymerase Chain Reaction (PCR) and Sanger sequencing in a cohort of 101 CVID patients. PCR reactions were performed using Go Taq Long Pcr Master Mix 2X (Promega) and primer pairs reported in Table S4. Sequence reactions were performed by Bio-Fab research Srl (Roma, Italy).
RNA purification and allele-specific real-time RT-PCR
Total RNA was extracted from Jurkat cells, healthy donors and T-CVID patients using RNeasy kit (Qiagen GmbH, Hilden, Germany) and reverse trascribed using iScript™ cDNA Synthesis Kit (Bio-Rad). Real-time quantitative PCR (qRT-PCR) was performed in triplicate on each cDNA on 96-well optical PCR plates (Sarstedt) using SSo Fast EvaGreenR SuperMix (Bio-Rad) according to the manufacturer’s instructions and a CFX96 Real-Time system (Bio-Rad). After an initial denaturation for 3 min at 95 °C, denaturation in the subsequent 39 cycles was performed for 10 s at 95 °C, followed by 30 s of primer annealing at 60 °C (or 70 °C for CCDC28B allele-specific primers). Results were processed and analyzed using CFX Manager Version 1.5 software (Bio-Rad). Transcript levels were normalized to HPRT1, which was used as a housekeeping gene. For cells transfected with GFP-tagged wild-type or mutant CCDC28B-encoding plasmid constructs, allele-specific real-time PCR data were normalized to the transfection efficiency using GFP-specific primers. The primers used to amplify the cDNA fragments are listed in Table S4.
Transfections and RNA interference
Jurkat cells or primary T-cells were transiently transfected by electroporation with human CCDC28B-specific esiRNAs (EHU082341) (KD) and unrelated control esiRNA (EHURLUC) (ctr) (150 ng/106 cells) (Sigma-Aldrich, Milan, Italy) and assays were carried out after 48 h. All samples were tested by immunoblotting to check the efficiency of CCDC28 knockdown.
Rescue experiments were performed by transfecting CCDC28B KD Jurkat cells with GFP-tagged wt CCDC28B (1 µg/106 cells) by electroporation and assays were carried out after 24 h. Transient transfection of Jurkat cells with GFP-tagged wt or mut CCDC28B (1 µg/106 cells, or dose response as specified in Fig. S10A) were performed by electroporation and assays were carried out after 24 h. All samples were resuspended in OPTI-MEM 1X solution (Gibco) before undergo electroporation using Gene Pulser II (Bio-Rad). Primary T cells from healthy donors or T-CVID patients were transiently transfected with GFP-tagged wt CCDC28B (1 µg/106 cells) or with pEGFP-N1, using the Human T-cell Nucleofector Kit (Amaxa Biosystems) and the conditions recommended by the manufacturer. Assays were carried out after 24 h.
IS formation and TCR recycling assay
Conjugates between Jurkat or primary T-cells (freshly purified or blasts, see Fig. S4A) and superantigen (SAg)-pulsed Raji B-cells were carried out as previously described [13]. In conjugates with Jurkat cells, Raji cells (used as APC) were pulsed for 2 h with 10 µg/ml SEE and labeled with 10 µM Cell Tracker Blue for the last 20 min, while for IS experiments with freshly purified or SAg-expanded T-cells Raji cells were pulsed with a mix of SEE, SEA and SEB (10 µg/ml each). Analyzes were carried out 15 min after conjugate formation, with the exception of the analyses shown in Figs. 2A and S2B, which included a shorter time (5 min) or longer times (30 and 60 min), respectively. Conjugates between T-cells and unpulsed B-cells, where no IS forms at the interface of the cell pairs, were used as negative controls. Samples were allowed to adhere for 15 min on polylysine-coated wells of diagnostic microscope slides (Erie Scientific Company), fixed by immersion in methanol for 10 min at −20 °C, or analyzed under non-permeabilizing conditions after fixation in 4% paraformaldehyde for 20 min at RT. Following fixation, samples were washed in PBS and incubated with primary antibodies (Table S3) overnight at 4 °C or 1 h at room temperature. After washing in PBS, samples were incubated for 1 h at room temperature with Alexa-Fluor-488- and −555-labeled secondary antibodies.
For the TCR recycling assays, cells were incubated with saturating concentrations of anti-human CD3ε mAb (clone OKT3) at 37 °C for 2 h, washed to remove excess mAb, acid-stripped and used in IS experiments as described above. Analyses were carried out 15 min after conjugate formation, with the exception of the analyses shown in Fig. S2D, which included longer time points (30 and 60 min).
Confocal and TIRF immunofluorescence, co-localization analyses
Confocal microscopy was carried out on a Zeiss LSM700 using a 63X objective. Images were acquired with pinholes opened to obtain 0.8-µm-thick sections. Detectors were set to detect an optimal signal below the saturation limits. Images were processed with Zen 2009 image software (Carl Zeiss, Jena, Germany). The quantitative co-localization analysis of CCDC28B with γ-tub, Rab5, Rab11, and GM130 was performed on median optical sections using ImageJ and JACoP plug-in to determine Mander’s coefficient M1 [44], which represents the percentage of CCDC28B pixels (red channel) that overlaps γ-tubulin, Rab5, Rab11 or GM130 pixels (green channel) (0 = no co-localization; 1 = 100% co-localization).
Scoring of conjugates for polarized TCR recycling to the IS in permeabilized or non-permeabilized cells as well as for PTyr, CD3ζ and γ-tub clustering at the IS in permeabilized cells was based on the concentration of the respective staining solely at the T-cell:APC contact. Centrosome polarization to the IS, assessed as the reduction of the distance of the centrosome from the APC contact site in T cells exposed to SAg-loaded versus unloaded APC [45], was measured using ImageJ. The recruitment index was calculated for each marker as the relative fluorescence at the T-cell:APC contact site compared to the remaining T-cell area. For co-localization in conjugates obtained following TCR recycling assays, analyses were performed on single tCD3 dots using ImageJ and JACoP plug-in to determine Manders’ coefficient M1. The same type of analysis was carried out on WASH/F-actin and WASH/Rab5 conjugates, where single WASH+ dots were analyzed, and FAM21/Rab5 conjugates, where FAM21+ dots were analyzed. The distance of F-actin+ or WASH+ dots from IS was measured using ImageJ.
In TIRF immunofluorescence experiments, F-actin ring formation was evaluated in primary T-cells, allowed to adhere for 15 min on cover glass, thickness 0.16 mm (Vwr International Srl, Milan, Italy) coated with purified anti-human CD3ε mAb, clone UCHT1 (Biolegend), fixed in PBS-4% paraformaldehyde methanol free for 15 min at room temperature, permeabilized with PBS-0.25% Triton X-100 for 15 min and stained with Alexa Fluor 488 phalloidin (Invitrogen) in PBS-1% BSA for 30 min. TIRF images were acquired on an inverted Leica DMi8 microscope (Leica Microsystem), with 63X objectives using a PCO Edge sCMOS camera (Leica Microsystems). Spreading areas were measured using ImageJ.
Flow cytometry
Protein tyrosine phosphorylation was analyzed by flow cytometry in conjugates of control or CCDC28B KD Jurkat cells and SEE-pulsed Raji cells (APC), at different time points. Conjugates were labeled with PE-labeled anti-human CD3 mAb (CD3ε, clone OKT3) (Biolegend) and anti-PTyr antibodies (Merk Millipore) and CD3+ cells were analyzed. F-actin was quantified by flow cytometry in primary T-cells stimulated with purified anti-human CD3ε mAb, clone UCHT1 (Biolegend) for 15 min at 37 °C, fixed and permeabilized using Cytofix/Cytoperm Plus kit (Becton Dickinson, San Jose, CA) and stained with Alexa Fluor 488 phalloidin (Invitrogen). F-actin content was calculated as fold increase of stimulated versus unstimulated samples. CD3 surface expression on Jurkat cells KD for CCDC28B expression, primary T-cells KD for CCDC28B and their respective controls, and PBMCs from CVID patients and healthy donors, was measured by flow cytometry using PE-labeled anti-human CD3 mAb (CD3ε, clone OKT3) (Biolegend). Flow cytometry was carried out using a Guava Easy Cyte cytometer (Millipore).
For proliferation assays, PBMCs from CVID patients and healthy donors were stimulated with plate-bound anti-CD3ε mAb (OKT3) and processed 16-48 h after activation. [3H]-thymidine (1 mCi) was added to each well for the last 18 h of culture. After harvesting the cells with an automatic harvester, proliferation was determined by measuring the [3H]thymidine incorporation in a liquid scintillation counter as described [8]. For IL-2 and IFNγ measurements, 106 cells were activated on plate-bound anti-CD3ε mAb, clone OKT3 (5 μg/ml) (BioLegend; #317302) and anti-CD28 mAb, clone CD28.2 (5 μg/ml) (BioLegend; #302902) for 6 h. Membrane permeabilization was carried out using the BD cytofix/cytoperm fixation/permeabilization kit (BD Bioscience; #554714) according to the manufacturer’s protocol prior to processing samples for flow cytometry.
Immunoprecipitations, immunoblots, and in vitro binding assays
For immunoprecipitation experiments, cells (5 × 107/sample) were pelleted, washed twice in ice-cold PBS and lysed in 500 μl lysis buffer (0.5% Triton X-100 in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl) in the presence of protease inhibitors (Calbiochem, 539,134) and the phosphatase inhibitor sodium vanadate (Sigma-Aldrich, S6508). Quantification was carried out using the BCA Assay kit (EuroClone, EMP014500). Postnuclear supernatants were immunoprecipitated using the appropriate antibodies (see Table S3) or mouse IgG isotype control (Invitrogen) and protein A Sepharose (GE Healthcare). PAS-antibody complexes were pelleted, washed 4X with 1 ml lysis buffer, then resuspended in 15 μl Laemmli buffer (LifeTechnologies, B0007), boiled for 5 min and subjected to SDS-PAGE. All gels included a sample of the lysates used for the immunoprecipitations (50 μg/sample). In vitro binding assays were carried using the GST-tagged proteins on GSH-Sepharose-precleared postnuclear supernatants from 5 × 107cells/sample, lysed in 1% Triton X-100 in the presence of protease inhibitors. Immunoblotting was carried out using peroxidase-labeled secondary Ig and a chemiluminescence detection kit (Pierce Rockford). Stripping was carried out by using ReBlot Plus Mild Antibody Stripping Solution, 10x (Chemicon). Blots were scanned using a laser densitometer (Duoscan T2500; Agfa, Milan, Italy) and quantified using ImageJ.
Statistical analysis
GraphPad (Prism Software) was used to calculate mean values, standard deviation values and statistical significance. Values with Gaussian distribution were analyzed using Student’s t test (paired or unpaired), one sample t test (theoretical mean = 1) or one-way ANOVA. Values without normal distribution were analyzed using Mann–Withney test. A level of P < 0.05 was considered statistically significant.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Resnick ES, Cunningham-Rundles C. The many faces of the clinical picture of common variable immune deficiency. Curr Opin Allergy Clin Immunol. 2012;12:595–601.
Yazdani R, Hakemi MG, Sherkat R, Homayouni V, Farahani R. Genetic defects and the role of helper T-cells in the pathogenesis of common variable immunodeficiency. Adv Biomed Res. 2014;3:2.
Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127:1360–1367 e1366.
Abolhassani H, Hammarstrom L, Cunningham-Rundles C. Current genetic landscape in common variable immune deficiency. Blood. 2020;135:656–67.
Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Drager R, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4:261–8.
Lougaris V, Baronio M, Gazzurelli L, Lorenzini T, Fuoti M, Moratto D, et al. A de novo monoallelic CTLA-4 deletion causing pediatric onset CVID with recurrent autoimmune cytopenias and severe enteropathy. Clin Immunol. 2018;197:186–8.
Fischer MB, Wolf HM, Hauber I, Eggenbauer H, Thon V, Sasgary M, et al. Activation via the antigen receptor is impaired in T cells, but not in B cells from patients with common variable immunodeficiency. Eur J Immunol. 1996;26:231–7.
Majolini MB, D’Elios MM, Boncristiano M, Galieni P, Del Prete G, Telford JL, et al. Uncoupling of T-cell antigen receptor and downstream protein tyrosine kinases in common variable immunodeficiency. Clin Immunol Immunopathol. 1997;84:98–102.
Boncristiano M, Majolini MB, D’Elios MM, Pacini S, Valensin S, Ulivieri C, et al. Defective recruitment and activation of ZAP-70 in common variable immunodeficiency patients with T cell defects. Eur J Immunol. 2000;30:2632–8.
Paccani SR, Boncristiano M, Patrussi L, Ulivieri C, Wack A, Valensin S, et al. Defective Vav expression and impaired F-actin reorganization in a subset of patients with common variable immunodeficiency characterized by T-cell defects. Blood. 2005;106:626–34.
Capitani N, Ariani F, Amedei A, Pezzicoli A, Matucci A, Vultaggio A, et al. Vav1 haploinsufficiency in a common variable immunodeficiency patient with defective T-cell function. Int J Immunopathol Pharmacol. 2012;25:811–7.
Dustin ML, Choudhuri K. Signaling and polarized communication across the T cell immunological synapse. Annu Rev Cell Dev Biol. 2016;32:303–25.
Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–9.
Finetti F, Patrussi L, Galgano D, Cassioli C, Perinetti G, Pazour GJ, et al. The small GTPase Rab8 interacts with VAMP-3 to regulate the delivery of recycling T-cell receptors to the immune synapse. J Cell Sci. 2015;128:2541–52.
Onnis A, Finetti F, Patrussi L, Gottardo M, Cassioli C, Spano S, et al. The small GTPase Rab29 is a common regulator of immune synapse assembly and ciliogenesis. Cell Death Differ. 2015;22:1687–99.
Vivar OI, Masi G, Carpier JM, Magalhaes JG, Galgano D, Pazour GJ, et al. IFT20 controls LAT recruitment to the immune synapse and T-cell activation in vivo. Proc Natl Acad Sci USA. 2016;113:386–91.
Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–30.
Cardenas-Rodriguez M, Osborn DP, Irigoin F, Grana M, Romero H, Beales PL, et al. Characterization of CCDC28B reveals its role in ciliogenesis and provides insight to understand its modifier effect on Bardet-Biedl syndrome. Hum Genet. 2013;132:91–105.
Soares H, Lasserre R, Alcover A. Orchestrating cytoskeleton and intracellular vesicle traffic to build functional immunological synapses. Immunol Rev. 2013;256:118–32.
Onnis A, Baldari CT. Orchestration of immunological synapse assembly by vesicular trafficking. Front Cell Dev Biol. 2019;7:110.
Das V, Nal B, Dujeancourt A, Thoulouze MI, Galli T, Roux P, et al. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity. 2004;20:577–88.
Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29.
Martin-Cofreces NB, Sanchez-Madrid F. Sailing to and docking at the immune synapse: role of tubulin dynamics and molecular motors. Front Immunol. 2018;9:1174.
Seaman MN, Gautreau A, Billadeau DD. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 2013;23:522–8.
Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711.
Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–23.
Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD. Endosomal receptor trafficking: Retromer and beyond. Traffic. 2018;19:578–90.
Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–61.
Harbour ME, Breusegem SY, Seaman MN. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem J. 2012;442:209–20.
Carter SP, Blacque OE. Membrane retrieval, recycling and release pathways that organise and sculpt the ciliary membrane. Curr Opin Cell Biol. 2019;59:133–9.
Radha Rama Devi A, Naushad SM, Lingappa L. Clinical and molecular diagnosis of joubert syndrome and related disorders. Pediatr Neurol. 2020;106:43–49.
Barroso-Gil M, Powell L, Sayer JA. RE: clinical and molecular diagnosis of joubert syndrome and related disorders. Pediatr Neurol. 2020;112:10.
Tsyklauri O, Niederlova V, Forsythe E, Prasai A, Drobek A, Kasparek P, et al. Bardet-Biedl Syndrome ciliopathy is linked to altered hematopoiesis and dysregulated self-tolerance. EMBO Rep 2021;22:e50785.
Romberg N, Lawrence MG. Birds of a feather: common variable immune deficiencies. Ann Allergy Asthma Immunol. 2019;123:461–7.
Capitani N, Baldari CT. F-actin dynamics in the regulation of endosomal recycling and immune synapse assembly. Front Immunol. 2021;9:670882. https://doi.org/10.3389/fcell.2021.670882.
Cassioli C, Baldari CT. A Ciliary View of the Immunological Synapse. Cells 2019;8:789.
Gawden-Bone CM, Frazer GL, Richard AC, Ma CY, Strege K, Griffiths GM. PIP5 kinases regulate membrane phosphoinositide and actin composition for targeted granule secretion by cytotoxic lymphocytes. Immunity. 2018;49:427–37.
Cassioli C, Onnis A, Finetti F, Capitani C, Brunetti J, Compeer EB. et al. The Bardet-Biedl Syndrome complex component BBS1 regulates proteasome-dependent F-actin clearance from the centrosome to enable its translocation to the T cell immune synapse. Pre-print bioRxiv https://doi.org/10.1101/2020.11.30.403980.
Finetti F, Patrussi L, Masi G, Lucherini OM, Onnis A, Pazour GJ, et al. Immune synapse targeting of specific recycling receptors by the intraflagellar transport system. J Cell Sci. 2014;127:1924–37.
Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol. 2020;40:24–64.
Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–8.
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103Pt 3:857–62.
Esquerre M, Tauzin B, Guiraud M, Muller S, Saoudi A, Valitutti S. Human regulatory T cells inhibit polarization of T helper cells toward antigen-presenting cells via a TGF-beta-dependent mechanism. Proc Natl Acad Sci USA. 2008;105:2550–5.
Acknowledgements
We wish to thank Claire Hivroz for critical reading of the manuscript and Vincenzo Sorrentino for useful advice.
Funding
This work was carried out with the support of Fondazione Telethon, Italy (Grant GGP16003) to CTB. The support of ERC Synergy (grant 951329) to CTB is also acknowledged.
Author information
Authors and Affiliations
Contributions
NC, AO, FF, CC, and CTB designed research and analyzed and interpreted data; NC, AO, FF, CC, JB, AT, and CDB carried out the experiments; AP, SDE, MB, LG, DDB, MMD, and VL contributed vital reagents; NC and CTB drafted the manuscript.
Corresponding authors
Ethics declarations
competing interests
The authors declare no competing interests.
Ethical approval
Experiments were performed after approval by the institutional review board of the Universities of Florence and Brescia. Peripheral blood was obtained after informed consent according to the Declaration of Helsinki, and sample size was kept small according to the guidelines of the ethics committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Supplementary information
Rights and permissions
About this article
Cite this article
Capitani, N., Onnis, A., Finetti, F. et al. A CVID-associated variant in the ciliogenesis protein CCDC28B disrupts immune synapse assembly. Cell Death Differ 29, 65–81 (2022). https://doi.org/10.1038/s41418-021-00837-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00837-5
This article is cited by
-
A novel coiled-coil domain containing-related gene signature for predicting prognosis and treatment effect of breast cancer
Journal of Cancer Research and Clinical Oncology (2023)