Abstract
Osteoporosis develops because of impaired bone formation and/or excessive bone resorption. Several pharmacological treatment of osteoporosis has been developed; however, new treatments are still necessary. Cholesterol and estrogen receptor-related receptor alpha (ERRα) promote osteoclasts formation, survival, and cellular fusion and thus become high risk factors of osteoporosis. In this study, we identified that carnosic acid (CA) suppressed bone loss by dual-targeting of sterol regulatory element-binding protein 2 (SREBP2, a major regulator that regulates cholesterol synthesis) and ERRα. Mechanistically, CA reduced nuclear localization of mature SREBP2 and suppressed de novo biogenesis of cholesterol. CA subsequently decreased the interaction between ERRα and peroxisome proliferator-activated receptor gamma coactivator 1-beta (PGC1β), resulting in decreased the transcription activity of ERRα and its target genes expression. Meanwhile, CA directly bound to the ligand-binding domain of ERRα and significantly promoted its ubiquitination and proteasomal degradation. Subsequently, STUB1 was identified as the E3 ligase of ERRα. The lysine residues (K51 and K68) are essential for ubiquitination and proteasomal degradation of ERRα by CA. In conclusion, CA dually targets SREBP2 and ERRα, thus inhibits the RANKL-induced osteoclast formation and improves OVX-induced bone loss. CA may serve as a lead compound for pharmacological control of osteoporosis.
Similar content being viewed by others
Introduction
Osteoclasts and osteoblasts continuously remodel bone tissue and maintain bone homeostasis by osteoclastic bone resorption and osteoblastic bone formation [1,2,3]. Osteoblasts are bone-forming cells that are differentiated from pluripotent mesenchymal stem cells [4]. Osteoblasts produce bone extracellular matrix, deposit calcium, and express osteoclastogenic factors to support bone formation [5]. Preosteoclasts differentiate from monocyte/macrophage lineage cells after stimulation by macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL). Subsequently, preosteoclasts fuse and differentiate into mature multinucleated osteoclasts, leading to bone resorption [6]. The imbalance in bone remodeling caused by abnormal osteoclast differentiation may result in skeletal diseases, such as osteoporosis and osteopetrosis [7]. Smoking, drinking alcohol, and lower estrogen levels in postmenopausal women all results in increased osteoclast activity, leading to bone loss and osteoporosis. Menopause increases total cholesterol (TC) and low density-cholesterol (LDL-c) increase substantially [8]. Many clinical studies have shown that high cholesterol level deteriorates bone health [9,10,11,12], therefore, women, especially those during menopausal stage, have much higher rates of osteoporosis than men at any given age [13, 14], at least partially due to increased cholesterol. Some in vitro and in vivo studies indicated that osteoclasts formation, survival, and cellular fusion depend on cholesterol [15, 16]. Cholesterol lowering drugs greatly mitigates osteoclasts differentiation and activity [17, 18], thus reduces osteoporosis-associated fracture risk [19], while the use of fibrates or other lipid-lowering drugs that mainly affect total triglycerides is not related to a significantly decreased bone fracture risk [20], suggesting selective reduction of cholesterol might be sufficient to delay bone loss. Sterol regulatory element-binding protein 2 (SREBP2), a key transcription factor that specifically regulates cholesterol synthesis, was proved to regulate osteoclasts differentiation [21,22,23]. Moreover, recent study reported that nuclear factor of activated T cells 1 (NFATc1), a key genes required for mature osteoclast formation, is one target gene of SREBP2 [24]. Pharmacological inhibition of SREBP2 by fatostatin or anhydroicaritin protects bone mass by suppression of RANKL-mediated osteoclasts differentiation [22, 23]. Together, SREBP2 is considered as a new therapeutic target in osteopenic disorders, such as osteoporosis.
Estrogen receptor-related receptor alpha (ERRα), one of ERR family members (ERRα, ERRβ, and ERRγ), is one of the first identified orphan nuclear receptors. ERRα and the ERs share high amino acid sequence similarity (68%) in the DNA-binding domain and a moderate similarity (36%) in other parts of the proteins, such as the ligand-binding E domain [25]. ERRα coordinates peroxisome proliferator-activated receptor gamma coactivator 1-beta (PGC1β) to enhance mitochondrial biogenesis and osteoclast function. In recent years, several groups have reported that ERRα regulates skeletal remodeling by controlling osteoclastogenesis [26,27,28]. In osteoclastogenesis, ERRα was identified as a pro-osteoclastic factor in vivo, and osteopetrosis was exhibited in the ERRα knockout mice. Concomitantly, osteoclastogenesis was significantly disturbed when ERRα was knocked down in vitro [29]. Moreover, ERRα was also implicated in osteoclasts mobility and actin cytoskeletal organization by regulating the osteopontin-integrin b3 chain-activated c-src (phosphorylated at the Tyr416) pathway causing the disruption of the specific actin structure implicated in osteoclast adhesion, migration, and invasion [30]. These reports suggest that ERRα is a therapeutic target for osteoporosis [26]. Interestingly, a recent study reports that cholesterol was identified as an endogenous ERRα agonist [31]. Considering the interdependence of cholesterol and ERRα, we reasoned that dual-targeting SREBP2 and ERRα might be an ideal strategy to treat osteoporosis.
Carnosic acid (CA) is a phenolic acid compound first found in Salvia officinalis L., with ~3% weight content in air-dried leaves. CA exhibited potent anti-oxidation and anti-ageing effects and is widely used for treating obesity, cardiovascular diseases, and cancer [32]. However, there is no report on the treatment of osteoporosis with CA. In this study, we found CA suppressed SREBP2 activity and inhibited the osteoclasts formation. Mechanistically, CA directly bound to ERRα and promoted its degradation. All together, we discovered a dual-targeting compound against SREBP2 and ERRα with excellent antiosteoporosis effect.
Materials and methods
Materials
CA was purchased from Shanghai U-sea Bio-tech co., Ltd (Shanghai, China). 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazoliumbromide (MTT), cholesterol, filipin, 25-hydroxycholesterol, and 4,6-diamidino-2-phenylindole (DAPI) were purchased from Keygen Biotech (Nanjing, China). RANKL, M-CSF were from R&D Biosystems (Minneapolis, USA). The TRAP staining kit was from Sigma-Aldrich (St. Louis, USA). DMEM, α-MEM and FBS (fetal bovine serum) was from GIBCO (Grand Island, New York, USA); rhodamine-labeled phalloidin (RP) from YEASEN (Shanghai, China); MG-132 was from MedChem Express (Shanghai, China). Cycloheximide was purchased from Selleckchem (Shanghai, China). Anti-ERRα (catalogue no. ab76228), anti-PGC1β (catalogue no. ab176328), and anti-SREBP2 (catalogue no. ab30682) antibodies were purchased from Abcam (Cambridge, UK), Anti-HA (catalogue no. AE008) and anti-Histone H3 (catalogue no. A2348) antibodies were purchased from ABclonal Technology (Wuhan, Hubei, China). SREBP1 (catalogue no. sc-8984) antibody was from Santa Cruz (Dallas, TX, USA). His-ERRα, His-ERRα (Q390A, G793A mutation), His-ERRα (6KA mutation), His-ERRα (K14A), His-ERRα (K24A), His-ERRα (K51A), His-ERRα (K68A), His-ERRα (K189A), His-ERRα (K208A), Flag-NEDD4, Flag-BARD1, Flag-MDM2, Flag-STUB1, Flag-SUMPF1 plasmids were constructed by gene synthesis and site-directed mutagenesis. siRNA MDM2#1 (CCUACUGAUGGUGCUGUAATT), MDM2#2 (CCUGCUUUACAUGUGCAAATT), STUB1#1 (GGAGAUGGAGAGCUAUG AUTT), STUB1#2 (CCAAGCACGACAAGUACAUTT) were purchased from GenePharma (Shanghai, China). Scrambled siRNA (UUCUCCGAACGUGUCACGUTT) was used as control.
Cell culture
The murine monocytic cell line RAW264.7, MC3T3-E1, HeLa, MDA-MB-231, MCF7, 293T (ATCC, VA) was cultured in a humid incubator with 5% CO2 at 37 °C, and maintained in DMEM containing 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. The primary mature osteoclasts were generated from BMMs (bone marrow-derived monocytes) as described [33]. Briefly, BMMs were isolated from tibiae and femurs from 6-week-old C57BL6 mice. BMMs (1–2 × 105) were seeded into a well of a 24-well plate and 1 × 106 BMMs were seeded into a well of a six-well plate. The cells were cultured in α-MEM containing 10% (v/v) FBS, 100 ng/ml RANKL and 30 ng/ml M-CSF at 37 °C in a humidified atmosphere containing 5% CO2 for 4–6 days.
Viability assay
Cell viability was detected by the MTT assay. Briefly, RAW264.7 cells were seeded at the density of 2 × 104 cells/well into 96-well plates. After 24 h, cells were treated with CA as indicated for 3 days. After the treatment, MTT (5 mg/ml) was added and cells were incubated for 4 h. The cytotoxicity of CA was determined by microplate reader (Multiskan FC).
Western blotting
RAW264.7 cells were washed with ice-cold PBS and dissolved with RIPA buffer (50 mM Tris –HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA and protease inhibitors). Whole-cell extracts were separated by SDS-PAGE and transferred into polyvinylidene fluoride membranes. The membranes were blocked with 5% skim milk in TBS with Tween-20 (PBST). Then, anti-SREBP2 antibody (Abcam, USA) or antiactin antibody (Beyotime Biotechnology, China) was bound overnight at 4 °C. After washing with TBST, HRP-conjugated secondary antibody (Beyotime Biotechnology, China) was bound for 1 h at room temperature. Immunoreactive signals were detected with Chemi-Lumi One Ultra (Tanon, China).
Quantitative RT-PCR
Total RNA was extracted from cells using TRIzol reagent (Vazyme, China) according to the manufacturer’s instructions. RNA concentrations were equalized and converted into cDNA using a kit (Hiscript II reverse transcriptase, Vazyme, Nanjing, China). Gene expression was measured in a LightCycler® 96 System (Roche, Basel, Switzerland) using SYBR-green dyes (Vazyme, China). Gene expression was normalized to GAPDH. The sequences of primers used in the experiments are listed in Supplemental Table 1.
Osteoclast formation and TRAP staining
Monocyte/macrophage cell line RAW264.7 (2 × 103 cells/well) were cultured in 96-well plates in DMEM with 10% FBS, 100 ng/ml of RANKL for 24 h. Then, cells were treated with DMEM containing 10% FBS, 100 ng/ml of RANKL in the presence or absence of CA for 4 days. After 4 or 5 days of culture, the cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP) activity. Multinucleated (>3 nuclei) TRAP-positive cells were calculated.
Actin-ring formation assay
After stimulation with M-CSF (30 ng/ml) and RANKL (30 ng/ml), we fixed the cells with 4% paraformaldehyde (PFA) for 10 min and washed three times with PBS. Then, the cells were incubated with TRITC–phalloidin (Sigma) for 1 h followed with DAPI staining (Sigma) for 10 min.
Filipin staining
Cells were seeded into 24-well plates at the density of 5 × 104 cells/well and maintained in 5% CO2 at 37 °C for 24 h. Then, cells were transfected with siRNA for 48 h. After that, cells were incubated in medium D for 24 h, or medium D with the indicated concentrations of CA for 16 h. After the treatment, 50 μg/ml fillipin were used to stain the treated cells for 30 min at 24 °C. The filipin stained cells were analyzed with EVOS FL Auto microscope (Life technologies). The quantification of the cellular neutral lipids or cholesterol was analyzed by Image-Pro Plus.
Triacylglycerol and cholesterol measurement
For measurement of intracellular total triacylglycerol (TG) and total cholesterol (TC), the cells were cultured in six-well plates and collected in 1 ml PBS. One hundred microliters of the total cell suspension was transferred into a new tube and centrifuged at 1000 × g for 5 min at 4 °C. Then, this portion of the cells were lysed in lysis buffer (RIPA lysis buffer) and used for protein quantification. The remaining cell suspension was used for lipid extraction. After centrifugation at 1000 × g for 5 min at 4 °C, the collected cells were mixed with 1 ml of chloroform/methanol (2:1, v/v) adequately on a shaker for 3 h at 24 °C. Then, 500 μl NaCl (0.1 M) was added into each reaction tube and mixed thoroughly, followed by centrifugation at 3700 rpm for 10 min. The lower organic phase was transferred and evaporated to dryness. The residual liquid was resuspended in 50 μl of 1% Triton X-100 in absolute ethanol, and the concentrations of lipids were measured using the TG and TC determination kit according to the manufacturer’s instructions, respectively (Kehua, Shanghai, China).
Microscale thermophoresis analysis
CA were titrated in different concentrations to purified recombinant human ERRα and mutation proteins. The reaction was performed in 50 mM Hepes, 50 mM NaCl, 0.01% Tween-20, and 2 mM MgCl2. Then, the samples were incubated at room temperature for 5 min before analyzing by microscale thermophoresis (MST). A NanoTemper Monolith Instrument (NT.115) was used for measuring thermophoresis. In this instrument, an infra blue Laser (IB Laser) beam couples into the path of light (i.e., fluorescence excitation and emission) with a dichroic mirror and is focused into the sample fluid through the same optical element used for fluorescence imaging. The IB laser is absorbed by the aqueous solution in the capillary and locally heats the sample with a 1/e2 diameter of 25 μm. Up to 24 mW of laser power where used to heat the sample, without damaging the biomolecules. To analyze the thermophoresis of a sample, 10 μl of sample solution were transferred in a glass capillary (NanoTemper, hydrophilic treated). Thermophoresis of the protein in presence of varying concentrations of compound was analyzed for 30 s. Measurements were performed at room temperature and standard deviation was calculated from three independent experiments.
Immunofluorescence
In brief, cells were fixed with 4% PFA for 15 min, and then permeabilized with 0.1% Triton X-100 in PBS for 15 min. after blocking in 3% BSA for 1 h, the cells were incubated with the primary and corresponding fluorophore-conjugated secondary antibodies. Confocal images were captured with an LSM 710 confocal microscope (ZEISS). The merged pictures were generated by LSM 7 IMAGE browser (Zeiss, Germany).
Reporter gene assay
RAW264.7 cells were transfected with luciferase reporter plasmids containing ERRα, ERRβ, ERRγ, or Nur77 enhancers respectively using X-tremeGENE HP DNA Transfection Reagent (Roche) for 24 h. β-galactosidase expression plasmid was co-transfected with luciferase reporters as an internal control. Cells were then exposed to different concentrations of CA. Cells were lysed in 100 μl lysis buffer sufficiently on a shaker for 40 min at room temperature. Fifty microliters of the total cell suspension was transferred into a 96-well white plate (Perkin Elmer) for luciferase activity detection. The remaining cell suspension was tested for β-galactosidase activity using a β-Gal reporter gene assay kit (Beyotime) according to instructions of the manufacturer. The luciferase activity was measured immediately after adding 50 μl luciferase substrates with a microplate reader and the intensity of β-Gal was also measured as the internal control.
Animal experiment
The laboratory animal facility in the animal center has been accredited by Association for Assessment and Accreditation of Laboratory Animal Care International. All experiments and animal care in this study were conducted in accordance with the Provision and General Recommendation of Chinese Experimental Animals Administration Legislation and approved by the Science and Technology Department of Jiangsu Province (SYXK (SU) 2016-0011). Female C57BJ/6L mice (SPF grade, 8 weeks old, 20–22 g) were purchased from Nanjing University (Nanjing, China). The animals were kept under a consistent temperature (24 °C) with 12 h light/dark cycle and fed standard food pellets with the access to sterile water ad libitum. Mice were ovariectomized (OVX) under ketamine (120 mg/kg)/xylazine (16 mg/kg) anesthesia. Seven days after ovariectomy, mice were randomly and blindingly divided into five groups (n = 12/group) and receive daily intragastric administration for 16 weeks: sham operated mice (Sham), ovariectomized mice treated with vehicle (OVX), OVX mice treated with 0.1 mg/kg E2 (OVX + E2 0.1 mg/kg), OVX mice treated with 30 mg/kg CA (OVX + CA 30 mg/kg), and OVX mice treated with 60 mg/kg CA (OVX + CA 60 mg/kg).
Microcomputed tomography (μCT) analysis
Tibias were fixed with 70% ethanol and subjected to μCT analysis using a SkyScan 1176 system (Bruker, Germany) with an isotropic voxel size of 6 μm for trabecular analyses according to the manufacturer’s instructions and the guidelines of the American Society for Bone and Mineral Research (ASBMR). Two hundred slices of proximal tibial metaphysis starting at 0.6 mm from the end of the growth plate were scanned and analyzed. Three-dimensional reconstructions were generated and analyzed according to the manufacturer’s instructions and the ASBMR guidelines.
Bone histomorphometry
For TRAP staining, tibias were fixed in 70% ethanol for 3 days, and the nondecalcified bones were embedded in methyl methacrylate. Longitudinal 5-μm-thick sections were cut on a microtome (Leica RM2255, Leica Microsystems, Germany) and subjected to TRAP staining using a TRAP Stain kit (Sigma, USA) according to the manufacturer’s protocol. Histomorphometry of the secondary spongiosa was performed with the OsteoMeasure analysis system (OsteoMetrics, USA) at 200-fold magnification according to the ASBMR guidelines.
Immunohistochemistry
Immunohistochemistry was carried out using 3-µm-thick sections fixed in 4% PFA. After deparaffinization, rehydration, and antigen retrieval, sections were incubated in blocking buffer containing 10% normal goat serum in PBS. Sections were incubated with described antibodies followed by washing and incubated with HRP-tagged goat anti-rabbit secondary antibody. Samples were subsequently rinsed in wash buffer and incubated in diaminobenzidine (Sigma, St. Louis, MO) for 5 min and counterstained in hematoxylin. Tissue slides were scored in a blinded fashion. No staining was observed with negative control rabbit anti-IgG antibody. The images were measured blindly by one observer using Image-Pro Plus (Media Cybernetics, Silver Spring, USA).
Statistical analysis
All data are expressed as the means standard error. The comparisons between groups were made using one-way ANOVA (Tukey’s multiple comparisons test). The significance of differences between two groups was determined using Student’s unpaired t test. Differences with p < 0.05 were considered to be statistically significant.
Results
CA is identified as an inhibitor for both SREBP and osteoclast formation
In our previous work, we screened a number of SREBPs inhibitors from a small molecule pool containing about 400 natural products using a reporter genes assay [8] (Fig. S1A). To identify whether these SREBPs inhibitor suppresses osteoclasts formation, RAW264.7 cells, a murine macrophage cell line, were treated with RANKL to differentiate into multinuclear TRAP-positive osteoclast-like cells [34]. Thirteen out of twenty-two SREBPs inhibitors suppressed RANKL-induced osteoclast-like cells formation (Fig. S1B, C). CA (Fig. S1D) exhibited the strongest activity to inhibit osteoclastogenesis (Fig. S1B, C). Meanwhile, CA did not obviously affect the cell viability of RAW264.7 cells or BMMs, even at the dose of CA up to 60 μM (Fig. S1E). Thus, CA was selected for further studies.
CA suppresses RANKL-induced osteoclast differentiation and function
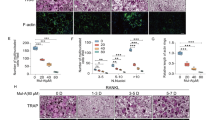
To determine the effects of CA on osteoclastogenesis, RAW264.7 and primary BMMs were used. In the solvent treated group, we observed the formation of TRAP-positive multinucleated osteoclasts in both BMMs (Fig. 1a, b) and RAW264.7 cells (Fig. S2A, B). The formation of osteoclasts was inhibited following the CA treatment in a concentration-dependent manner (Figs. 1a, b and S2A, B). The osteoclasts area and TRAP intensity were also dose dependently decreased (Figs. 1a, b and S2A, B) treated with CA. We next detected the differentiation process of BMMs or RAW264.7 cells with RANKL stimulation. Treatment with CA during the 1–3 days of differentiation obviously inhibited RANKL-induced osteoclasts differentiation (Figs. 1c, d and S2C, D). In comparison, if CA was added at later stage (day 5), it only slightly reduced the osteoclasts number, osteoclasts area and TRAP intensity (Figs. 1c, d and S2C, D). These data suggested that CA inhibited RANKL-induced osteoclasts differentiation at early stage.
a TRAP-positive osteoclast treated with indicated concentrations of CA followed by stimulation with 100 ng/ml RANKL and 30 ng/ml M-CSF for 7 days. b Quantification of area of osteoclasts, TRAP-positive multinuclear cells, and TRAP intensity. c BMMs were seeded and treated with 15 μM CA for the indicated days during osteoclastogenesis. d Quantification of area of osteoclasts, TRAP-positive multinuclear cells, and TRAP intensity. e BMMs were incubated with or without 100 ng/ml RANKL and 30 ng/ml M-CSF, followed by treatment with indicated concentration of CA. Cells were fixed and stained for F-actin (left). Osteoclasts having actin rings were counted (right). f BMMs cells were incubated with or without 100 ng/ml RANKL and 30 ng/ml M-CSF, followed by treatment with indicated concentration of CA for 96 h. RT-qPCR (n = 3 biological replicates) was used to assess relative mRNA expression levels of indicated genes. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
To further assess the effects of CA on osteoclastogenesis, differentiated BMMs and RAW264.7 cells by RANKL treatment formed obvious actin-ring structures as stained with TRITC-phalloidin (Figs. 1e and S2E). In comparison, the number and size of actin ring were obviously reduced by CA treatment dose dependently (Figs. 1e and S2E). The expression of the osteoclastogenesis marker genes, such as acid phosphatase (Acp5), cathepsin K, TRAP, Nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), and cellular oncogene fos (C-Fos), was significantly inhibited by CA (Figs. 1f and S2F). Next, we assessed the effect of CA on osteoblasts formation and mineralization in vitro. The results showed that CA treatment did not affect the cell viability of osteoblasts (Fig. S3A) and did not influence the expression of the osteogenesis-related genes, such as alkaline phosphatase, type 1 collagen, osteocalcin, runt-related transcription factor 2, and Osteoprotegerin (Fig. S3B–F). Taken together, these in vitro data suggested that CA inhibited osteoclastogenesis.
CA improves ovariectomy-induced bone loss by inhibiting osteoclast activity
To evaluate the antiosteoporosis effect of CA in vivo, mice were either sham operated or OVX, and then were treated with CMC-Na, estradiol valerate (E2, 0.1 mg/kg) or CA (30, 60 mg/kg) for 16 weeks. There were no obvious adverse events recorded after the OVX procedure or the subsequent treatment with CA. The body weight was significantly decreased in CA and E2 treatment group (Fig. 2a). After 16 weeks of CA administration, the levels of serum alkaline phosphatase, bone gla protein, serum 1 collagen crosslinked c-terminal peptide (CTX-1), and TRAP (strACP) increased in the ovariectomized mice, similar to the effect of E2 (Fig. 2b–e). The level of the blood calcium did not change in the ovariectomized and CA-treated mice (Fig. 2f). Treatment with CA (60 mg/kg) or E2 significantly reversed the blood phosphorus level in ovariectomized mice (Fig. 2g).
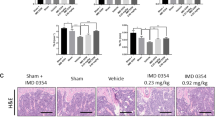
The fixed vertebral bodies of OVX mice were divided into four groups, 12 mice/group. The mice were treated E2 (0.1 mg/kg) or CA (30, 60 mg/kg) for 16 weeks. a Body weight. b–g Effect of CA on blood ALP, BGP, CTX-1, strACP, calcium, and phosphorus. h CA suppressed bone loss in an OVX-induced bone loss model. Representative reconstructed 3D μCT images of proximal tibia in mice treated with CMC-Na, or E2 (0.1 mg/kg) or CA (30 or 60 mg/kg). i–n Quantitative μCT analysis of trabecular structure in mice of each group. o H&E and TRAP staining, respectively, of the vertebral body sections (black arrows, TRAP-positive cells). p OcS/BS (%) in treatment groups. Bars represent means ± S.D., n = 12/group, *p < 0.05, **p < 0.01, and ***p < 0.001 vs control.
To evaluate the effects of CA on bone mass loss in ovariectomized mice, the tibiae of mice were then collected for μCT analysis. Representative 3D reconstructions of proximal tibial trabecular bone showed that CA treatment protected the OVX mice from bone loss (Fig. 2h), as shown by increased bone mineral density (BMD), bone volume/tissue volume, trabecular thickness, trabecular number (Fig. 2i–l). Furthermore, trabecular spacing and structure model index were significantly reduced in the OVX + CA group compared with the OVX control group (Fig. 2m, n). These results indicated that CA protected against ovariectomy-induced bone loss. Then, the histomorphometric analyses of proximal tibial trabecular bones by TRAP staining were also performed to further confirm the effect of CA. Consistent with the μCT results, the osteoclasts surface/bone surface was decreased in CA-treated OVX mice (Fig. 2o, p). These results suggest that CA protects against OVX-induced bone loss by inhibiting osteoclasts formation.
CA decreases cholesterol level by inhibiting SREBP2 activity in RANKL-induced osteoclasts
It has been demonstrated that CA promotes the degradation of mature form of SREBP2 via the 26S proteasome in liver cells, thus inhibits the hepatic de novo lipogenesis, especially, the synthesis of cholesterol [35]. Next, we examined whether CA inhibits the activation of SREBPs in osteoclasts. The reporter assay indicated that RANKL increased the transcription activity of SREBPs (Fig. 3a). The mature SREBP2 (mSREBP2) level was increased by RANKL stimulation and was significantly inhibited by CA (Figs. S4A and 3b). In comparison, RANKL and CA did not induce the mSREBP1 level (Fig. S5A, C). Furthermore, CA significantly inhibited RANKL-induced nuclear localization of SREBP2 (Fig. 3c). Similar results were observed in vivo (Fig. 3d). The expression of the SREBP2 target genes, such as hydroxymethylglutaryl coenzyme A reductase, hydroxymethylglutaryl coenzyme A synthase, low-density lipoprotein receptors, mevalonate kinase, and farnesyl diphosphate synthase, was significantly decreased by CA treatment (Figs. S4B–F and 3e–i). In comparison, the gene expression of SREBP1c was not influenced by CA treatment (Figs. 3j and S4G). These results indicated that CA specifically inhibited SREBP2 activity and may inhibit de novo synthesis of cholesterol and intracellular transport of LDL-c in osteoclasts. Next, we detected the level of TC and TG in osteoclasts formed from either BMMs (Figs. S4H and S5D) or RAW264.7 cells (Figs. 3k and S5A). It was shown that CA significantly decreased TC content in a dose-dependent manner (Figs. 3k and S4H) but it did not affect cellular TG level (Fig. S5B, D). The effect of CA on cellular TC content was further confirmed by filipin staining (Figs. 3l and S4I). Taken together, these results demonstrated that CA decreased the level of cholesterol by inhibiting SREBP2 activity in RANKL-induced osteoclasts.
a The SRE luciferase reporter plasmid was co-transfected into the RAW264.7 cells with expression plasmid for β-gal for 24 h. The cells then were incubated with or without the indicated concentrations of CA and stimulated with 100 ng/ml of RANKL for another 24 h. Cell supernatants were collected and assayed for luciferase activity. b–c RAW264.7 cells were incubated with or without 100 ng/ml RANKL, followed by treatment with indicated concentration of CA for 48 h. b The cells were lysed and the mature SREBP2 were detected by western blot (WB) (left) and quantification of normalized mature SREBP2 levels to actin controls (right). c The localization of SREBP2 in cells was visualized by immunofluorescence. d The localization of SREBP2 in osteoclast of treated mice by immunochemistry. e–j RT-qPCR was used to assess relative mRNA expression levels of indicated genes. k–l RAW264.7 cells were incubated with or without 100 ng/ml RANKL, followed by treatment with indicated concentration of CA for 5 days. k The cellular TC was detected by the detection kit. l Cholesterol was stained by Filipin. All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
CA disrupts the formation of ERRα/PGC1β complex and inhibits the ERRα activity partially depend on reduction of cellular cholesterol in osteoclasts
In recent studies, it has been shown that the nuclear receptor ERRα regulates skeletal remodeling by controlling osteoclastogenesis, a key cellular differentiation process essential for bone resorption. Wei et al. demonstrated that cholesterol is an endogenous ligand of ERRα and promotes osteoclastogenesis by increasing the interaction of ERRα and PGC1β [31]. As CA suppressed de novo cholesterol biosynthesis and reduced intracellular cholesterol, we reasoned that the interaction between ERRα/PGC1β would be weakened. First, we found that CA significantly suppressed RANKL-induced ERRα transactivation in a dose-dependent manner (Fig. 4a), while the transcriptional activity of other members of ERR family including ERRβ, ERRγ or Nur77 were not affected (Fig. 4b–d). The interaction between ERRα and PGC1β was largely disrupted after CA treatment (Fig. 4e, f). RANKL stimulated the expression of ERRα target genes that promote mitochondrial and osteoclast activity, such as Aconitate hydratase 2, Isocitrate Dehydrogenase 3 (NAD (+)) alpha (IDH3α), Acp5, Short-chain specific acyl-CoA dehydrogenase (SCAD), Medium-chain specific acyl-CoA dehydrogenase (MCAD), ATP synthase subunit beta (ATP5b), and Ndg2 (Figs. S6A–F and 4g–m). CA significantly decreased these ERRα target genes expression (Figs. S6A–F and 4g–m). These results demonstrated that CA significantly disrupted the RANKL-induced activity of ERRα.
a–d The indicated luciferase reporter plasmid was co-transfected into the RAW264.7 cells with expression plasmid for β-gal for 24 h. The cells then were incubated with or without the indicated concentrations of CA and stimulated with 100 ng/ml of RANKL for another 24 h. Cell supernatants were collected and assayed for luciferase activity. e Co-immunoprecipitation analysis of the effects of CA on ERRα and PGC1β interaction. RAW264.7 cells were treated with 100 ng/ml RANKL for 5 days in the presence of 15 μM CA. f Quantification of indicated protein levels of Fig. 5e. g–m RAW264.7 cells were incubated with or without 100 ng/ml RANKL, followed by treatment with indicated concentration of CA for 5 days. RT-qPCR was used to assess relative mRNA expression levels of indicated genes. All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
Surprisingly, when exogenous cholesterol was added to CA-treated cells, the inhibition of ERRα activity was only partially reversed (Fig. 5a), although the ERRα and PGC1β binding were completely recovered (Fig. 5b). Consistently, there was only a partial reversal of the suppressed ERRα target gene expression (Fig. 5c–j). Similarly, the inhibition effect of osteoclasts formation by CA was also partially mitigated in cholesterol replenishment experiment (Fig. 5k–l). In addition, exogenous overexpression of mSREBP2 reproduced the cholesterol replenishment experiment (Fig. S7A–D). These results indicated that CA suppressed osteoclastogenesis only partially depending on reducing the cholesterol content and ERRα/PGC1β complex formation.
a The ERRα luciferase reporter plasmid was transfected into the RAW264.7 cells with expression plasmid for β-gal for 24 h. Cells then were incubated with or without 15 μM CA or 10 μM cholesterol and stimulated with 100 ng/ml of RANKL for another 24 h. Cell supernatants were collected and assayed for luciferase activity. b–j RAW264.7 cells were treated with 100 ng/ml RANKL for 5 days in the presence of 15 μM CA or 10 μM cholesterol. b The cells were lysed and the proteins were detected by WB (left), quantification of indicated protein levels (right). c–j RT-qPCR was used to assess relative mRNA expression levels of indicated genes. k TRAP-positive osteoclast treated with 15 μM CA or 10 μM cholesterol followed by stimulation with 100 ng/ml RANKL for 5 days. l Quantification of area of osteoclasts, TRAP-positive multinuclear cells and TRAP intensity. All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
CA promotes the ubiquitination-proteasomal degradation of ERRα
Next, we found that CA significantly decreased the protein level of ERRα (Figs. 6a and S8A) without affecting its mRNA expression (Figs. 6b and S8B). This effect was comparable with XCT790, an inverse antagonist of ERRα [36]. Consistently, the protein level of ERRα was also obviously decreased in CA-treated OVX mice (Fig. 6c). Although XCT790 obviously promotes ERRα degradation, it significantly exhibited cytotoxicity at 20 μM concentration (Fig. S9A). Moreover, XCT790 greatly increased mSREBP2 proteins (Fig. S9B), subsequently increases expression of SREBP2 target genes (Fig. S9C–H). As CA reduced both SREBP2 and ERRα protein levels without causing cytotoxicity, CA has certain therapeutic advantages compared with XCT790. As shown in Fig. 6d, the reduction of ERRα was caused by the increased turnover of ERRα. CA substantially increased the ubiquitination of ERRα (Fig. 6f). MG-132, a proteasome inhibitor, abolished the effect of ERRα depletion by CA (Fig. 6e). These results indicated that CA promoted ERRα proteasomal degradation.
RAW264.7 cells were treated with 100 ng/ml RANKL for 5 days in the presence of CA (7.5, 15 μM) or XCT790 (10 μM). a The cells were lysed and the proteins were detected by WB (left), quantification of indicated protein levels (right). b RT-qPCR was used to assess relative mRNA expression levels of ERRα genes. c The protein level of ERRα in mice was detected by immunohischemistry. d 293T cells were transfected with His-ERRα plasmid for 24 h, and then incubated with 100 µM cycloheximide for 1 h, afterwards, the cells were supplemented with 50 µM cycloheximide plus vehicle, or 7.5 µM CA for indicated time. His-ERRα was detected by WB (left), quantification of His-ERRα protein levels (right). e RAW264.7 cells were treated with 100 ng/ml RANKL for 5 days in the presence of CA (7.5 μM) or MG-132 (10 μM). The cells were lysed and the proteins were detected by WB (left), quantification of indicated protein levels (right). f 293T cells were transfected with His-ERRα and HA-ubiquitin plasmids for 24 h, the cells then were supplemented with or without 7.5 µM CA for another 24 h, the cells were lysed and proteins were detected by WB (left), quantification of indicated protein levels (right). All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
STUB1 is identified as the E3 ligase of ERRα and mediates its ubiquitylation and proteasomal degradation
To identify the E3 ligase (E3s) of ERRα, we employed UbiBrowser25, an integrated bioinformatics platform to predict the proteome-wide E3-substrate network based on naive Bayesian network (http://ubibrowser.ncpsb.org). We queried ERRα as substrate, 108 predicted E3 ligases in UbiBrowser are presented in Fig. 7a. These E3s were classified into different families and shown in the E3 family hierarchical tree (Fig. 7a, right panel). Five E3 ligases with top confidence scores among 108 predicted E3s were filtered and listed (Fig. 7b). To further confirm the E3 ligase of ERRα, we separately overexpressed these five E3s (NEDD4, BARD1, MDM2, STUB1, and SMUPF1) and examined the ERRα level accordingly. Only MDM2 and STUB1 overexpression led to decreased ERRα protein level (Fig. 7c). Further, we assessed protein level of ERRα in cells transfected with either MDM2 or STUB1 siRNAs. STUB1 knockdown significantly increased the ERRα protein level (Fig. 7d), while MDM2 knockdown did not affect ERRα protein level (Fig. S10). These data suggest that only STUB1 is sufficient and required for ERRα degradation. We then validated the interaction between ERRα and its potent E3 ligase (STUB1). STUB1 was co-immunoprecipitated with ERRα (Fig. 7e). Taken together, these data suggested that STUB1 as a bona fide E3 ligase of ERRα. Consistently, in STUB1 knockdown cells, CA no longer reduced ERRα protein level (Fig. 7f), suggesting CA promotes ERRα proteasomal degradation via STUB1 mediated ubiquitination.
a A network view of E3-ERRα interactions (left panel) and the E3 hierarchical tree for ERRα (right panel). UbiBrowser was employed to explore the E3 ligases for ERRα. The representative predicted E3 ligases surround ERRα. The node colors and characters reflect the E3 type. The edge width, the node size, and the edge shade are corrected with the confidence score. The predicted E3s and their position in the E3 family hierarchical tree was presented. In this tree, texts in each circle (just like “U,” “R,” and “H”) represent the E3 family. The number in the bracket following each E3 family represents the number of corresponding predicted E3-ERRα interaction. b The information of top five confidence of ERRα E3 ligases. c 293T cells were transfected with indicated plasmids for 24 h. Cells were then lysed and the proteins were detected by WB (left), quantification of His-ERRα protein levels (right). d 293T cells were transfected with indicated siRNAs for 72 h. Cells were then lysed and the proteins were detected by WB (left), quantification of His-ERRα protein levels (right). e The interaction of ERRα with STUB1 was detected by co-immunoprecipitation. f 293T cells were transfected with indicated siRNAs and His-ERRα plasmids for 72 h. The cells then treated with CA for 24 h. Cells then lysed and the proteins were detected by WB (left), quantification of His-ERRα protein levels (right). g Molecular docking of CA to ERRα. h The interaction between ERRα and CA was detected by MST. i The interaction between a mutant (Q390A, G793A) form of ERRα and CA was detected by MST. j 293T cells were transfected with indicated plasmids for 24 h. Cells were then treated with CA for another 24 h. the cells then lysed and the interaction of ERRα with STUB1 and Ubiquitin was detected by co-immunoprecipitation. k 293T cells were transfected with indicated plasmids for 24 h. the cells then treated with CA for another 24 h. the cells then lysed and the proteins were detected by WB. All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
CA binds directly to ERRα and promotes its proteasomal degradation
Based on the above results, we reasoned that CA might be an ERRα inverse agonist that reduces the stability of ERRα. In order to verify this conjecture, we first assessed whether CA binds to ERRα through a virtual docking assay. The results showed that the binding energy between ERRα and CA is −11.27 kJ/mol. More specifically, two hydrogen bonds are formed between the two phenolic hydroxyl groups of CA and Gln390 and Gly393 in the ligand-binding domain of ERRα (Fig. 7g). Afterwards, a direct binding between CA and ERRα was detected by the MST assay, the result showed that the KD between CA and ERRα was 1.63 ± 0.88 μM (Fig. 7h). After the residues Gln390 and Gly393 of ERRα were mutated to alanine (Q390A, G793A) to prevent hydrogen bond formation, the binding between CA and the mutated ERRα protein got lost (Fig.7i). CA increased the binding ERRα and STUB1, and increased the ubiquitination of ERRα (Fig. 7j). CA neither increased the interaction of mutated ERRα (Q390A, G793A) with STUB1 nor upregulated the ubiquitination of mutated ERRα (Q390A, G793A) (Fig. 7j). Consequently, CA did not lead to the mutated ERRα (Q390A, G793A) degradation (Fig. 7k). In conclusion, these results suggest that CA directly binds to ERRα, promoting its interaction with its E3 ligase STUB1. Then ERRα undergoes ubiquitination and proteasomal degradation.
CA promotes STUB1 mediated ERRα ubiquitination at K51 and K68
We then used the UbPred database (http://www.ubpred.org) to predict the ubiquitination site of ERRα. As shown in Fig. S11A, B, six lysine residues were predicted to be ERRα ubiquitination sites. When all six lysine residues were mutated to alanines, the mutated ERRα (6KA) were resistant to CA-mediated proteasomal degradation (Fig. 8a). We further mutated each of these six lysines to alanines. It turned out that K51A and K68A mutations got resistance against CA induced proteasomal degradation (Fig. 8b). Thus, the ubiquitination sites of ERRα were successfully identified. In conclusion, CA bound to ERRα, leading to STUB1 mediated ubiquitination at K51 and K68, causing ERRα proteosomal degradation. Next, we co-transfected PGC1β and ERRα wild type or mutant (6KA) overexpression plasmids in RAW264.7 cells. The ERRα target genes suppressed by CA were reversed (Fig. 8c, d) by mutant ERRα (6KA), but not wild-type ERRα, consistent with results in Fig. 8a. Meanwhile, the inhibition effect of osteoclasts formation by CA was also obviously reversed in cells overexpressed ERRα (6KA), but not wild-type ERRα (Fig. 8e, f). These data suggest that ERRα degradation is essential for CA efficacy in inhibiting osteoclasts differentiation.
a 293T cells were transfected with indicated plasmids for 24 h. The cells then treated with CA for 24 h, the cells then lysed and the proteins were detected by WB (left), quantification of His-ERRα protein levels (right). b 293T cells were transfected with indicated ERRα mutation plasmids for 24 h, the cells then treated with CA for 24 h, the cells then lysed and the proteins were detected by WB. c–e The wild-type ERRα and mutated ERRα (6KA) plasmids were co-transfected with PGC1β into the RAW264.7 cells for 24 h, the cells then were incubated with or without CA and stimulated with 100 ng/ml of RANKL for another 5 days. c–d qRT-PCR was used to assess relative mRNA expression levels of indicated ERRα target genes. e TRAP-positive osteoclasts were stained by kit. f Quantification of area of osteoclasts, TRAP-positive multinuclear cells and TRAP intensity. All experiments were repeated three times. Bars represent means ± SD. *p < 0.05, **p < 0.01, ***p < 0.005.
Discussion
Bone maintenance depends on a balance between bone formation and bone resorption that implicates osteoblasts and osteoclasts and the osteocytes that modulate response of bone mechanical stress [37, 38]. Many studies have identified a link between dyslipidemia and low BMD for postmenopausal women [39]. Interestingly, osteoporosis is mainly due to an excess of osteoclasts since the amount of bone resorbed by the osteoclasts is not restored with the new bone deposited by the osteoblasts, suggesting that cholesterol may directly act through osteoclasts to induce bone loss in postmenopausal women [40]. Recently, cholesterol was shown to be the exogenous ligands of ERRα, and has the ability to recruit coactivators PGC1β to ERRα in osteoclasts. ERRα mediated the effect of cholesterol on bone resorption and skeletal remodeling [31]. Therefore, inhibition of intracellular cholesterol level may be a strategy to inhibit the osteoclasts formation. In previous studies, it has been demonstrated that SREBP2 is associated to osteoclast differentiation and SREBP2 inhibitors, such as fatostatin and anhydroicaritin inhibit osteoclast formation [21,22,23]. However, the physiological functions of SREBP2 in osteoclast remain unclear. Here, we reported that RANKL activates SREBP2 transcription activity, increases the content of cholesterol, which is the endogenous agonist of ERRα, therefore, accelerates osteoclasts formation. CA, a potent inhibitor of SREBP2, reduced the ERRα activity and inhibits osteoclasts formation through decreasing the level of cholesterol.
In view of the important role in the pathogenesis of osteoporosis, SREBP2, and ERRα have been considered as therapeutic targets for the treatment of osteoporosis. In this study, for the first time, we found CA as a dual-targeted drug against SREBP2 and ERRα with a potent effect to treat osteoporosis. Although SREBP2 and ERRα also have a correlation with the regulation of osteoblasts, CA has no significant effect on osteoblast formation. Moreover, there was no significant effect on SREBP1c expression and fatty acid metabolism in osteoclasts. These data show that CA has a good specific inhibitory effect of cholesterol metabolism.
Currently, the drugs for clinical treatment of osteoporosis are mainly divided into basic supplements, bone resorption inhibitors, and bone formation accelerating agents. Hormone replacement therapies (HRT) preserve BMD and prevent fractures during menopause [41]. However, estrogen exposure is suspected to have an association with increased risk of breast cancer, endometrial cancer, and cervical cancer [42,43,44]. Therefore, safer treatment regimens are needed for postmenopausal women with osteoporosis. Cholesterol promotes breast tumor growth and metastasis in murine models [45, 46]. SREBP2, the major transcription factor that control cholesterol biosynthesis, was found to be highly expressed in breast cancer tissues and correlated with a poor prognosis. SREBP2 was similarly under the transcriptional control of CREB and its induction regulates the expression of matrix metalloproteinases, key degradative enzymes involved in bone metastases by breast cancer cells. Accordingly, targeting of SREBP2 with fatostatin attenuated breast cancer-induced osteolysis in vivo [24]. Meanwhile, ERRα was reported to be a potent diagnostic and prognostic factor in human breast carcinoma [47, 48]. The overexpression of ERRα stimulates VEGF expression in primary tumors, promoting angiogenesis [49]. ERRα also supports breast cancer bone metastasis by increasing RANK expression [50]. In conclusion, SREBP2 and ERRα are considered to be important targets for the treatment of breast cancer. In our study, CA exerted both SREBP2 and ERRα inhibitory effects, thus leading to suppress breast cancer and cervical cancer progression (Fig. S12A–F). Taken together, our strategy by dual targeting of SREBP2 and ERRα has significant security advantages over traditional HRT. Therefore, inhibition of SREBP2 and ERRα may become a new generation of therapeutic strategy for osteoporosis in menopausal women.
PROteolysis-TArgeting Chimeras (PROTACs) technology has been used for eliminating “undruggable” proteome for targeted therapies [51, 52]. ERRα has receiving much attention in developing PROTACs molecules [53, 54]. These compounds are all designed via von Hippel–Lindau protein-based ubiquitination and proteasomal degradation [55]. The ERRα E3 ligases, as well as its ubiquitination sites, are yet to be clarified. In this study, we found that CA-promoted ERRα ubiquitination and proteasomal degradation by directly interacting with ERRα. STUB1 was discovered to regulate ERRα protein stability in osteoclasts. After CA binds to ERRα, STUB1 was recruited to bind and ubiquitinate ERRα at K51 and K68. Thus, CA promoted ERRα degradation (Fig. 9). In a former report, STUB1 was shown to regulate osteoclasts formation by promoting tumor necrosis factor receptor associated factor 6 degradation [56]. The bone remodeling phenotype of STUB1−/− mice might also attribute to decreased ERRα ubiquitination and degradation.
a Binding of RANKL to its cognate receptor RANK leads to activation of SREBP2 and de novo cholesterol synthesis. Cholesterol promotes ERRα and PGC1β binding and increases osteoclastogenesis gene expression. b CA prevents SREBP2 nucleus translocation and reduced cholesterol level in osteoclasts. Meanwhile, CA binds to ERRα and leads to K51 and K68 ubiquitination via STUB1. ERRα then undergoes proteosomal degradation and osteoclastogenesis gene expression is suppressed.
In conclusion, we have discovered that CA functions as the first dual-targeted small molecule against SREBP2 and ERRα. CA inhibits RANKL-induced osteoclast formation and ovariectomy-induced bone loss by inhibiting the SREBP2/cholesterol/ERRα signal pathway. This discovery enriches the pharmacological activity of CA and provides an experimental evidence for the diversity function of rosemary as a food additive. CA can serve as a lead compound for pharmacological control of osteoporosis.
References
Manolagas SC, Jilka RL. Bone marrow, cytokines, and bone remodeling—emerging insights into the pathophysiology of osteoporosis. N Engl J Med. 1995;332:305–11.
Prideaux M, Findlay DM, Atkins GJ. Osteocytes: the master cells in bone remodelling. Curr Opin Pharmacol. 2016;28:24–30.
Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–64.
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7.
Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–96.
Ikebuchi Y, Aoki S, Honma M, Hayashi M, Sugamori Y, Khan M, et al. Coupling of bone resorption and formation by RANKL reverse signalling. Nature. 2018;561:195.
Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791.
Zheng Z-G, Zhou Y-P, Zhang X, Thu PM, Xie Z-S, Lu C, et al. Anhydroicaritin improves diet-induced obesity and hyperlipidemia and alleviates insulin resistance by suppressing SREBPs activation. Biochem Pharmacol. 2016;122:42–61.
Derby CA, Crawford SL, Pasternak RC, Sowers M, Sternfeld B, Matthews KA. Lipid changes during the menopause transition in relation to age and weight: the Study of Women's Health Across the Nation. Am J Epidemiol. 2009;169:1352–61.
Jeong T-D, Lee W, Choi S-E, Kim JS, Kim H-K, Bae SJ, et al. Relationship between serum total cholesterol level and serum biochemical bone turnover markers in healthy pre-and postmenopausal women. BioMed Res Int. 2014;2014:398397.
Makovey J, Chen JS, Hayward C, Williams FM, Sambrook PN. Association between serum cholesterol and bone mineral density. Bone. 2009;44:208–13.
Yamaguchi T, Sugimoto T, Yano S, Yamauchi M, Sowa H, Chen Q, et al. Plasma lipids and osteoporosis in postmenopausal women. Endocr J. 2002;49:211–7.
Curry SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, et al. Screening for osteoporosis to prevent fractures: US Preventive Services Task Force recommendation statement. J Am Med Assoc. 2018;319:2521–31.
Mandal CC. High cholesterol deteriorates bone health: new insights into molecular mechanisms. Front Endocrinol. 2015;6:165.
Luegmayr E, Glantschnig H, Wesolowski G, Gentile M, Fisher J, Rodan G, et al. Osteoclast formation, survival and morphology are highly dependent on exogenous cholesterol/lipoproteins. Cell Death Differ. 2004;11:S108.
Sato T, Morita I, Murota S. Involvement of cholesterol in osteoclast-like cell formation via cellular fusion. Bone. 1998;23:135–40.
Grasser W, Baumann A, Petras S, Harwood H, Devalaraja R, Renkiewicz R, et al. Regulation of osteoclast differentiation by statins. J Musculoskelet Neuronal Interact. 2003;3:53–62.
Ruan F, Zheng Q, Wang J. Mechanisms of bone anabolism regulated by statins. Biosci Rep. 2012;32:511–9.
Mundy G, Garrett R, Harris S, Chan J, Chen D, Rossini G, et al. Stimulation of bone formation in vitro and in rodents by statins. Science. 1999;286:1946–9.
Meier CR, Schlienger RG, Kraenzlin ME, Schlegel B, Jick H. HMG-CoA reductase inhibitors and the risk of fractures. J Am Med Assoc. 2000;283:3205–10.
Inoue K, Imai Y. Identification of novel transcription factors in osteoclast differentiation using genome‐wide analysis of open chromatin determined by Dnase-seq. J Bone Miner Res. 2014;29:1823–32.
Inoue K, Imai Y. Fatostatin, an SREBP inhibitor, prevented RANKL-induced bone loss by suppression of osteoclast differentiation. Biochim Biophys Acta. 2015;1852:2432–41.
Zheng Z-G, Zhang X, Zhou Y-P, Lu C, Thu PM, Qian C, et al. Anhydroicaritin, a SREBPs inhibitor, inhibits RANKL-induced osteoclastic differentiation and improves diabetic osteoporosis in STZ-induced mice. Eur J Pharmacol. 2017;809:156–62.
Jie Z, Xie Z, Xu W, Zhao X, Jin G, Sun X, et al. SREBP-2 aggravates breast cancer associated osteolysis by promoting osteoclastogenesis and breast cancer metastasis. Biochim Biophysa Acta. 2019;1865:115–25.
Eichner LJ, Giguère V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–52.
Gallet M, Vanacker J-M. ERR receptors as potential targets in osteoporosis. Trends Endocrinol Metab. 2010;21:637–41.
Wan Y. PPARγ in bone homeostasis. Trends Endocrinol Metab. 2010;21:722–8.
Wei W, Wang X, Yang M, Smith LC, Dechow PC, Wan Y. PGC1β mediates PPARγ activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–16.
Bonnelye E, Aubin JE. An energetic orphan in an endocrine tissue: a revised perspective of the function of estrogen receptor‐related receptor alpha in bone and cartilage. J Bone Miner Res. 2013;28:225–33.
Bonnelye E. Estrogen Receptor Related Receptor Alpha (ERRα) in skeletal tissues. Endocrinol Metab Syndr. 2016;5:2161–1017.1000244.
Wei W, Schwaid AG, Wang X, Wang X, Chen S, Chu Q, et al. Ligand activation of ERRα by cholesterol mediates statin and bisphosphonate effects. Cell Metab. 2016;23:479–91.
Birtic S, Dussort P, Pierre FX, Bily AC, Roller M. Carnosic acid. Phytochemistry. 2015;115:9–19.
Marino S, Logan JG, Mellis D, Capulli M. Generation and culture of osteoclasts. BoneKEy Rep. 2014;3:570.
Hotokezaka H, Sakai E, Kanaoka K, Saito K, Matsuo K-i, Kitaura H, et al. U0126 and PD98059, specific inhibitors of MEK, accelerate differentiation of RAW264. 7 cells into osteoclast-like cells. J Biol Chem. 2002;277:47366–72.
Xie Z, Wan X, Zhong L, Yang H, Li P, Xu X. Carnosic acid alleviates hyperlipidemia and insulin resistance by promoting the degradation of SREBPs via the 26S proteasome. J Funct Foods. 2017;31:217–28.
Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Martin R, et al. Regulation of PPARγ coactivator 1α (PGC-1α) signaling by an estrogen-related receptor α (ERRα) ligand. Proc Natl Acad Sci USA. 2004;101:8912–7.
Tanaka Y, Nakayamada S, Okada Y. Osteoblasts and osteoclasts in bone remodeling and inflammation. Curr Drug Targets-Inflamm Allergy. 2005;4:325–8.
Sims NA, Martin TJ. Coupling the activities of bone formation and resorption: a multitude of signals within the basic multicellular unit. BoneKEy Rep. 2014;3:481.
Ghadiri-Anari A, Mortezaii-Shoroki Z, Modarresi M, Dehghan A. Association of lipid profile with bone mineral density in postmenopausal women in Yazd province. International. J Reprod Biomed. 2016;14:597.
Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–45.
Lindsay R, Gallagher JC, Kleerekoper M, Pickar JH. Effect of lower doses of conjugated equine estrogens with and without medroxyprogesterone acetate on bone in early postmenopausal women. J Am Med Assoc. 2002;287:2668–76.
Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–85.
Chung S-H, Franceschi S, Lambert PF. Estrogen and ERα: culprits in cervical cancer? Trends Endocrinol Metab. 2010;21:504–11.
Adami HO, Persson I, Hoover R, Schairer C, Bergkvist L. Risk of cancer in women receiving hormone replacement therapy. Int J Cancer. 1989;44:833–9.
Nelson ER, Chang C-y, McDonnell DP. Cholesterol and breast cancer pathophysiology. Trends Endocrinol Metab. 2014;25:649–55.
Llaverias G, Danilo C, Mercier I, Daumer K, Capozza F, Williams TM, et al. Role of cholesterol in the development and progression of breast cancer. Am J Pathol. 2011;178:402–12.
Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, et al. Estrogen-related receptor α in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–6.
Jarzabek K, Koda M, Kozlowski L, Sulkowski S, Kottler M-L, Wolczynski S. The significance of the expression of ERRα as a potential biomarker in breast cancer. J Steroid Biochem Mol Biol. 2009;113:127–33.
Stein RA, Gaillard S, McDonnell DP. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol. 2009;114:106–12.
Vargas G, Bouchet M, Bouazza L, Reboul P, Boyault C, Gervais M, et al. ERRα promotes breast cancer cell dissemination to bone by increasing RANK expression in primary breast tumors. Oncogene. 2019;38:950.
Pei H, Peng Y, Zhao Q, Chen Y. Small molecule PROTACs: an emerging technology for targeted therapy in drug discovery. RSC Adv. 2019;9:16967–76.
An S, Fu L. Small-molecule PROTACs: an emerging and promising approach for the development of targeted therapy drugs. EBioMedicine. 2018;36:553–62.
Peng L, Zhang Z, Lei C, Li S, Ren X, Chang Y, et al. Identification of New Small-Molecule Inducers of Estrogen-related Receptor α (ERRα) degradation. ACS Medicinal Chem Lett. 2019;10:767–72.
Bondeson DP, Mares A, Smith IE, Ko E, Campos S, Miah AH, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611.
Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, Noblin DJ, et al. Targeting the von Hippel–Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1α interaction. J Am Chem Soc. 2012;134:4465–8.
Li S, Shu B, Zhang Y, Li J, Guo J, Wang Y, et al. CHIP regulates osteoclast formation through promoting TRAF6 protein degradation. Arthritis Rheumatol. 2014;66:1854.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2019YFC1711000), the National Natural Science Foundation of China (81773957, 81421005, 81903871), the National Key Subject of Drug Innovation (No. 2017ZX09301012003), China Postdoctoral Science Foundation (2018M642380), the 111 Project (B16046). This project was also supported by the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (SKLNMZZCX201820), the “Double First-Class” University Project (CPU2018GF04).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Zheng, ZG., Cheng, HM., Zhou, YP. et al. Dual targeting of SREBP2 and ERRα by carnosic acid suppresses RANKL-mediated osteoclastogenesis and prevents ovariectomy-induced bone loss. Cell Death Differ 27, 2048–2065 (2020). https://doi.org/10.1038/s41418-019-0484-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0484-5
This article is cited by
-
HSP90β promotes osteoclastogenesis by dual-activation of cholesterol synthesis and NF-κB signaling
Cell Death & Differentiation (2023)
-
A 3D-printed molybdenum-containing scaffold exerts dual pro-osteogenic and anti-osteoclastogenic effects to facilitate alveolar bone repair
International Journal of Oral Science (2022)
-
Cholesterol inhibits autophagy in RANKL-induced osteoclast differentiation through activating the PI3K/AKT/mTOR signaling pathway
Molecular Biology Reports (2022)
-
Site-1 protease controls osteoclastogenesis by mediating LC3 transcription
Cell Death & Differentiation (2021)