Abstract
Cancer cells undergo complex metabolic alterations. The mechanisms underlying the tuning of cancer metabolism are under active investigation. Here, we identify the uncharacterized deubiquitinase JOSD2 as a positive regulator of cancer cell proliferation by displaying comprehensive effects on glucose catabolism. We found that JOSD2 directly controls a metabolic enzyme complex that includes Aldolase A, Phosphofructokinase-1 and Phosphoglycerate dehydrogenase, in vitro and in vivo. Further, JOSD2 expression, but not a catalytically inactive mutant, deubiquitinates and stabilizes the enzyme complex, thereby enhancing their activities and the glycolytic rate. This represents a selective JOSD2 feature that is not shared among other Machado–Joseph disease DUBs or observed in nontransformed cells. JOSD2 deficiency displays cytostatic effects and reduces glycolysis in a broad spectrum of tumor cells of distinct origin and its expression correlates with poor prognosis in non-small cell lung cancer. Overall, our study provides evidence for a previously unknown biological mechanism in which JOSD2 integrates glucose and serine metabolism with potential therapeutic implications.
Similar content being viewed by others
Introduction
Cancer cells undergo robust and complex metabolic rearrangements leading to changes in pathways critical for energy production and anabolic processes to support unrestrained cell proliferation [1, 2]. A variety of both intrinsic and extrinsic signals have been shown to alter the core metabolism of cells in a tumor context-dependent manner [2]. This is often mirrored by the aberrant signaling networks downstream of tumor suppressors and oncogenes, such as phosphoinositide 3-kinase, hypoxia-inducible factor, MYC proto-oncogene bHLH transcription factor (MYC) and mutant TP53. The tumor microenvironmental cues, including nutrient availability and interactions with stromal cells can further influence the metabolic phenotype of cancer cells [1,2,3,4]. In addition, metabolic enzymes can harbor mutations that can lead to altered catalytic activities. Enzymes in metabolic pathways have further been shown to display copy-number gains or losses that contribute to different stages of cancer development, such as tumor growth, metastasis and resistance to therapies [3, 5,6,7].
Beyond the genetic alterations, the abundance of metabolic enzymes has been suggested to be directly controlled by ubiquitin conjugations, which highlights the importance of proteolytic regulation in shaping metabolic processes [8]. Ubiquitin posttranslational modifications play a vital role in the ubiquitin-proteasome system (UPS) in maintaining protein quality control and homeostasis by regulating the proteasomal degradation, thus controlling the abundance and function of proteins [9]. Ubiquitin conjugation is accomplished by the sequential actions of ubiquitin‐activating (E1), ubiquitin‐conjugating (E2), and ubiquitin‐ligating (E3) enzymes that are responsible for regulating the specificity of the ubiquitination [10, 11]. Deubiquitinases or deubiquitinating enzymes (DUBs) are also critical components of the UPS. This subfamily of proteases possesses reversible abilities against E3 ligases, in which they enzymatically remove ubiquitin molecules from the target proteins [11], thus consequently, prevent substrate degradation leading to protein stabilization. However, whether deubiquitination processes are relevant for the direct regulation of metabolic enzyme stability to potentially shape metabolic programs in cancer cells is currently not fully elucidated.
DUBs play a major role in multiple cellular processes, including cell cycle regulation, cell growth, and proliferation [12,13,14] and can harbor cancer-specific mutations suggesting a role as tumor suppressors or oncogenes [15,16,17,18,19,20]. As dysregulation of DUB activities is associated with many pathologies, including human malignancies, they represent attractive drug targets for small-molecule discovery [17]. Still, many DUBs are poorly understood when it comes to their precise functions thus the identification and characterization of DUB target proteins is crucial to understand their physiological and pathological role.
Here, through an integrative effort employing systematic co-expression profiling, quantitative proteomics, isotopomer tracing and biochemical analyses, we unveil a new layer of metabolic regulation in cancer cells. Our findings provide a novel mechanism for the regulation of glucose metabolism through the control of protein stability of glycolytic enzymes.
Material and methods
Cell culture
All NSCLC cancer cell lines (NCI-H2087, NCI-H1792, NCI-H1437, U1810, NCI-H1755, NCI-H838, NCI-H3122, and A549) and ovarian cancer SK-OV-3 were grown in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% (v/v) heat-inactivated FBS (Gibco, Waltham, MA, USA), 100 U/ml penicillin and streptomycin (Sigma-Aldrich), 2 mM glutamine (Sigma-Aldrich, St. Louis, MO, USA). Breast adenocarcinoma MDA-MB-231, cervical HeLa cells, and human embryonic kidney HEK293T cells were grown in Dulbecco′s Modified Eagle′s medium (DMEM, Gibco) with the same supplements as RPMI and SH-SY5Y were maintained in DMEM:F12 with the same supplements as the RPMI. The nontransformed (normal) lung cells (Wi-38, IMR-90) were grown in DMEM supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin, 2% (w/v) glutamine and 1% nonessential amino acids. The chronic myelogenous leukemia cell line HAP Parental (HAP-P) and Josephin Domain-containing protein 2 (JOSD2) knockout cell line HAP (HAP-KO) (Horizon Discovery) were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco) with the same supplements as the RPMI. JOSD2 knockout cell line HAP-KO was generated by a 343 bp insertion in exon 2 using CRISPR/Cas9-editing system (Guide RNA Sequence: GCGCTGCCGTTCGTGGTACA). All cells were maintained in a logarithmic growth phase at 37 °C in a humidity atmosphere with 5% CO2.
Analysis of primary NSCLCs
Normalized gene expression values from the previous study (GSE31210) [21] were retrieved from Gene Expression Omnibus (GEO) using the R package GEOquery [22] to examine the correlation between DUBs and glycolysis-related genes. For gene symbols with multiple probe set assignments, the probe set was selected based on the probe with the highest absolute value of the differential expression.
Overall survival was analyzed using the web-based software program Kaplan–Meier Plotter (https://kmplot.com/analysis/). For JOSD2 and STAM binding protein (STAMBP), 673 patients could be included in the analyses and for Ubiquitin Specific Peptidase 5 (USP5), Proteasome 26S Subunit, Non-ATPase 7 (PSMD7), and Proteasome 26S Subunit, Non-ATPase 14 (PSMD14), 720 patients. The cutoff to divide patients into expression high and low groups was determined by the performance of all possible cutoff values between the lower and upper quantiles. Survival curves and the hazard ratio plot were generated using the R package Survminer version 0.4.8.
Metabolome and isotopomer analyses
To measure the levels of intracellular metabolites, extracts were prepared from 1 × 106 cells per sample in three biological replicates and analyzed using a capillary electrophoresis (CE)-connected ESI-TOF MS system as previously described [23]. Three 10-cm petri dishes were incubated for 24 h after transfection with either siRNA JOSD2 or nontargeting control siRNA prior to extraction. To trace glucose-derived carbons, cells were washed three times with PBS and incubated at 37 °C in glucose-free RPMI medium supplemented with 10% dialyzed FBS, 4 mM glutamine and 10 mM U-13C-glucose (Cambridge Isotope Laboratories) for 24 h. Intracellular metabolites (for both the metabolite and isotopomer analyses) were extracted with methanol per Human Metabolome (HMT) metabolite extraction protocol for adherent cells. Briefly, 5% mannitol was used to wash cells three times and extracted in 1.3 ml methanol containing 10 μM Internal Standard Solution (Human Metabolome Technologies). The extracted metabolites were centrifuged at 2300 × g at 4 °C for 5 min. The supernatant was centrifugally filtered through a Millipore 5-kDa cutoff filter to remove proteins at 9100 × g at 4 °C for 3 h. The filtrate was lyophilized, suspended in Milli-Q water and analyzed using Capillary-Electrophoresis Time of Flight Mass Spectrometer (CE-TOF MS, Human Metabolome Technologies, Boston, MA, USA).
Data processing
Raw data were obtained by CE-TOF MS and peaks were extracted using MasterHands automatic integration software [24]. The peak detection limit was determined by the signal-to-noise ratio (3:1). The peak area for each metabolite was normalized with respect to the area of the internal standard for the absolute quantitation of metabolites.
Quantitative multiplex proteomics
Sample preparation
All reagents were prepared in 50 mM HEPES, pH 8.5. Cysteines were reduced using 10 mM dithiothreitol (DTT) (56 °C, 30 min). Samples were cooled to 24 °C and alkylated with 10 mM iodoacetamide (room temperature, in the dark, 30 min). Subsequently, the samples were prepared for LC–MS/MS using the SP3 protocol [25], digested with trypsin (enzyme to protein ratio, 1:50) at 37 °C overnight. TMTplex™ Isobaric Label Reagent (Thermo Fisher, Waltham, MA, USA) was added to the samples per the manufacturer’s instructions. Labeled peptides were cleaned up using OASIS® HLB µElution Plate (Waters). Offline high pH reverse phase fractionation was performed using an Agilent 1200 Infinity high-performance liquid chromatography (HPLC) system, equipped with a Gemini C18 column (3 μm, 110 Å, 100 × 1.0 mm, Phenomenex). The solvent system consisted of 20 mM ammonium formate (pH 10.0) as mobile phase (A) and 100% acetonitrile as mobile phase (B).
Fractionation
Offline high pH reverse phase fractionation was performed using an Agilent 1200 Infinity HPLC system equipped with a quaternary pump, degasser, variable wavelength UV detector (set to 254 nm), Peltier-cooled autosampler, and fraction collector (both set at 10 °C for all samples). The column was a Gemini C18 column (3 μm, 110 Å, 100 × 1.0 mm, Phenomenex) with a Gemini C18, 4 × 2.0 mm SecurityGuard (Phenomenex) cartridge as a guard column. The solvent system consisted of 20 mM ammonium formate (pH 10.0) as mobile phase (A) and 100% acetonitrile as mobile phase (B). The separation was accomplished at a mobile phase flow rate of 0.1 mL/min using the following linear gradient: 100% A for 2 min, from 100% A to 35% B in 59 min, to 85 % B in a further 1 min, and held at 85% B for an additional 15 min, before returning to 100% A and re-equilibration for 13 min. Thirty-two fractions were collected along with the LC separation that were subsequently pooled into 15 fractions. Pooled fractions were dried under vacuum centrifugation, reconstituted in 4% acetonitrile/ 1% formic acid.
LC–MS/MS
Peptides were separated using the UltiMate 3000 RSLC nano-LC system (Dionex) fitted with a trapping cartridge (µ-Precolumn C18 PepMap 100, 5 µm, 300 µm i.d. × 5 mm, 100 Å) and an analytical column (Acclaim PepMap 100 75 µm × 50 cm C18, 3 µm, 100 Å). The analytical column outlet was coupled directly to a QExactive plus (Thermo Fisher, Waltham, MA, USA) using the Proxeon nanoflow source in positive ion mode. Solvent A was water, 0.1% formic acid and solvent B was acetonitrile, 0.1% formic acid. Trapping time was 6 min with a constant flow of solvent A at 30 µL/min onto the trapping column. Peptides were eluted via the analytical column a constant flow of 0.3 µL/min. During the elution step, the percentage of solvent B increased in a linear fashion from 2% to 4% B in 4 min, from 4% to 8% in 2 min, then 8% to 28% for a further 96 min, and finally from 28% to 40% in another 10 min. Column cleaning at 80% B followed, lasting 3 min, before returning to initial conditions for the re-equilibration, for 10 min.
The peptides were introduced into the mass spectrometer (QExactive plus, Thermo Fisher, Waltham, MA, USA) via a Pico-Tip Emitter 360 µm OD × 20 µm ID; 10 µm tip (New Objective) and a spray voltage of 2.3 kV was applied. The capillary temperature was set at 320 °C. Full scan MS spectra with mass range 350–1400 m/z were acquired in profile mode in the FT with a resolution of 70,000. The filling time was set at a maximum of 100 ms with a limitation of 3 × 106 ions. DDA was performed with the resolution of the Orbitrap set to 35,000, with a fill time of 120 ms and a limitation of 2 × 105 ions. Normalized collision energy of 32 was used. A loop count of 10 with count 1 was used and a minimum AGC trigger of 2e [2] was set. Dynamic exclusion time of 30 s was applied. The peptide match algorithm was set to ‘preferred’ and charge exclusion ‘unassigned’, charge states 1, 5–8 were excluded. Isolation window was set to 1.0 m/z and 100 m/z set as the fixed first mass. MS/MS data were acquired in profile mode.
Data analysis
Acquired data were processed by IsobarQuant (https://doi.org/10.1038/nprot.2015.101) and Mascot (v2.2.07) and scored against a Uniprot Homo sapiens proteome database (UP000005640) containing common contaminants and reversed sequences. The data were searched with the following modifications: Carbamidomethyl (C) and TMT10 (K) (fixed modification), Acetyl (N-term), Oxidation (M) and TMT10 (N-term) (variable modifications). The mass error tolerance for the full scan MS spectra was set to 10 ppm and for the MS/MS spectra to 0.02 Dalton. A maximum of two missed cleavages was allowed. For protein identification a minimum of two unique peptides with a peptide length of at least seven amino acids and an false-discovery rate (FDR) <0.01 were prerequisites on both the peptide and protein level. After the raw data were log2 transformed, Batch cleaned, and the data from different groups were then normalized using the Quantile method and variation across the samples were analyzed by principal component analysis (PCA). The significance (adj. p < 0.05) between different groups were calculated using the limma package. The TMT signals were normalized using the JOSD2-downregulated proteins were analyzed for enrichment using the Reactome and Gene Ontology (GO) databases using R package ReactomePA [26].
Xenografts and immunoprecipitation
A total of 5 × 106 cells were subcutaneously implanted into female athymic (NCI-H2087, with PBS:matrigel 1:1 and NCI-H1792) and female NMRI nude mice (NCI-H1437) and the different tumors were grown for 28–50 days and the tumors were snap-frozen in liquid nitrogen. No randomization was needed as mice consisted of one experimental group per xenograft. Animal experiments was blinded to the authors as it was performed at ProQinase (Freiburg, Germany) as fee-for-service according to the guidelines of the Federation for Laboratory Animal Science Associations, EU. Tumor sizes were calculated usirng a caliper and the formula W2 × L/2 (L = length and W = the perpendicular width of the tumor, L > W). Tumors were subsequently homogenized and lysed in the IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40). Lysates were then cleared by centrifugation (12,000 × g, 20 min, 4 °C) and protein concentration was determined by BCA assay. Ten percent of total protein per sample was mixed with Laemmli loading buffer, boiled and saved as the input sample. The remaining lysates were diluted to 1 μg/μl and a total of 1 mg protein was used further to incubated with 2 μg of the indicated antibody and IgG control overnight at 4 °C. Protein G-agarose beads (Roche) were then added and incubated for 3 h. Immuno complexes were washed three times with 0.1% NP-40 lysis buffer and Laemmli sample buffer was added. Samples were then boiled and subjected to western blot analysis. The anti-PHGDH (sc-100317, 6B2, Santa Cruz Biotech., Inc.) was used for immunoprecipitations followed by immunoblotting with anti-PHGDH (HPA021241, Sigma-Aldrich), anti-Aldolase A (#8060, D73H4, Cell Signaling), or anti-PFK-1 (PA5-29336, Thermo Scientific).
Determination of glycolytic function and mitochondrial respiration
The extracellular acidification rate (ECAR) and the mitochondrial oxygen consumption rates (OCR) were measured in real-time using the XFp Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). Cells were seeded in an XFp miniplate at a density of 10,000 cells per well in their respective growth medium. Cells were subsequently transfected on the following day with either nontargeting siRNA or two independent siRNA targeting JOSD2. On the next day, cells were washed twice with 200 µl of XF Base medium (Seahorse Bioscience, North Billerica, MA, USA) followed by the incubation with 175 µl of the same medium. Cells were either analyzed using XFp Glycolysis Stress Test Kit (Seahorse Bioscience) or XF Cell Mito Stress Test Kit (Seahorse Bioscience) following the manufacturer’s instruction. Briefly, after the baseline measurement, the following subsequent injections were made, for glycolysis (ECAR) measurements 10 mM glucose, 1 μM Oligomycin and 50 mM 2-deoxyglucose, and for the respirometry (OCR) analyses 1 μM oligomycin (a mitochondrial ATP synthase inhibitor), 0.5 μM FCCP (a mitochondrial uncoupler), and 0.5 μM Rotenone/Antimycin A (Complex I/Complex III inhibitors). Protein concentration was determined by the bicinchoninic acid (BCA) assay (Pierce, Thermo Fisher Scientific, Walthman, MA, USA) and used to normalize the Seahorse data.
RNAi
One hundred thousand cells per well were forward transfected in 0.5 ml of growth medium in six-well plates. Interferin (Polyplus transfection, New York, NY, USA) was mixed with 40 nM siRNA targeting JOSD2 (ON-TARGETplus Human JOSD2 siRNA, J-015500-05 (#1) and J-015500-07) (#2) or JOSD1 (J-017674-05) (#1), J-017674-06 (#2), or ATAXIN3 (J-012013-07) (#1), J-012013-08 (#2), or siRNA pool for JOSD2 (SMARTpool: ON-TARGETplus JOSD2 L-015500-00-0005) or USP5 (SMARTpool: ON-TARGETplus USP5 L-006095) in a final volume of 150 µl/well and added to the cells. The control siRNA (ON-TARGETplus nontargeting siRNA D-001810-10) from (Dharmacon, Lafayette, CO, USA) was used. The knockdown was analyzed by qPCR 24–72 h post transfection.
Transfections
Plasmid transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher, Waltham, MA, USA) in a 10-cm petri dish per manufacturer’s protocol with a cell confluency at 70–80%. For each transfection final reaction mix contained: 1000 µl Opti-MEM medium, 15 µl Lipofectamine 2000, and 6 µg plasmid. Protein expression was assessed 48 h after transfection. The following Origene plasmids were used: HA-Ubiquitin, pCMV-PFK-1 (RC230492, Origene), pCMV- Aldolase A (RC), pCMV-PHGDH, pCMV-DDK-tagged wild-type JOSD2, pCMV-DDK-tagged C24A mutant JOSD2.
Ubiquitination assay
HEK293T cells were transfected as described above with PHGDH, Aldolase A or PFK-1, HA-Ubiquitin, with wild-type or the C24A JOSD2 mutant. Cells were exposed to 5 µM of the proteasomal inhibitor MG132 (Sellechem) for the last 4 h prior to harvesting of cells. Lysates were subjected to immunoprecipitation (IP) with anti-PHGDH (sc-100317, 6B2, Santa Cruz Biotech., Inc.), anti-Aldolase A (#LS-B1461, LifeSpan Biosciences Inc.), or anti-PFK-1 (HPA002117, Sigma-Aldrich) antibodies followed by immunoblotting with anti-PHGDH (HPA021241, Sigma-Aldrich) anti-Aldolase A (#8060, D73H4, Cell Signaling) anti-Ubiquitin (BML-PW8810, Enzo Life Sciences) antibodies to examine the ubiquitination status of these three metabolic enzymes.
DUB assay
Deubiquitination assay was performed using recombinant His-JOSD2 (Ubiquigent) and polyubiquitin chains (BostonBiochem). JOSD2 were freshly prepared in assay buffer (40 mM Tris-HCl, pH 7.5, 5 mM DTT, 0.005 % BSA) and activated for 10 min at 30 °C. Subsequently, 1 μg of JOSD2 was mixed with 200 ng of Lys48 polyubiquitin (Ubi1-Ubi7) chains and incubated at 30 °C. The reactions were stopped after 24 h by addition of SDS sample buffer (Bio-Rad) containing 100 mM DTT. Ubiquitin cleavage was detected using immunoblotting to PVDF membrane and anti-ubiquitin antibodies (Novus Biologicals, NB300).
Glucose uptake assays
The uptake of glucose was measured using the Glucose-Uptake-Glo Assay kit (Promega, J1343) following the manufacturer’s instructions. Briefly, JOSD2 was silenced using siRNA for in A549 cells and the glucose uptake was subsequently examined in triplicates for up to 60 min. Luminescence was measured using the GloMax discover (Promega) after 60 min incubation with 2-deoxyglucose 6-phosphate.
Site-directed mutagenesis
The JOSD2C24A mutant was generated using QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) and the following oligos:
JOSD2C24A Forward: 5′-tggagctggctgctgtccacgccctcaacaacgttctgc-3′
JOSD2C24A Reverse: 5′-ggacagcagccagctccaggcgctgccgttcgtggtaca-3′
Mutagenesis was verified by DNA sequencing.
cDNA synthesis and qPCR
RNA was isolated using the PureLink™ RNA Mini Kit (Thermo Fisher, Waltham, MA, USA) and residual DNA was removed using PureLink™ DNase Set (Thermo Fisher, Invitrogen). mRNA (1 µg) was synthesized to cDNA with the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed with 200 ng cDNA using Maxima qPCR SYBR green master mix (Thermo Fisher Scientific, Waltham, MA) and amplified with a 7500 real-time PCR system (Applied Biosystems, Foster City, CA). The ΔΔCT method was applied to determine the relative mRNAs expression, Tubulin was used as the reference genes.
The following primers were used:
JOSD2 (Fwd) 5′-CACCCACCGTGTACCACGAA -3′
JOSD2 (Rev) 5′-GCTGCGATGAGGGTTCAGC-3′
ATXN3 (Fwd) 5′-TCGGAAGAGACGAGAAGCCTAC-3′
ATXN3 (Rev) 5′-AAGTGCTCCTGAACTGGTGGCT-3′
JOSD1 (Fwd) 5′-TTGCCACTTCCCCAAACATCTC-3′
JOSD1 (Rev) 5′-AGAACTCCTACCTTCCTTGCCC-3′
USP5 (Fwd) 5′-GAAGTGTTCCGCTTCTTGGTGG-3′
USP5 (Rev) 5′-TTGCCGCTTCTTCTCCTCGTAC-3′
Aldolase A (Fwd) 5′-CAGGGACAAATGGCGAGACTAC -3′
Aldolase A (Rev) 5′-TCCCCAATCTTCAGCACACAAC-3′
PFK-1 (Fwd) 5′ACCCATGAAGAGCACCATGCAG-′3
PFK-1 (Rev) 5′-TGGACAAAGAAGACACGGGCAC -3′
PHGDH (Fwd) 5′-GTCATCAACGCAGCTGAGAA-3′
PHGDH (Rev) 5′ - AACTTCTTCCGCTCCCATTT-3′
Tubulin (Fwd) 5′-TCTACCTCCCTCACTCAGCT-3′
Tubulin (Rev) 5′-CCAGAGTCAGGGGTGTTCAT-3′
Western blot
Immunoblotting was performed as described before [27]. Briefly, cells were lysed using Complete Lysis-M buffer with protease inhibitors (Roche Diagnostics, Risch-Rotkreutz, Switzerland). The lysates were cleared by centrifugation, and the protein concentration was determined in the supernatant by the BCA assay. Equal amounts of the protein/sample were mixed with Laemmeli’s loading buffer, heated and separated on 15% SDS–PAGE followed by transfer to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk, 0.01% Tween in PBS at room temperature and subsequently probed with the primary antibodies (diluted in PBS containing 1% BSA and 0.1 % NaN3) overnight, at 4 °C. The JOSD2 antibody was diluted at 1 µg/µl in 5% nonfat milk in PBS. Horseradish peroxidase-conjugated secondary antibodies (goat anti-mouse or goat anti-rabbit, Thermo Fisher, Waltham, MA, USA) were diluted in 2.5% nonfat milk in PBS with 0.005% Tween. Clarity TM Western ECL (BioRad, Hercules, CA, USA) was used to develop membranes. The following primary antibodies were used: anti-ubiquitin (Enzo, BML-PW8810) and (Novus Biologicals, NB300), anti-PHGDH (6B2, Santa Cruz Biotech., Inc., sc-100317), anti-Aldolase A (D73H4) (Cell Signalling, #8060), anti-JOSD2 (LS Biosciences, LS-C102435) anti-DDK (FLAG) (OriGene, TA50011-100), anti-TPI (Abcam, ab96696), anti-β-Actin (Santa Cruz Biotech., Inc., sc-81178), anti-PFK-1 (Sigma-Aldrich, HPA002117), anti-Porin 3 (#529532, Calbiochem).
Co-immunoprecipitation
A549 cells were either grown in regular RPMI media (25 mM glucose) or RPMI glucose-free media supplemented with 5 mM glucose for 16 h and cells were subsequently lysed with 1% NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 % NP-40). Lysates were cleared by centrifugation and protein concentration was determined by BCA assay. Ten percent of total protein per sample was mixed with Laemmli loading buffer, boiled and saved as the input sample. The remaining lysates were diluted to 1 μg/μl, and a total of 1 mg protein was used further to incubated with 2 μg of the indicated antibody and IgM control (for PHGDH) or IgG control (for Aldolase a or PFK-1) overnight at 4 °C. Protein G-agarose beads (Roche) were then added and incubated for 3 h. Immuno complexes were washed three times with 0.1% NP-40 lysis buffer and Laemmli sample buffer was added. Samples were then boiled and subjected to western blot. The following antibodies were used for immunoprecipitations: anti-PHGDH (sc-100317, 6B2, Santa Cruz Biotech., Inc), anti-Aldolase A (#LS-B1461, LifeSpan Biosciences Inc.), or anti-PFK-1 (HPA002117, Sigma-Aldrich), followed by immunoblotting with anti-PHGDH (HPA021241, Sigma-Aldrich), anti-Aldolase A (#8060, D73H4, Cell Signaling) or anti-PFK-1 (PA5-29336, Thermo Scientific).
Enzymatic activity assays
All activity kits were performed per each manufacturer’s instructions.
PFK (MAK093, Sigma-Aldrich)
Briefly, cells were harvested, washed with PBS and lysed using douncer in 200 µl of ice-cold PFK Assay Buffer followed by centrifugation at 13,000 × g for 10 min. The supernatant was used for further analysis. The reaction mix for one reaction included: 42 µl PFK assay buffer, 2 µl PFK enzyme mix, 2 µl PFK developer, 2 µl ATP, and 2 µl PFK substrate.
Aldolase (ab196994, Abcam)
Cells were harvested, washed with PBS and lysed using ice-cold Aldolase Assay Buffer, followed by centrifugation at 10 000 × g, 4 °C, 5 min. The collected supernatant was used for further analysis. The reaction mix for one reaction included 44 µl aldolase assay buffer, 2 µl, aldolase substrate, 2 µl aldolase enzyme mix and 2 µl aldolase developer.
PHGDH (K569-100, Biovision)
Cells were harvested, washed with PBS and lysed using ice-cold PHGDH Assay Buffer, followed by centrifugation at 10,000 × g, 4 °C, 10 min and the supernatant was used for the assay. The reaction mix for one reaction included: 46 µl PHGDH Assay Buffer, 2 µl PHGDH developer and 2 µl PHGDH substrate.
All activity assays were read measured at 450 nm using a GloMax Discover Microplate Reader (Promega, Madison, WI, USA). The enzyme activities were normalized to protein concentration.
Clonogenic and cell growth assays
Indicated cell lines were transfected with control siRNA or two independent siRNA targeting JOSD2 for 24 h and cultured in replicates of three in 96-well plates at 1000 cells/well in 200 µl of RPMI medium. Cell growth was assessed every 24 h for up to 120 h by Cell Titer Glo reagent (Promega) and the luminescent signal was read on a GloMax Discover Microplate Reader (Promega, Madison, WI, USA). For each well, the fold change in luminescence was calculated relative to its respective control (nontargeting control siRNA).
In the clonogenic growth assays, A549 and NCI-H1792 cells were seeded (1000 cells/dish) in a 6-cm petri dish in three replicates and on the following day transfected with either nontargeting siRNA, siRNA targeting JOSD2 or siRNA targeting JOSD2 supplemented with 1 mM pyruvate. Cells were cultured for 7 days and subsequently washed with PBS and fixed in 4% paraformaldehyde for 20 min. The cells were then stained with 0.1% Crystal Violet for 15 min and washed five times in distilled H2O. Images were acquired using the EVOS XL Core Imaging system (Thermo Fisher Scientific), and the cell colonies were counted for each condition.
Cellular fractionation
Cells were harvested, washed in ice-cold PBS and lysed by increasing the detergent strength of lysis buffers to obtain proteins from cellular fractions. To obtain cytosolic proteins, cells were lysed in buffer A (150 mM NaCl, 50 mM Hepes, pH 7.4, 0.05 % Digitonin, 1 M Hexylene glycol and completeTM protease inhibitor cocktail) and incubated for 20 min on ice. The obtained extract was centrifuged at 2000 × g for 10 min and the supernatant was collected as the cytosolic fraction. The remaining pellet was then solubilized in Lysis Buffer B (150 mM NaCl, 50 mM Hepes, pH 7.4, 1 % Igepal, 1 M Hexylene glycol and completeTM protease inhibitor cocktail) followed by incubation for 20 min on ice followed by centrifugation at 7000 × g for 10 min. The collected supernatant containing organelles and membrane-bound proteins were discarded and the residual pellet containing nuclei was lysed in Lysis Buffer C (150 mM NaCl, 50 mM Hepes, pH 7.4, 0.5 % (w/v) Sodium deoxycholate, 0.1 % (w/v) SDS, 1 M Hexylene glycol and completeTM protease inhibitor cocktail) for 20 min on ice and centrifuged for 10 min 12,000 × g to obtain nuclear proteins. The cell fractions were analyzed by western blot.
Cycloheximide treatment
A549 cells were seeded in 6-well plates, JOSD2 was downregulated by siRNA (as described above), 24 h later, fresh media with 10 µM cycloheximide (CHX, Sigma-Aldrich) added. Cells were treated for up to 48 h and 5 µM MG132 (Sellechem) was added during the last 4 h prior to cell harvesting and western blot.
Statistics
All graphs were designed in Grap Pad Prism and all statistics were calculated using Graph Pad Prism Software 8, San Diego, USA. The specifics for each panel are described in the respective figure legend.
Results
Identification of the deubiquitinase JOSD2 as a potential regulator of glucose metabolism
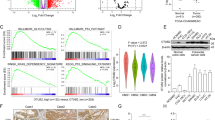
To identify DUB(s) with potential regulatory metabolic functions, we hypothesized that responsible DUB(s) should be co-elevated with a high expression signature of the glycolytic enzyme(s). This warranted a systematic interrogation of the co-expression of all human DUBs with transcripts encoding glycolysis enzymes. We reasoned that such a scenario may be most relevant for tumors with high rates of glycolysis, including NSCLCs. Our initial analysis revealed normalized gene expression levels of 77 DUBs in 226 NSCLC patients among all encoded active DUBs in the human genome [28, 29]. These 77 DUBs were subsequently correlated with the expression of enzymes related to glucose metabolism in lung cancer patients [30], covering distinct reaction steps of glycolysis. The Pearson correlation coefficient revealed a positive co-expression profile of five DUBs including, JOSD2, USP5, PSMD7, PSMD14, and STAMBP with 14 out of 15 glycolytic enzymes (Fig. 1A). To probe the potential clinical relevance of these DUBs, we examined their expression in nine previously generated data sets with clinical follow-up information (GSE14814, GSE19188, GSE29013, GSE30219, GSE31210, GSE3141, GSE31908, GSE37745, and GSE50081). A total of 673 and 720 patients, expressing either JOSD2, STAMBP, or USP5, PSMD7, PSMD14, respectively, were included in the Kaplan–Meier analyses. In order to categorize patients into low- and high-expressing groups, the performance of all possible cutoff values were calculated between the upper and lower quantiles. Colored dashed lines indicate the confidence interval (Fig. 1B). The Kaplan–Meier analyses showed that high expression of USP5 and JOSD2, but not PSMD7, PSMD14, and STAMBP significantly (p < 0.05) predicted poor prognosis (Fig. 1B). In parallel, the hazard ratios were calculated for each of the five DUB candidates. This further linked JOSD2 and USP5 to be significantly (p < 0.05) associated with increased risk (Fig. 1C). To functionally test the potential role of JOSD2 and USP5 on glycolysis, the DUBs were silenced (Supplementary Fig. 1A) and the ECAR analysis was used as a proxy measure of glycolysis. We found that only knockdown of JOSD2 displayed substantial suppressive effects on glycolysis (Fig. 1D, E). To exclude that the effect of JOSD2 on glycolysis is due to off-target effect by the siRNAs, we assessed glycolysis in HAP-JOSD2 knockout cells (HAP-KO) compared to HAP parental (HAP-P). Consistently, both glycolysis and the glycolytic capacity were profoundly reduced upon JOSD2 deficiency (Fig. 1F and Supplementary Fig. 1A). Combined, these data uncover a prognostic value of JOSD2 in NSCLC and suggest a functional role for JOSD2 protein in promoting glycolysis.
A The heat map depicts the Pearson correlation coefficient between all expressed human deubiquitinases with indicated glycolytic enzymes in 226 NSCLC patients. A positive correlation is indicated in blue. B Kaplan–Meier analysis for JOSD2, STAMBP, USP5, PSMD7, and PSMD14. The patients were separated into low and high expressing by calculating performance of all possible cutoff values between the upper and lower quantiles. The best performing threshold was used as cutoff. Dashed colored lines indicates the confidence interval (95%). C The hazard ratios calculated for each of the five candidate DUBs as indicated. Hazard ratios >1.0 equals increased risk markers, whereas ratios <1 represent markers of decreased risk. The p value was calculated using adjusted Cox proportional hazards mode, **p < 0.01, ***p < 0.001. The glycolytic function of A549 (D) and NCI-H2087 (E) cell lines after JOSD2 and USP5 knockdown by pool siRNAs compared to nontargeting siRNA (siRNA control). F The glycolytic function in HAP cells comparing JOSD2 knockout (HAP-JOSD2-KO) to the parental (HAP-P). Glycolysis analysis was performed using an Extracellular Flux Analyzer. Glucose was injected followed by oligo (oligomycin) and 2-DG (2-deoxyglucose) as indicated and data normalized to protein concentration. n = 3–5 independent ECAR measurements per cell line. Error bars, ± SEM. **p < 0.01, ***p < 0.001; two-tailed student’s t test.
Metabolomics approach identifies JOSD2 as a positive regulator of glycolysis
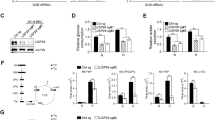
To investigate the role of JOSD2 in metabolism, we examined the effect of knockdown of JOSD2 (Fig. 2A) on the cellular glucose uptake (a key limiting step for glycolysis) [31], the total intracellular pool of lactate (the terminal product of glycolysis) and ATP as measured by CE-TOF MS, in three independent biological replicates. These analyses revealed that while JOSD2 does not affect the glucose uptake markedly across 60 min (Fig. 2B), its silencing caused a significant reduction of the intracellular pools of both lactate and ATP (Fig. 2C).
A Immunoblot showing the knockdown efficiency by two independent siRNAs targeting JOSD2, compared to nontargeting control siRNA. Actin was used as loading control. The effect of siRNA silencing of JOSD2 on, B glucose uptake in A549 cells, and C the intracellular amount of lactate and ATP as analyzed in three independent biological replicates (indicated by black dots) from A549 cells by CE-TOF/MS analysis. D Schematic picture of amino acids originating from glucose carbons. Purple circles indicate the expected carbon labeling per amino acid from uniformly labeled glucose. E A549 cells treated with two independent siRNAs targeting JOSD2 (si1 or si2) or a control siRNA (siCtrl) and cultured with uniformly labeled glucose (U13C6-glucose) for 24 h and analyzed by CE-TOF/MS. Data are from three independent biological replicates and presented as percentage of total pool per indicated metabolite (Mean ± SD). Statistics are shown for the significantly reduced metabolites. F The proportional effect (Percent change±SD) of JOSD2 silencing compared to control siRNA on the indicated isotopologues with statistics. G CE-TOF/MS analysis showing the effect of JOSD2 knockdown on the extracellular amino acid pools in matched cell culture media from cells as in (E). Data are presented as percentage of total pool per amino acid. Dashed line indicates no change. Error bars, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ns nonsignificant, Welch’s t-test.
To more comprehensively investigate the metabolic effects of JOSD2 and to provide independent and parallel evidence, we employed a mass spectrometry-based isotopomer analysis, as stable isotopes are powerful tools to investigate metabolism, where the 13C-enrichment patterns of downstream metabolites provide information about the pathway activities [32]. To this end, A549 cells were provided with uniformly labeled glucose (U-13C6 glucose) and analyzed by a targeted CE-TOF/MS in three independent biological replicates. We focused on the relevance of JOSD2 for the synthesis of amino acids, ATP and lactate. A schematic overview of the contribution of glucose-derived carbons to amino acids is depicted in Fig. 2D. Incubation with the glucose tracer revealed a substantial labeling of metabolites related to glycolysis including serine, glycine, alanine, lactate, and ATP. Metabolites more related to mitochondrial function such as aspartate, arginine, asparagine, glutamate, and glutamine remained largely unlabeled (>90% of the total pool per metabolite) suggesting that these amino acids mainly originate from nonglucose carbon sources in these cells (Supplementary Fig. 1B). We observed that the m + 3 isotopologues of serine and lactate as well as of m + 2 glycine were markedly suppressed upon JOSD2 knockdown (Fig. 2E, F). In particular, ~50% of de novo serine biosynthesis (Ser m + 3) was affected by JOSD2 (Fig. 2F). This biochemical reaction is limited by the phosphoglycerate dehydrogenase (PHGDH) activity, an enzyme shown to be a prognostic factor and to provide pro-survival benefits including the production of nucleotides and glutathione [27, 33] in lung cancer. Moreover, while a large proportion of the total alanine pool was labeled, the alanine isotopologue m + 3 was significantly suppressed by JOSD2 RNAi, consistent with reduced glycolysis. We also detected that pyruvate carboxylation, a feature previously observed in NSCLC patients [34] is moderately yet significantly sensitive to JOSD2 knockdown, as evident from a reduced Asp m + 3 isotopologue, a marker for pyruvate carboxylase activity (Fig. 2E). We then analyzed whether glucose can contribute to the synthesis of ATP (composed of a five-carbon adenine ring and a ribose sugar of five carbons, totaling ten carbon atoms). We observed that glucose contributes to ~80% of the total ATP pool in the control samples compared to only ~30% upon silencing of JOSD2 (Supplementary Fig. 1B). Of note, several ATP isotopologues (m + 2, m + 3, m + 4, m + 6, m + 7 and m + 8) were present in the control but remained undetected in the two siRNA-treated samples. Further, we observed a significant decrease in the m + 5 isotopologue of ATP, consistent with a reduced flux through glycolysis (Fig. 2F and Supplementary Fig. 1B). Combined, these results suggest that JOSD2 can control the contribution of glycolysis to the intracellular ATP pool.
CE-TOF/MS analyses of the extracellular amino acid pools, in the matched cell culture media (from Fig. 2E, 2F), confirmed that the JOSD2-mediated metabolic changes indeed reflect the intracellular metabolite pools as no major alterations occurred in cell media (Fig. 2G and Supplementary Fig. 1C).
Collectively, these data demonstrate a pronounced impact of the deubiquitinase JOSD2 on glycolysis and as well as the synthesis of select amino acid pools in NSCLC cells.
JOSD2 silencing reduces glycolysis in a broad range of tumor cells of distinct origin and displays cytostatic effects
JOSD2 belongs to the Machado–Joseph disease (MJD) DUB family, known to display both Lys48 and Lys63 in vitro activities [28, 35], yet a defined cellular function of JOSD2 remains unidentified. To broaden and substantiate our findings, we analyzed the effect of JOSD2 knockdown (Supplementary Fig. 1E, F) on glycolysis as assessed by the Seahorse technology in a larger cell line panel comprising of several cancer types of distinct origin including NSCLCs (Fig. 3A), carcinomas from breast, cervix and ovary, and neuroblastoma (Fig. 3B), as well as normal (nontransformed) cells (Fig. 3C). We uncovered that JOSD2 silencing resulted in a marked reduction of glycolysis in the tested cancer cell lines, revealing a common effect of JOSD2 shared by diverse carcinomas (Fig. 3A, B), while no significant effect was observed in nontransformed cells (Fig. 3C). Collectively, these data consolidate the functional relevance of JOSD2 as a regulator of glycolysis and suggest that this may be a cancer-specific feature.
Glycolysis was measured by an Extracellular Flux Analyzer upon JOSD2 knockdown by two independent siRNAs compared to nontargeting siRNA (siControl) in (A) NSCLC cell line panel (B) cancer cell lines of distinct origin: neuroblastoma (SH-SY5Y), breast cancer (MDA-MB-231), cervical cancer (HeLa), and ovarian cancer (SK-OV-3) and in (C) nontransformed (normal) cells. Three independent ECAR measurements per cell line were performed and the data were normalized to protein concentration. D The effect of JOSD2 knockdown on mitochondrial oxygen consumption rates (OCR) in A549 cells after sequential injection of Oligomycin, FCCP, and Rotenone/Antimycin A. Results were normalized to protein concentration. E Calculations of the indicated mitochondrial parameters from (D). F OCR of JOSD2 knockout (HAP-JOSD2-KO) cells compared to parental cell line (HAP-P) and normalized to protein concentration. G Calculations of mitochondrial parameters from (F). H Cell proliferation assay assessed over 5 days showing the effect of JOSD2 knockdown in the indicated cell lines after JOSD2 knockdown by two independent siRNAs. Normalized to data obtained by using the nontargeting siRNA. Statistics are color coded per sample group. I Clonogenic growth and quantification of NCI-H1792 and A549 cells investigated under similar conditions as in (H) but with the addition of an experimental rescue sample group containing media supplemented with 1 mM pyruvate. J JOSD1 and ATXN3 were genetically silenced in A549 cells by two independent siRNAs targeting each DUB and glycolysis were measured (n = 3) and results were normalized to protein concentration. Error bars, ± SD, *p < 0.01; **p < 0.001; ***p < 0.0001, ns nonsignificant, two-tailed Students’ t test.
Beyond a high glycolytic activity, rapidly proliferating cancer cells often display a sustained oxidative phosphorylation, since the tricarboxylic acid (TCA) cycle intermediates functions as precursors for the synthesis of amino acids, lipids and nucleotides [5, 36, 37]. Therefore, we tested if JOSD2 silencing (in A549 cells) or knockout of JOSD2 (in HAP cells) may influence mitochondrial respiration and observed that it was reduced upon both JOSD2 deficiencies (Fig. 3D–G). However, the effect of JOSD2 on the mitochondrial function is likely to be secondary as the metabolomics data suggests that pyruvate oxidation (Asp m + 2, a marker for pyruvate dehydrogenase activity) remain marginal in these cells (Fig. 2E and Supplementary Fig. 1C).
We next determined the potential pro-growth benefits JOSD2 may provide cancer cells. By genetic silencing of JOSD2 in four NSCLC cell lines, we monitored cell growth and clonogenicity and found that JOSD2 deficiency led to a significant growth retardation compared to control cells (Fig. 3H, I). As our metabolomics data suggest that JOSD2 affects glycolysis primarily upstream of pyruvate, we reasoned that pyruvate supplementation may blunt the effect of JOSD2 deficiency. The addition of pyruvate partially, yet significantly, reversed the effects of JOSD2 knockdown (Fig. 3I) arguing that JOSD2 sustain cell proliferation and growth by glucose catabolism.
To address whether the JOSD2-mediated metabolic changes are JOSD2-specific, we investigated two additional MJD DUB family members, Josephin domain-containing protein 1 (JOSD1) and Ataxin 3 (ATXN3). We observed that knockdown of JOSD1 or ATXN3 (Supplementary Fig. 1D) did not affect glycolysis (Fig. 3J), highlighting the selectivity of JOSD2’s function in glycolysis.
Identification of potential JOSD2 substrates by quantitative multiplexed proteomics
To determine if JOSD2 might impact the metabolic profile by controlling the stability of enzymes in central metabolic pathways, we undertook an global quantitative proteomics approach amenable to multiplexing that could be instrumental as a basis for our hypothesis. The prediction is that the genetic silencing of JOSD2 should cause a decrease in any potential target protein(s). We employed tandem mass tag (TMT)-based quantitative mass spectrometry approach to determine relative changes in the proteome. This method is suitable for simultaneous quantifiable comparison of multiple unique tryptic peptides per candidate protein across independent biological repeats. We compared the effect of JOSD2 silencing to a control siRNA in A549 cells (Fig. 4A). The detected TMT signals were normalized by the Quantile method (Fig. 4B) and principal component analysis (PCA) confirmed a clear partitioning of the sample groups (Fig. 4C). Only proteins with >2 peptides per quantified protein, across all four biological replicates were considered. We then performed several unbiased biological function distribution analyses using the Reactome (Fig. 4D) and the GO (Fig. 4E) databases of the significantly JOSD2-downregulated proteins (adj. p value of <0.05). Beyond confirming an enrichment of proteins in carbohydrate and amino acid metabolic pathways, importantly, our analyses additionally highlighted ubiquitination-dependent protein catabolism and protein polyubiquitination among the top significantly (adjust. p < 0.1) enriched cellular processes linked to JOSD2 (Fig. 4D, E). Given that our data so far links JOSD2 with glucose metabolism, to identify potential candidate substrate(s), we turned our attention to glycolytic proteins. Consistent with the biological function distribution analyses, multiple glycolysis-related proteins were found to be affected upon JOSD2 knockdown in the detected proteome, with a reduced steady-state abundance of metabolic enzymes, including Aldolase A (Aldo A), phosphofructokinases (PFK-1 and PFKL), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucose-6-phosphate isomerase (GPI), lactate dehydrogenase (LDHA and LDHB) and Phosphoglycerate dehydrogenase (PHGDH), with a FDR less than 5% (Fig. 4F). Notably, five out of these candidate proteins (PFK-1, PFKL, GPI and Aldolase A) were detected as significantly co-expressed with JOSD2 in lung cancer patients, as shown in Fig. 1A. Functionally, these enzymes account for six biochemical reactions in glycolysis and the serine biosynthesis (Fig. 4F, right panel) suggesting that JOSD2 impacts both simultaneously without affecting the mRNA expression (Supplementary Fig. 1G, H).
Multiplex quantitative TMT proteomic approach of JOSD2 silenced A549 cells. A Schematic overview of the experimental strategy, four biological replicates of control and JOSD2 knockdown sample groups were analyzed using 8-plex mass spectrometry. Western blot showing the knockdown efficiency. B The TMT signal distribution after normalization of the sample sets following LC–MS/MS. C Score plot of principal components analysis (PCA) of siControl and siJOSD2 samples. PC1–PC3: principal component 1–3. Each point represents a biological replicate. Biological function analyses performed using (D) the Reactome and (E) the Gene Ontology (GO) databases. Pathways related to ubiquitination, glucose, and amino acid metabolism are highlighted in red. F Relative TMT signal distribution (normalized to the control siRNA) of the proteins (green) affected by JOSD2 silencing. Proteins related to glucose catabolism (middle panel). The biochemical enzymatic reactions comprising the identified metabolic proteins are color coded as indicated in the list, to the right. Fructose 1,6-bisphosphate (F16BP) dihydroxyacetone phosphate (DHAP), glyceraldehyde 3-phosphate (G3P), 1,3-bisphosphoglycerate (13-BPG), glucose-6-phosphate (G6P) frutcose-6-phosphate (F6P) 3-phosphoglycerate (3PG) 3-phosphopyruvate (3P Pyr).
JOSD2 deficiency inhibits the catalytic activities of enzymes involved in glucose catabolism
A reduction of the JOSD2 target proteins may translate into a suppression of their catalytic activities. We then determined whether the activity of reversible and irreversible enzymes of glycolysis along with the glycolytic side-branch driven by PHGDH are dependent on JOSD2 in four NSCLC cell lines with high glycolytic rates [27, 38] and in HAP-P and HAP-JOSD2-KO cell lines. A significant inhibition of Aldolase, PFK and PHGDH, to various extent, were observed in all tested cancer cells upon both JOSD2 deficiencies (Fig. 5A–C). To further substantiate these findings, we undertook a CE-TOF/MS analysis and uncovered that both the intracellular serine pool and the product of PFK-1 (F16BP) were reduced upon siJOSD2 (Fig. 5D). This is coherent with the fact that PHGDH drives the first committed step in de novo serine biosynthesis. Aldolase A, on the other hand, converts fructose 1,6-bisphosphate (F16BP) to glyceraldehyde-3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP). Notably, the product of PFK (F16BP) is the substrate for Aldolase. Combined, this suggests that JOSD2 silencing concurrently reduces PFK and Aldolase beyond PHGDH. In line with this, we observed a similar reduction of the metabolites in JOSD2 knockout cells (Fig. 5E). Our quantitative metabolite data further supports a scenario in which F16BP and DHAP may limit glycolysis, which is consistent with previous literature on mammalian cells and yeast [31, 39, 40]. In particular, F16BP production was recently identified as a key control step in regulating flux through glycolysis [31]. Collectively, these findings reveal that JOSD2 can reduce select metabolic protein levels, restraining their catalytic activities.
The effect of JOSD2 RNAi on enzymatic activities of A Aldolase, B PHGDH and, C PFK, in six independent cell lines (A549, NCI-H1437, NCI-H2087, NCI-H1792, HAP-KO and HAP-P), Results were normalized to protein concentration (n = 4–6 per cell line and enzyme). Data are presented as mean ± SEM. CE-TOFMS analysis of the enzymatic products of PHGDH (Serine), PFK-1 (F16BP) and Aldolase A (DHAP) from three independent biological replicates (n = 3) D comparing the effect of two independent siRNAs targeting JOSD2 normalized to siRNA control or E JOSD2 knockout metabolites proportional to the total pool in the parental line. Metabolites are presented as the percent of the total pool. Error bars, ± SD. *p < 0.01, **p < 0.001, ***p < 0.0001; two-tailed student’s t test.
JOSD2 deubiquitinates and stabilizes a metabolic enzyme complex that promotes glucose and serine metabolism
Despite the suggested JOSD2 Lys48 activity toward a ubiquitin linkage that commonly results in proteasomal degradation of proteins, no target substrates of JOSD2 has been previously identified [35]. We hypothesized that JOSD2 may directly deubiquitinate and stabilize the identified select metabolic enzymes. Hence, we examined the protein half-life upon JOSD2 silencing of PHGDH, PFK-1, and Aldolase A and found a significant shortening of half-life upon knockdown of JOSD2 (Fig. 6A, B), which was blunted by the proteasome inhibitor, MG132 (Fig. 6C). In order for JOSD2 to directly regulate the stability of the afore mentioned proteins it requires that JOSD2 should be able to cleave Lys48-linked ubiquitin chains, a prerequisite for substrate proteins to evade proteasomal degradation [11]. In line with a previous study [35], we show that recombinant JOSD2 efficiently cleaved Lys48-linked ubiquitin chains as observed by a concurrent decrease of Ub2–Ub7 and appearance of Ub1 (Fig. 6D). Another prerequisite for the regulation of metabolic enzymes by JOSD2 is its presence in the cytosol, i.e., where glycolysis occurs, to allow access and substrate binding. Since the intracellular localization of endogenous JOSD2 is not known, we first performed a prediction analysis based on its amino acid sequence to obtain information on its potential subcellular localization (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). While this analysis predicted both nuclear/cytosolic localization, our cellular fractionation, confirmed JOSD2 to be cytosolic (Fig. 6E), as the glycolysis-related enzymes.
A Western blot analysis showing cycloheximide (CHX) chase experiments of JOSD2, PHGDH, Aldolase A, and PFK-1 protein degradation up to 48 h, in A549 cells following transfection with siRNA JOSD2 or control siRNA for 24 h prior CHX addition. MG132 (5 µM) was supplied for the last 4 h. The indicated enzyme levels were normalized to porin. B Image J quantification of PHGDH, Aldolase A and PFK-1 protein level normalized to Porin (n = 3). C Image J quantification of the effect of MG132 at 48 h. D In vitro Lys48 DUB activity assay showing the cleavage of polyubiquitin chains in the presence of recombinant JOSD2 after 24 h incubation time. E Cellular fractionation of A549 cells into cytosol and nucleus, LDHA and Lamin B were used as a marker for each compartment, respectively. F–H Immunoblots of ubiquitin showing the effect of wild-type JOSD2 or a catalytically inactive mutant JOSD2C24A on PHGDH, PFK-1 or Aldolase A ubiquitination patterns in HEK293T cells. Cells were treated with the proteasomal inhibitor, MG132 (5 μM) for 4 h, prior to harvesting, to prevent substrate degradation.
Next, to directly assess the deubiquitinase activity of JOSD2, we generated a catalytically inactive JOSD2 mutant by replacing a cysteine residue at position 24 with alanine (JOSD2C24A) as previously described [35]. We then probed whether JOSD2 can directly modulate the polyubiquitination pattern on the three metabolic enzymes by expressing and comparing a control vector, wild-type JOSD2 or the JOSD2C24A mutant in HEK293T cells for 48 h in the presence of the proteasome inhibitor MG132 for the last 6 h, to prevent potential degradation. Indeed, the wild-type JOSD2, but not the JOSD2C24A mutant, caused substantial deubiquitination of PHGDH, PFK-1, and Aldolase A, respectively (Fig. 6F–H), emphasizing that the regulatory effect on glycolysis-related enzymes is dependent of direct deubiquitination activity of JOSD2.
To determine if JOSD2 could interact with the metabolic proteins and whether such interaction is dependent on its DUB activity, we compared a DDK-tagged JOSD2C24A mutant with DDK-tagged wild-type JOSD2 in immunoprecipitation of PHGDH, PFK-1, and Aldolase A (Fig. 7A–C) individually and could show that JOSD2 could directly interact with PHGDH, PFK-1, and Aldolase A, independently of its DUB activity (Fig. 7A–C).
A–C Western blots of immunoprecipitation of either PHGDH, PFK-1 or Aldolase A, from HEK293T cell lysates transfected with either wild-type or JOSD2C24A (DDK-tagged) for 48 h. D–F Immunoprecipitation of either PHGDH, PFK-1, Aldolase A, or TPI from A549 cell lysates, after 16 h incubation in normal growth media (25 mM glucose) or media containing 5 mM glucose. Mouse-IgG was used as a control for PHGDH and rabbit-IgG for Aldolase A and PFK-1. G Tumor volumes (red dots) of NCI-H1792, NCI-H2087, and NCI-H1437 xenografts. H Immunoblot analysis of JOSD2, PHGDH, PFK-1, Aldolase A in three normal lungs (1–3 gray font) compared to six (1–6 black font) NCI-H1437 tumors. Actin was used to ensure equal loading. I–K Immunoprecipitation of PHGDH, and membranes were stained for PHGDH, Aldolase A, and PFK-1 from the tumor tissue lysates.
As enzymes carrying high metabolic loads often exhibit physical clustering, we reasoned whether the JOSD2-regulated metabolic enzymes might form a complex that integrates both glucose and serine metabolism. We further hypothesized that such a complex formation might depend on the cellular glucose availability. To examine this, A549 cells were grown under normal culture media conditions for cancer cells (25 mM glucose) or in low glucose (5 mM) and subsequent immunoprecipitation of endogenous PHGDH, Aldolase A, or PFK-1 were performed. We scored for any reciprocal interaction between the three proteins and found that independently of which metabolic enzyme that was immunoprecipitated, these enzymes were captured in all tested pulldowns, while another glycolytic enzyme, triose-phosphate isomerase (TPI), was not (Fig. 7D–F). This is consistent with previous literature showing that Aldolase A and PFK-1 can interact [41, 42]. However, in contrast to Aldolase A and PFK-1, low glucose levels resulted in markedly less binding of PHGDH, suggesting that the complex might be nutrient sensitive.
We then investigated whether such a protein complex could exist in vivo in NSCLC tumors and if these proteins are co-expressed compared to normal lung tissue. To this end, we implanted three distinct NSCLCs cell lines to mice (Fig. 7G). Protein levels of Aldolase A, PFK-1, PHGDH, and JOSD2 were first investigated in NCI-H1437 tumor lysates (Fig. 7H) before PHGDH immunoprecipitation experiments from lysates of three distinct types of tumors (Fig. 7I–K). These analyses confirmed that the enzymes are largely co-expressed with JOSD2 (Fig. 7H and Supplementary Fig. 1I) and interacts in vitro and in vivo (Fig. 7A–C, H–K).
To functionally determine the JOSD2 variants on glycolysis, we reintroduced the JOSD2 variants (as in 7A–C) in A549 (Fig. 8A, B) and HAP-JOSD2-KO cells (Fig. 8C, D). These data revealed that the expression of the wild-type JOSD2, but not the catalytically inactive DUB, significantly stimulated glycolysis (Fig. 8B, D). Altogether, our findings indicate that JOSD2 can interact, deubiquitinate to stabilize three metabolic target proteins that exist in a complex to positively regulate glucose catabolism.
A ECAR analysis of A549 cells expressing a control empty vector (Cv), wild-type JOSD2 or JOSD2C24A mutant protein for 48 h. Expression was confirmed by western blot using anti-DDK antibody. B Calculations of the indicated glycolytic parameters for A549 ECAR (A). C Comparison of glycolytic function in HAP-KO cell line after transfection with control empty vector (Cv), wild-type JOSD2 or JOSD2C24A mutant protein for 48 h. Expression was confirmed by western blot using anti-DDK antibody. D Calculations of the indicated glycolytic parameters for HAP ECAR (C).
Discussion
As malignancies are disorders characterized by unrestrained cell proliferation, cancer cells undergo robust and complex metabolic rearrangements to support bioenergetics and biosynthetic nodes [1, 2]. The complex regulatory mechanisms of cancer metabolism are not fully uncovered and are under current vigorous investigation. Here, through an integrative effort we unveil a new layer of metabolic regulation in cancer cells that may provide novel therapeutic opportunities.
DUBs are critical components of protein turnover, demonstrated to participate in regulating many biological processes [9]. Hence, the identification of target proteins for DUBs is essential for our understanding of the select cell signaling events individual DUBs may shape.
Despite our proposed link for JOSD2 in the regulation of PHGDH three years ago [27], a precise cellular role of this DUB and any metabolic evidence remained elusive. Further, regulation by ubiquitination mechanisms also requires E3 ligases that can add ubiquitin to PHGDH. However, not until recently the first E3 ligase for PHGDH was identified when Parkin was shown to regulate PHGDH stability and serine metabolism in lung and breast cancer [43]. Even more recently RNF5, an E3 ligase was suggested to be important to control PHGDH degradation in breast cancer [44]. It remains to be determined whether the activities of these E3 ligases are directly counter JOSD2’s deubiquitination.
Our observation that deficiency in JOSD2 heavily limits PFK-1 product formation (F16BP), may be of particular importance to resolve the central role of JOSD2 to regulate glycolysis. F16BP has been recognized as a key control step in regulating flux through glycolysis [31] and PFK-1 is often considered the most important control site in glycolysis.
Remarkably, we found that many of the uncovered JOSD2-linked pathways have been shown to be co-expressed with PHGDH in primary lung adenocarcinomas [27]. This highlights that multiple JOSD2-related pathways are mirrored when probing PHGDH-associated gene transcripts in lung adenocarcinoma patients, comprising amino acid metabolism, DNA synthesis, and cell cycle progression.
While our findings demonstrate significant inhibitory effects on glycolysis upon JOSD2 deficiency, our analyses of the glycolytic function in cells expressing either wild-type JOSD2 or the catalytically inactive JOSD2C24A variant show the most prominent effect of JOSD2 on maximal glycolysis, when the mitochondria are inhibited by oligomycin. These data support the hypothesis that metabolic regulation by JOSD2 may be, in particular, relevant for highly glycolytic cancer types preferentially relying on glucose catabolism for their proliferative state and survival. Consistent with this prediction, we show that JOSD2 promotes growth and proliferation. Thus, JOSD2 may display similar functions in cancer types, beyond NSCLCs, that display a dependency on glycolysis.
Our identification of a protein complex of glycolysis-related enzymes is consistent with previous findings showing that glycolysis enzymes tend to engage in various complexes to control glycolysis [45, 46].
Deubiquitination processes relevant to cancer progression and metabolism primarily suggested that DUBs may regulate the stability of classical metabolic regulators including oncogenes, their signaling network or glucose uptake [47,48,49,50,51,52,53]. Further studies are required to identify whether other DUBs may display metabolic effects and if these findings may have therapeutic implications.
References
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47.
Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between metabolism and cancer biology. Cell. 2017;168:657–69.
Locasale JW, Vander Heiden MG, Cantley LC. Rewiring of glycolysis in cancer cell metabolism. Cell Cycle. 2010;9:4253.
Metallo CM, Vander, Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–98.
Stanley IA, Ribeiro SM, Gimenez-Cassina A, Norberg E, Danial NN. Changing appetites: the adaptive advantages of fuel choice. Trends Cell Biol. 2014;24:118–27.
Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–77.
Queiroz AL, Vakifahmetoglu-Norberg H, Norberg E. Resistant to targeted therapy—aim for metabolic liabilities. Theranostics. 2018;8:2061–3.
Nakatsukasa K, Okumura F, Kamura T. Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: lessons from yeast. Crit Rev Biochem Mol Biol. 2015;50:489–502.
Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513.
Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–44.
Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63.
Ge Z, Leighton JS, Wang Y, Peng X, Chen Z, Chen H, et al. Integrated genomic analysis of the ubiquitin pathway across cancer types. Cell Rep. 2018;23:213–26.e213.
Leznicki P, Kulathu Y. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J Cell Sci. 2017;130:1997–2006.
Zhang X, Smits AH, van Tilburg GB, Jansen PW, Makowski MM, Ovaa H, et al. An interaction landscape of ubiquitin signaling. Mol Cell. 2017;65:941–55.e948.
Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–86.
Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–88.
Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78.
He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol Ther. 2017;177:96–107.
Ambroise G, Yu TT, Zhang B, Kacal M, Hao Y, Queiroz AL, et al. Systematic analysis reveals a functional role for STAMBPL1 in the epithelial-mesenchymal transition process across multiple carcinomas. Br J Cancer. 2020;123:1164–77.
Ouchida AT, Kacal M, Zheng A, Ambroise G, Zhang B, Norberg E, et al. USP10 regulates the stability of the EMT-transcription factor Slug/SNAI2. Biochem Biophys Res Commun. 2018;502:429–34.
Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–11.
Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7.
Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, et al. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst. 2008;4:135–47.
Sugimoto M, Hirayama A, Robert M, Abe S, Soga T, Tomita M. Prediction of metabolite identity from accurate mass, migration time prediction and isotopic pattern information in CE-TOFMS data. Electrophoresis. 2010;31:2311–8.
Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757.
Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9.
Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, et al. PHGDH defines a metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep. 2017;19:2289–303.
Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat Commun. 2014;5:4763.
Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403.
Giatromanolaki A, Sivridis E, Arelaki S, Koukourakis MI. Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp Lung Res. 2017;43:167–74.
Tanner LB, Goglia AG, Wei MH, Sehgal T, Parsons LR, Park JO, et al. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 2018;7:49–62.e48.
Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201.
DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–81.
Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Investig. 2015;125:687–98.
Seki T, Gong L, Williams AJ, Sakai N, Todi SV, Paulson HL. JosD1, a membrane-targeted deubiquitinating enzyme, is activated by ubiquitination and regulates membrane dynamics, cell motility, and endocytosis. J Biol Chem. 2013;288:17145–55.
DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200.
Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun. 2017;482:426–31.
Zhang B, Tornmalm J, Widengren J, Vakifahmetoglu-Norberg H, Norberg E. Characterization of the role of the malate dehydrogenases to lung tumor cell survival. J Cancer. 2017;8:2088–96.
Peeters K, Van Leemputte F, Fischer B, Bonini BM, Quezada H, Tsytlonok M, et al. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat Commun. 2017;8:922.
Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548:112–6.
Marcondes MC, Sola-Penna M, Torres Rda S, Zancan P. Muscle-type 6-phosphofructo-1-kinase and aldolase associate conferring catalytic advantages for both enzymes. IUBMB Life. 2011;63:435–45.
Rais B, Ortega F, Puigjaner J, Comin B, Orosz F, Ovadi J, et al. Quantitative characterization of homo- and heteroassociations of muscle phosphofructokinase with aldolase. Biochim Biophys Acta. 2000;1479:303–14.
Liu J, Zhang C, Wu H, Sun XX, Li Y, Huang S, et al. Parkin ubiquitinates phosphoglycerate dehydrogenase to suppress serine synthesis and tumor progression. J Clin Investig. 2020;130:3253–69.
Wang C, Wan X, Yu T, Huang Z, Shen C, Qi Q, et al. Acetylation stabilizes phosphoglycerate dehydrogenase by disrupting the interaction of E3 ligase RNF5 to promote breast tumorigenesis. Cell Rep. 2020;32:108021.
Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–6.
Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc Natl Acad Sci USA. 2005;102:2402–7.
Arceci A, Bonacci T, Wang X, Stewart K, Damrauer JS, Hoadley KA, et al. FOXM1 deubiquitination by USP21 regulates cell cycle progression and paclitaxel sensitivity in basal-like breast cancer. Cell Rep. 2019;26:3076–86.e3076.
Kim SR, Kim JO, Lim KH, Yun JH, Han I, Baek KH. Regulation of pyruvate kinase isozyme M2 is mediated by the ubiquitin-specific protease 20. Int J Oncol. 2015;46:2116–24.
Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, et al. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–95.
Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, et al. USP7 Is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating beta-catenin deubiquitination. Cell Rep. 2017;21:612–27.
Schulein-Volk C, Wolf E, Zhu J, Xu W, Taranets L, Hellmann A, et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 2014;9:1099–109.
Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–94.
Zhao Y, Wang X, Wang Q, Deng Y, Li K, Zhang M, et al. USP2a supports metastasis by tuning TGF-beta signaling. Cell Rep. 2018;22:2442–54.
Acknowledgements
This work was supported by grants from The Swedish Research Council (VR), The Swedish Cancer Society (Cancerfonden), Ragnar Söderbergs Stiftelse, Stiftelsen Lars Hiertas Minne, Stiftelsen Längmanska Kulturfonden, and the Karolinska Institute Tenure Track Research grants. We thank Dr. Yuqing Hao for advice on the mutagenesis. We thank Drs. Tsutomu Hoshiba and Laura Shelton (Human Metablome Technilogies) for the isotopomer and metabolome studies. We thank Drs. Mandy Rettel and Frank Stein (EMBL proteomics facility) for the quantitative proteomics.
Author information
Authors and Affiliations
Contributions
LK performed isotopomer labeling, ECAR analyses, OCR for HAP and A549 cell lines. BZ performed bioinformatics, pathway, and the statistical analysis of proteomics data. LK and DN performed the western blots, qPCR, and enzymatic activity assays. DN performed immunoprecipitations, deubiquitination assays, cell growth, and experiments with HAP cells. ALQ performed OCR of A549 cells for the initial submission, MK performed mutagenesis and qPCR, and ES performed fractionation. HVN provided critical advice and reagents. EN, HVN, LK, BZ, and DN wrote the original manuscript with input from all authors. LK, BZ, DN, and EN prepared the figures. All authors analyzed and interpreted data. LK, DN, and MK performed the revision and editing. EN designed, supervised, and financed the research.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by L. Scorrano
Supplementary information
Rights and permissions
About this article
Cite this article
Krassikova, L., Zhang, B., Nagarajan, D. et al. The deubiquitinase JOSD2 is a positive regulator of glucose metabolism. Cell Death Differ 28, 1091–1109 (2021). https://doi.org/10.1038/s41418-020-00639-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-00639-1
This article is cited by
-
Josephin domain containing 2 (JOSD2) promotes lung cancer by inhibiting LKB1 (Liver kinase B1) activity
Signal Transduction and Targeted Therapy (2024)
-
JOSD2 mediates isoprenaline-induced heart failure by deubiquitinating CaMKIIδ in cardiomyocytes
Cellular and Molecular Life Sciences (2024)
-
Deubiquitinase JOSD2 improves calcium handling and attenuates cardiac hypertrophy and dysfunction by stabilizing SERCA2a in cardiomyocytes
Nature Cardiovascular Research (2023)
-
Deubiquitinating enzyme Josephin-2 stabilizes PHGDH to promote a cancer stem cell phenotype in hepatocellular carcinoma
Genes & Genomics (2023)
-
The emerging role of deubiquitylating enzymes as therapeutic targets in cancer metabolism
Cancer Cell International (2022)