Abstract
Polarization of naive T cells into interferon (IFN)-γ-producing T helper 1 (Th1) cells is an essential event in the inflammatory response to pathogens. Herein, we identify the RNA binding protein Sam68 as a specific modulator of Th1 differentiation. Sam68-knockout (ko) naive T cells are strongly defective in IL-12-mediated Th1 polarization and express low levels of T-bet and Eomes. Consequently, Sam68-ko Th1 cells are significantly impaired in IFN-γ production. Moreover, we found that Sam68 is required for the induction of an inflammatory Th1 response during Mycobacterium bovis Bacillus Calmette–Guerin (BCG) infection, thus limiting bacterial dissemination in the lungs. Mechanistically, Sam68 directly binds to the microRNA miR-29, a negative regulator of Th1 response, and inhibits its expression during BCG infection. These findings uncover a novel post-transcriptional mechanism required for the Th1-mediated defense against intracellular pathogens and identify a new function for Sam68 in the regulation of the immune response.
Similar content being viewed by others
Introduction
The immune response is orchestrated by CD4-positive (CD4+) T cells that are activated by antigen presenting cells and differentiate into distinct subsets of effector T helper (Th) cells [1]. Depending on the initial antigen eliciting the response, Th cells polarize into specific subtypes that are characterized by the array of cytokines they produce to mediate the immune response. For example, interleukin (IL)-12 promotes polarization of Th1 cells, which produce interferon (IFN)-γ and participate to clearance of intracellular pathogens [2]. On the other hand, Th2 cells produce IL-4, IL-5 and IL-13 which mediate the immune response against helminths [3]. More recently, a distinct subset of pro-inflammatory Th cells has been identified, which produce IL-17 (Th17 cells) and display specific functions in antifungal immunity [4,5,6].
Th1 polarization is orchestrated by a precise gene expression program set in motion by signaling pathways and executed by specific transcription factors, such as the T-box transcription factor expressed in T cells (T-bet) [1]. In addition, post-transcriptional regulation of gene expression also contributes to the Th1-specific differentiation program. For instance, microRNAs (miRNAs) regulate several aspects of Th cell biology [7] and global deficiency in miR expression causes aberrant production of IFN-γ by Th cells. In particular, expression of miR-29 in Th1 cells directly targets the transcripts encoding IFN-γ [7] and two transcription factors known to induce IFN-γ production (T-bet and Eomes) [8]. Notably, miR-29 expression is repressed in IFN-γ-producing T cells during infection of mice with the Mycobacterium bovis Bacillus Calmette–Guérin (BCG). Conversely, genetic interference with miR-29 expression enhances Th1-mediated responses and resistance to BCG infection [7]. These studies clearly demonstrated a role for miR-29 in suppressing Th1-mediated immune responses. Nevertheless, how expression of miR-29 is regulated upon infection of intracellular pathogens remains unknown.
MiRNAs are small noncoding RNAs that generally repress gene expression at the post-transcriptional level. They are transcribed by the RNA polymerase II (RNAPII) as longer primary transcripts (pri-miR) that are processed by the RNase III enzyme Drosha and the double-stranded RNA binding cofactor DGCR8 into shorter nucleotide hairpin precursors (pre-miR) [9]. Pre-miRs are then cleaved by Dicer to form ~22 base pair RNA duplexes, of which one strand forms the mature miR that is loaded onto the miRNA-induced silencing complex (RISC) and mediates repression of messenger RNAs (mRNAs) through complementary base pairing [9]. In addition to these core proteins, miR biogenesis and function can be modulated by various RNA binding proteins (RBPs). For instance, LIN28 binds pre-let-7 and denatures the pre-miR stem, thus impairing cleavage by Dicer [10]. Furthermore, competitive binding of antagonistic RBPs can determine the efficiency of pri-miR processing [11]. In this regard, it is currently unknown whether modulation of miR expression by specific RBPs also takes part in Th1-mediated immune responses. However, previous work indicated that miR-29 expression in male germ cells is repressed by Sam68 [12], a multifunctional RBP involved in multiple aspects of RNA synthesis and processing [13,14,15].
Sam68 contains a KH-type RNA binding domain flanked by regulatory regions involved in protein–protein interactions and post-translational modifications, including a carboxyl-terminal region that is phosphorylated by tyrosine kinases [13]. Sam68 participates to regulation of RNA metabolism and signaling pathways evoked by extracellular cues [13,14,15] and plays physiological roles in osteogenesis [16], gametogenesis [17, 18], adipogenesis [19] and brain development and function [20,21,22]. In most cases, Sam68 involvement in these physiological processes has been related to regulation of specific RNA targets [17,18,19,20,21,22,23].
Pioneer studies on Sam68 function suggested a role for this RBP in T-cell activation. Upon stimulation of the T-cell receptor (TCR), Sam68 is phosphorylated in the carboxyl-terminal tyrosine-rich domain by Src-family kinases [24, 25] and acts as an adaptor molecule in the early signaling cascade of T-cell activation [25]. Furthermore, Sam68 is required for the tumor necrosis factor (TNF)-induced signaling pathway in T cells, where it takes part to the large complex leading to activation of a pro-apoptotic signaling cascade [26]. More recently, it was proposed that Sam68 promotes activation of nuclear nuclear factor-κB complexes and enhances transcription of CD25 [27], the receptor of IL-2 associated with T-cell activation and proliferation [28]. Although these findings clearly implicate Sam68 in specific immunological processes, the broad impact of Sam68 function in the activation of immune responses in vivo has not been investigated yet, nor it is known whether lack of Sam68 impairs specific immunological responses to pathogens.
Herein, we have explored the potential role of Sam68 in the polarization of mouse Th cells. We found that Sam68 is specifically required for Th1 polarization following IL-12 stimulation in vitro, or BCG infection in vivo. Expression of Th1-specific transcription factors and of IFN-γ were significantly reduced in Sam68-knockout (ko) T cells. Notably, Sam68 binds in vivo to the precursor of miR-29, and it is required for miR-29 repression upon infection. These results uncover a previously unknown post-transcriptional checkpoint in the Th1 differentiation program during infection and highlight Sam68 as a novel regulator of IFN-γ production in response to intracellular pathogens.
Results
Th1 polarization is impaired in Sam68-ko mice
To investigate the possible role of Sam68 in Th polarization, we differentiated naive CD4+ T cells from wild-type (wt) and Sam68-ko mice into Th1 and Th2 profiles by anti-CD3/28-mediated induction and co-treatment with IL-12 or IL-4, respectively [2]. The analysis of typical Th1 (IFN-γ) and Th2 (IL-4) cytokine production after 7 days of polarization indicated a strong and significant reduction in the percentage of IFN-γ-producing cells derived from Sam68-ko T cells, whereas the percentage of IL-4-producing cells was not significantly affected (Fig. 1a, b). Likewise, expression of IFN-γ transcript and protein was significantly lower in Sam68-ko Th1 cells than in wild-type cells (Fig. 1c, d). Importantly, the mitogenic response to TCR activation, reported as fold expansion of CD4+ T cells and IL-2 production, was comparable in wild-type and knockout cells (Supplementary Figure 1a,b). Moreover, the altered response to IL-12 did not depend on defective expression of IL-12 receptor (IL12R) 1 and 2, as their levels were not significantly altered in Sam68-ko naive CD4+ T cells (Supplementary Figure 2a). Likewise, we found no significant difference in expression of IFN-γ receptors (IFNGR1 and IFNGR2) (Supplementary Figure 2b), which may have accounted for defective IFN-γ-induced positive feedback loop during Th1 polarization [29]. Lastly, specificity in regulation of Th1 polarization was not due to the expression level of Sam68, as this did not significantly differ in Th1- and Th2-polarized cells (Supplementary Figure 3). Notably, Sam68 expression did not affect IFN-γ production by memory CD4+ stimulated with anti-CD3/28 (Supplementary Figure 4). These findings indicate that Sam68 is specifically required during the polarization of naive CD4+ T cells into Th1 cells by acting on processes upstream of IFN- γ production and downstream of IL-12 and IFN- γ receptors.
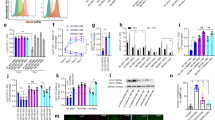
Th1 polarization is impaired in Sam68-ko mice. a Representative flow cytometry plots of intracellular IFN-γ and IL-4 in naive T cells, from wt and Sam68-ko mice, stimulated in the presence of anti-CD3 plus anti-CD28 and polarizing cytokines for 7 days, assessed after 5 h of restimulation with PMA and Ionomycin in the presence of Brefeldin-A. Percentage of IL-4+ IFN-γ– cells (top left), IL-4+IFN-γ+ cells (top right) or IL-4–IFN-γ+ cells (bottom right) is reported. b Bar graphs show data from seven independent experiments performed as in a (mean and s.e.m). c RT-PCR analysis of the expression of IFN-γ mRNA in naive CD4+ T cells, from wt and Sam68-ko mice, differentiated in vitro for 7 days in the presence of anti-CD3 plus anti-CD28 and polarizing cytokines. Cycling threshold values are normalized to those of mRNA encoding ribosomal protein L-34. Data are the mean and s.e.m. of six independent experiments. d–f Bar graphs show the results of ELISA (d) or multiplex assay (e) of IFN-γ in supernatants of naive T cells, from wt and Sam68-ko mice, differentiated as described in a. Data (mean and s.e.m.) are representative of four independent experiments. Pearson’s correlation analysis (f) between the levels of IFN-γ determined by ELISA and the levels of IFN-γ determined by multiplex assay (Luminex). g Analysis of IL-17, IL-21, IL-22, and GM-CSF by multiplex assay in supernatants of naive T cells, from wt and Sam68-ko mice, differentiated as described in a. Data (mean and s.e.m.) are representative of four independent experiments. Th0: no polarizing cytokines; Th1: anti-IL-4; Th2: IL-4 and anti-IFN-γ; Th17: IL-6, TGF-β, anti-IL-4, anti-IFN-γ, anti-IL-2; *p < 0.05 (Student’s t-test)****p<0,001
Next, we asked whether Sam68 was also required for polarization of Th17 cells, another pro-inflammatory Th cell subset [4]. To test this hypothesis, we used a standard Th17 differentiation assay and analyzed by multiplex assay a broad array of Th17 cytokines, including IL-17, IL-21, IL-22 and granulocyte-macrophage colony-stimulating factor (GM-CSF) [5]. Reliability of the multiplex assay was validated by the significant correlation with IFN-γ expression detected by enzyme-linked immunosorbent assay (ELISA; Fig. 1e, f). Nevertheless, we found no significant changes in Th17 cytokines between wild-type and knockout cells (Fig. 1g). Collectively, these results indicate that Sam68 specifically regulates Th1 cell polarization.
Sam68 regulates expression of T-bet and Eomes during Th1 differentiation
The transcription factors T-bet (encoded by Tbx21) and Eomes independently induce IFN-γ expression and production [30,31,32]. We observed that both Tbx21 and Eomes expression are reduced in Th1 cells derived from Sam68-ko mice (Fig. 2a, b). Accordingly, the percentage of T-bet- and Eomes-positive cells was drastically reduced in Sam68-ko Th1 cells (Fig. 2c, d). Thus, Sam68 is required for efficient expression of T-bet and Eomes during polarization of mouse Th1 cells. Similar to what was observed for IFN- γ production, this crucial function of Sam68 is strictly related to polarization of Th1 cells from naive CD4+ T cells, as the percentage of T-bet- and Eomes-positive cells within memory CD4+ T cells was not affected by its ablation (Supplementary Figure 5). Together, these results confirm a key role for Sam68 in the Th1 cell differentiation program.
Sam68 regulates T-bet and Eomes during Th1 cell differentiation. a, b RT-PCR analysis of the expression of T-bet (a) and Eomes (b) mRNA in naive CD4+ T cells, from wt and Sam68-ko mice, differentiated in vitro for 7 days in the presence of anti-CD3 plus anti-CD28 and polarizing cytokines. Cycling threshold values are normalized to those of mRNA encoding ribosomal protein L-34. Data are the mean and s.e.m. of six independent experiments. c, d Flow cytometry of intracellular T-bet (c), Eomes (d), and IFN-γ in naive T cells, from wt and Sam68-ko mice, stimulated in the presence of anti-CD3 plus anti-CD28 and polarizing cytokines for 7 days, assessed after 5 h of restimulation with PMA and Ionomycin in the presence of Brefeldin-A. One representative and cumulative data from 11n independent experiments are reported. Th0: no polarizing cytokines; Th1: IL-12; *p < 0.05 **p < 0.01; ***p < 0.005(Student’s t-test)
Ablation of Sam68 function impairs the Th1 response to BCG infection
The impact of Sam68 expression on Th1 polarization in vivo was further assessed upon infection by BCG, an intracellular bacteria that induces a Th1-mediated immune response [33]. Sam68 wild-type and knockout mice were infected by intravenous BCG injection and the immune response was evaluated 28 days afterwards [7]. Since IFN-γ producing cells from unstimulated splenocytes were barely detected in all conditions (Fig. 3a, b), we challenged splenocytes from BCG-infected mice, or phosphate-buffered saline (PBS) control mice, with agents that stimulate all T cells (i.e., anti-CD3/28 and phorbol 12-myristate 13-acetate (PMA)) and with the purified protein derivative of M. tuberculosis (PPD), which selectively stimulates BCG-specific T cells (Fig. 3a). Stimulation was assessed as the percentage of IFN-γ-producing cells detected by intracellular staining (Fig. 3b) and by measuring the levels of IFN-γ in the culture supernatants (Fig. 3c). As previously reported [7], CD4+ T cells from the spleen of BCG-infected wild-type mice significantly expressed more IFN- γ than cells from uninfected mice. Remarkably, this induction was largely ablated in Sam68-ko mice (Fig. 3a–c), indicating a key requirement for Sam68 in this response.
Th1 immune response induced by BCG in vivo is affected in Sam68-ko mice. a Flow cytometry analysis of IFN-γ-producing CD4+ T cells in splenocytes from wt and Sam68-ko mice treated with PBS or infected with BCG and, 28 days later, restimulated with anti-CD3-28, PMA and Ionomycin, or PPD for 5 h in the presence of Brefeldin-A. b Bar graphs show data from seven independent experiments performed as in a (mean and s.e.m). c ELISA of IFN-γ in supernatants of splenocytes wt and Sam68-ko mice treated with PBS or infected with BCG and, 28 days later, restimulated for 24 h with PMA and Ionomycin, or PPD. Data are the mean and s.e.m. of 11 independent experiments. d BCG burden in lungs of wt and Sam68-ko mice 28 days after intravenous infection of BCG. Data are expressed as CFU/g of tissue (mean and s.e.m. of eight independent experiments); *p < 0.05; **p < 0.01; ***p < 0.005 (Student’s t-test)
Th1 responses promote host resistance to infection by intracellular bacteria. Thus, we investigated whether Sam68-ko mice were more susceptible to BCG infection. On day 28 after intravenous injection, bacterial burden was analyzed by measuring colony-forming live bacteria extracted from lungs [7]. Consistent with the lower IFN-γ response, we found more pulmonary BCG colony-forming units in Sam68-ko mice than in wild-type mice (Fig. 3d), indicating the pathological relevance of the impaired Th1 response in vivo.
To investigate quantitatively and qualitatively the global T helper cytokine profile regulated by Sam68 during BCG infection, we then analyzed a broad array of Th cytokines produced by PPD-specific lymphocytes. The reliability of the multiparametric technology in this experimental setting was first validated by correlating the levels of IFN-γ (Fig. 4a, b) with those detected by conventional ELISA (Fig. 3c). Notably, the multiparametric analysis also revealed that BCG infection induces IL-2, TNF-α, IL-6 and low levels of IL-10 in wild-type mice and that induction of all these cytokines is reduced in Sam68-ko mice (Fig. 4c). By contrast, IL-17, IL-21, IL-22, GM-CSF, IL-4 and IL-13 expression was not induced upon infection and was not affected by Sam68 ablation (Fig. 4c). These results strongly indicate that Sam68 plays a key role in the orchestration of a global Th1-specific response to BCG infection.
Sam68 regulates a broad immune response induced against BCG infection. a IFN-γ analyzed by multiplex assay in supernatants of splenocytes wt and Sam68-ko mice treated with PBS or infected with BCG and, 28 days later, restimulated, or not, for 24 h with PMA and Ionomycin, or PPD. Data are the mean and s.e.m. of four independent experiments. b Pearson’s correlation analysis between the levels of IFN-γ determined by ELISA and the levels of IFN-γ determined by multiplex assay (Luminex). c Analysis of IL-2, IL-4, IL-5, IL-6, IL-10, IL-13, IL-17, IL-21, IL-22, TNF-α, GM-CSF and IFN-γ by multiplex assay in supernatants of splenocytes wt and Sam68-ko mice treated with PBS or infected with BCG and, 28 days later, restimulated for 24 h with PPD. Data are the mean and s.e.m. of four independent experiments; *p < 0.05; **p < 0.01; (two-way ANOVA test)
Sam68 regulates miR-29 expression during BCG infection
Expression of IFN- γ, T-bet and Eomes is kept under repression by miR-29a/b in T cells and this repression is partially relieved upon polarization into the Th1 profile [7, 8]. Previous evidence indicated that Sam68 modulates expression of selected microRNAs, including miR-29 [12]. Thus, we asked whether Sam68 is involved in the regulation of miR-29 expression during Th1 polarization. To address this question, we analyzed miR-29a and miR-29b expression in splenocytes from BCG-infected and uninfected mice. As expected, miR-29a and miR-29b were significantly downregulated in T cells of wild-type mice after BCG infection. By contrast, this regulation was ablated in Sam68-ko mice (Fig. 5a), suggesting that Sam68 function is required for repression of miR-29 expression during Th1 polarization. We also tested whether other miRNAs involved in mycobacterial infection control (i.e., miR-155, miR-125b-5p and miR-337-3p) [34] were affected. However, while miR-337-3p was not detected, the other two miRNAs were neither modulated by BCG infection nor by Sam68 expression under our experimental conditions (Supplementary Figure 6a). Likewise, long noncoding RNAs (lncRNAs) previously shown to be regulated by Sam68 [35] were either not expressed in splenocytes (Hotair and SR-lncRNA-2) or not significantly modulated by BCG infection or Sam68 expression (SR-LncRNA-1 and Mir155hg; Supplementary Figure 6b). Furthermore, we also tested expression of Tet3, a recently described target of miR-29b in neurons [36]. Although Tet3 expression is slightly upregulated during BCG infection, this trend was not significant and it was also observed in Sam68-ko cells. Thus, the effect of Sam68 on the BCG-mediated modulation of miR-29a/b appears specific.
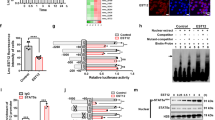
Sam68 regulates miR-29 expression during BCG infection. a Expression of miR-29a (left panel) and miR-29b (right panel) in Sam68 wt and ko mice upon treatment with PBS (white bars) and BCG (black bars). Histograms represent RT-qPCR analysis from 6 independent experiments (mean and s.e.m). MiR-29 expression was normalized to U6 snRNA. b CLIP assay was performed to monitor Sam68 binding to miR-29 precursors. Splenocytes were UV crosslinked and immunoprecipitated with anti-Sam68 IgGs. The bar graph shows qPCR signals amplified from the CLIP assays expressed as percentage of amplification from the input RNA. PBS condition is expressed in white, BCG condition is expressed in black, control CLIP from Sam68-ko splenocytes is expressed in gray. Histograms represent RT-qPCR analysis from 6 independent experiments (mean and s.e.m.). c Schematic representation of Sam68 regulation of miR-29 expression upon BCG infection in mouse splenocytes. On the left, Sam68 inhibits miR-29 expression during BCG infection by directly binding to miR-29 precursors (pre-miR). This inhibition is necessary to allow the expression of Tbet and Eomes, which in turn drive Ifng transcription and clearance of infectious BCG. On the right, in the absence of Sam68 (Sam68-ko mice), miR-29 expression is not downregulated upon BCG infection and, consequently, Tbet, Eomes and Ifng transcripts can be targeted and inhibited by miR-29, and bacteria growth is not controlled; *p < 0.05; **p < 0.01 (Student’s t-test)
To investigate whether miR-29a and miR-29b are directly regulated by Sam68 in T cells, we performed ultraviolet light cross-link immunoprecipitation (CLIP) experiments to covalently link RBPs to their cellular targets in live cells [37, 38]. CLIP assays revealed that binding of Sam68 to miR-29a and miR-29b precursors is significantly increased in wild-type splenocytes upon BCG infection (Fig. 5b). Notably, the low levels of binding in splenocytes from PBS-injected wild-type mice were comparable to those detected in Sam68-ko splenocytes, indicating that Sam68 is specifically recruited to its target pre-miRs only during BCG infection. These results indicate that Sam68 directly binds to miR-29a/b, or their precursors, in T cells from infected mice, thus exerting an inhibitory role on their expression and/or processing (Fig. 5c).
Discussion
Our study uncovers a novel layer in the Th1 differentiation program by identifying Sam68 as a key regulator of miR-29 expression. The lack of Sam68 significantly impaired IFN-γ production by Th1 cells polarized from naive CD4+ T cells in the presence of IL-12, or by CD4+ T cells upon in vivo infection with BCG, a potent inducer of IL-12 production. These results indicate that Sam68 is involved in IL-12-mediated IFN-γ induction. However, Sam68 expression did not affect IFN-γ production by unstimulated memory CD4+ T lymphocytes and, similarly, no differences were observed in the basal production of IFN-γ by Th0, or memory T lymphocytes obtained from PBS-injected mice. This could be explained by the evidence that, under a basal condition, low production of IFN-γ is provided by a default Th1-type action that occurs independently on the IL-12 pathway, unlike the functional Th1 response [39].
The production of IFN-γ by Th1 cells is regulated by the transcription factors T-bet and Eomes. Notably, expression of both these Th1-related transcription factors, as well as that of IFN-γ itself, are negatively regulated by miR-29 [7, 8]. Such repression is relieved upon bacterial infections through inhibition of miR-29 expression [7]. However, which factor(s) is involved in Th1-specific reduction of miR-29 expression has remained unknown. Herein, we propose Sam68 as a key player in this response. Indeed, Sam68 was specifically recruited to miR-29a/b pre-miR upon BCG infection and ablation of its function in vivo abolished the reduction in miR-29a/b expression in differentiating Th1 cells. Thus, our study highlights a novel role for Sam68 in Th1-mediated immune responses and suggests a model where binding of Sam68 to miR-29 precursors during BCG infection interferes with their maturation, thus lowering miR-29a/b expression levels and relieving repression of IFN-γ, T-bet and Eomes transcripts to warrant efficient differentiation of Th1 cells and control of infection (Fig. 5c). Interestingly, the immune response triggered by BCG infection reveals that Sam68 regulates not only production of IFN-γ by Th1 cells, but also of other pro-inflammatory cytokines like IL-6, TNF-α, IL-2 [1, 2] as well as factors controlling the inflammation, such as IL-10 [40]. Thus, Sam68 orchestrates the global Th1 response in vivo and this mechanism is crucial for the clearance of infectious bacteria (Fig. 5c). Moreover, since miR-29 was also shown to inhibit IFN-γ induction in innate immune cells during Listeria monocytogenes infection [9], Sam68 may be more generally involved in the immune responses against bacteria. It is also important to note that Sam68 is directly involved in infections from enterovirus, foot-and-mouth disease virus (FMDV) and HIV virus by promoting their replication cycle [41,42,43]. Nevertheless, since the immune response to these pathogens was not investigated, future studies using other pathogenic agents will be required to test this hypothesis.
MiRNAs are small, endogenous RNAs that regulate gene expression, including genes involved in several immune processes [44]. The expression of miRNAs is regulated at several stages during their biogenesis, including the interaction with the enzymes Drosha and Dicer, responsible of the conversion of pri-miRNA in pre-miRNA, and pre-miRNA in mature miRNA, respectively [44, 45]. Sam68 was reported to regulate miRNA expression in male germ cells, including miR-29b, and to interact with Drosha and Dicer [12]. Furthermore, Sam68 was recently confirmed to regulate miR-29b biogenesis in mouse neuroblastoma cells [36]. Our findings now indicate that Sam68 is directly involved in miR-29 regulation in T lymphocytes, thus contributing to the control of pathogenic infections. These observations suggest a conserved function for Sam68 in the biogenesis of this physiologically important microRNA. Notably, lack of Sam68 was reported to strongly suppress differentiation of neural progenitor cells that express Eomes (i.e., also named Tbr2) during cortical neurogenesis [22]. Although the implication of miR-29 expression in this process was not investigated, our findings suggest that Sam68 may utilize common mechanisms, such as the miR-29/Eomes axis, to modulate gene expression during multiple cell differentiation programs. In this scenario, it will be of interest to dissect the specific molecular mechanism by which Sam68 regulates miR-29 expression in future studies.
Regulation of miRNA function represents a flexible system particularly relevant for the Th cell differentiation process, where a fine-tuned modulation of cell diversity and plasticity is required [46]. Plasticity of Th1 cells is essential during the immune response against intracellular bacteria such as mycobacteria [47]. Th1 cells produce high levels of IFN-γ, thus inducing a wide array of molecules that stimulate macrophages [48] and activate cytotoxic properties of CD8+ T cells until complete clearance of the pathogens is fulfilled [49,50,51,52]. Notably, Sam68-ko mice display high lethality soon after birth, in the absence of overt phenotypes [17]. Thus, it is possible that such perinatal weakness results from predisposition to infection and defective immune responses to pathogens and future studies will be aimed at investigating this possibility. Likewise, Sam68 could also regulate chronic inflammation and autoimmunity, where a persistent or uncontrolled Th1 immune response causes the pathological state [2]. Moreover, given the broad spectrum of diseases in which miR-29 is involved, including fibrosis [53], HIV latency [54], leukemia [55] and multiple sclerosis [56], fine-tuning of the Sam68/miR-29 axis may also have implications in the immune responses during these pathologies.
Materials and methods
Mice
C57BL/6 Sam68-ko mice were generated by replacing exon 4 and part of exon 5 with a neomycin-resistant gene cassette as previously characterized [16]. Breeding, maintenance and animal procedures were conducted as described in the project authorized by Ministry of Health (protocol number 510/2017-PR), in accordance with institutional guidelines of the Interdepartmental Service Centre–Station for Animal Technology, University of Tor Vergata, and of the Fondazione Santa Lucia, and in accordance with national and international laws and policies (Directive 2010/63/EU of the European Parliament and of the Council, Italian Legislative Decree 26/2014). Genomic DNA was isolated from the tail, amplified by PCR (Fwd GATATGATGGATGATATCTGTCAG; Rev AAATCCTAACCCTCCTCAGTCAG) and genotyped as described previously [57]. Female animals at 6–8 weeks of age were used for the experiments.
Preparation of cells
For differentiation of T helper cells, naive CD4+ T cells were sorted from the spleens of mice with high sped cell sorter MoFlo (Coulter) for CD44 low and CD62L high CD4+ T cells; memory CD4+ T cells were sorted as CD44 high and CD62L low CD4+ T cells; cell purity was >97%. Then, cells were cultured in Iscove's modified Dulbecco's medium (Life Technologies), and stimulated with beads anti-CD3/28 (Life Technologies) (ratio 2 beads/1 cell) in the presence of polarizing cytokines as follows: IL-12 (5 ng/ml) (Miltenyi) for Th1 cells; anti-IFN-γ (10μγ/ml) (eBioscience), IL-4 (10 ng/ml) (Miltenyi) for Th2 cells; IL-6 (25 ng/ml), tTransforming growth factor (TGF)-β (2.5 ng/ml) (Miltenyi), anti-IL-4, anti-IFN-γ, anti-IL-2 (10 μg/ml) (eBioscience) for Th17 cells. After 7 days of differentiation, T cells were washed and restimulated for 5 h with PMA and ionomycin (both 500 ng/ml) (both from Sigma) in the presence of Brefeldin-A (1 μg/ml) (Sigma) for the last 4 h of incubation. For analysis of Th1 response in vivo by intracellular staining, splenocytes from infected and control mice, 28 days later, were restimulated for 5 h with PMA and ionomycin (both 500 ng/ml) (both Sigma), purified protein derivative (PPD) (300IU/ml) (Life Technologies), or anti-CD3/28 (ratio 2 beads/1 cell) (Life Technologies) in presence of Brefeldin-A (1μg/ml) (Sigma) for the last 4 h of incubation. For analysis of Th1 response in vivo by ELISA, splenocytes or total CD4 T cells sorted by high sped cell sorter MoFlo (Coulter) were restimulated for 5 h with PMA and ionomycin (both 200 ng/ml) (both from Sigma), or anti-CD3/28 (ratio 2 beads/1 cell) (Life Technologies) for 24 h. Splenocytes were also restimulated with PPD (300 IU/ml) (Life Technologies).
Flow cytometry and intracellular cytokine staining
All fluorescein-conjugated monoclonal antibodies were from BD Pharmingen or Miltenyi. For intracellular cytokine staining (IFN-γ, IL-4, T-bet and Eomes), cells were stained in the Foxp3 buffer kit according to the manufacture’s instructions (eBioscience).
Real-time quantitative PCR
Total RNA was extracted by RNeasy Microkit (Qiagen). A mix containing random hexamers, Oligo dT15m (Promega) and SuperScript II Reverse Transcriptase (Life Technologies) were used for complementary DNA (cDNA) synthesis. Transcripts were quantified by real-time quantitative PCR (RT-PCR) on an LC480 (Roche) with Applied Biosystems predesigned TaqMan Gene Expression Assays and Taqman Gene expression Master Mix (Life Technologies). For transcriptional analysis the following probes were used (identified by Applied Biosystems assay identification number): Mm00504390_m1(RPL34); Mm00434189_m1 (IL12RB1); Mm00434200_m1 (IL12RB2); Mm00599890_m1 (IFNGR1); Mm00492626_m1 (IFNGR2); Mm00450960_m1 (TBX21); Mm01168134_m1 (IFNG); Mm01351985_m1 (EOMES). For each sample, mRNA abundance was normalized to the amounts of Ribosomal Protein L-34 (RPL34).
For miRNA detection, total RNA was extracted using Trizol (Life Technologies) according to the manufacturer’s instructions and subjected to DNase digestion (Roche). For miRNA expression analysis TaqMan method was employed. Briefly, 10 ng of total RNA was reverse transcribed using TaqMan miRNA reverse transcription kit (Applied Biosystems, 4366596) following the manufacturer’s instructions. Then, 1.3 µl of each miR-specific cDNA was submitted to PCR amplification using Taqman universal PCR master mix II (Applied Biosystems, 4440044). The following TaqMan miRNA assays were used as probes: hsa-miR-29a (002112), hsa-miR-29b (0004139) and U6 small nuclear RNA (snRNA; 001973). The comparative cycle threshold (Δ ΔCt) method was used to analyze the relative expression levels using U6 snRNA as internal controls.
The oligonucleotides used to detect the binding of Sam68 to miR-29 precursors originated from chromosome 6 (Chr6) and chromosome 1 (Chr1) loci were: Fw premiR29a-Chr6 accccttagaggatgactgat, Rv premiR29a-Chr6 aaccgatttcagatggtgct; Fw premiR29b1Chr6 ggaagctggtttcatatggtgg, Rv premiR29b1-Chr6 agaacactgatttcaaatggtgct; Fw premiR29b2-Chr1 tggaagctggtttcacatggt, Rv premiR29b2-Chr1 aaaacactgatttcaaatggtgct.
Cytokine quantification
IFN-γ in culture supernatant was quantified with an ELISA kit (R&D Systems). Other cytokines were quantified using the Mouse High Sensitivity T Cell Magnetic Bead Panel (IL-2, IL-4, IL-5, IL-6, IL-10, IL-13, IL-17, TNF-α, GM-CSF, IFN-γ), and Mouse Th17 Magnetic Bead Panel (IL-21, IL-22) (Millipore), following the manufacturer’s protocol, and analyzed by Luminex.
Bacterial culture, infection and quantification
Mycobacterium bovis BCG, Pasteur substrain (TMC1011), was grown in Middlebrook 7H9 (Difco) broth supplemented with 10% albumin, dextrose and catalase (ADC), and 0.05% Tween-80. Bacteria were then aliquoted in equivalent medium containing 10% glycerol and stored at −80 °C, until use [58]. Mice were inoculated intravenously with 5 × 106 colony-forming units of BCG. At 28 days, bacterial burden was assessed by homogenization of the organs in 0.05% (vol/vol) Tween-80, followed by plating serial dilutions onto Middlebrook 7H10 agar plates supplemented with 10% (vol/vol) oleic acid–OADC (Difco) and 0.05% /(vol/vol) glycerol. Bacterial concentration was measured after incubation of plates for 3 weeks at 37 °C and 5% CO2.
CLIP assay
CLIP assays were performed as previously described [37]. In brief, splenocytes were irradiated once with 400 mJ/cm2 in a Stratlinker 2400 at 254 nm. Cell suspension was centrifuged at 4000 rpm for 3 min, and the pellet was incubated for 10 min on ice in lysis buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 1% Igepal CA-630 (Sigma-Aldrich I8896), 0.1% SDS, 0.5% sodium deoxycholate, 0.5 mM Na3VO4, 1 mM dithiothreitol, protease inhibitor cocktail (Sigma-Aldrich), and RNase inhibitor (Promega)). Samples were briefly sonicated and incubated with 10 μl of 1:1000 RNase I (Ambion, AM2295) dilution and 2 μl Turbo DNase (Ambion, AM2238) for 3 min at 37 °C shaking at 1100 rpm, and then centrifuged at 15,000 × g for 10 min at 4 °C. Then, 1 mg of extract was immunoprecipitated using anti-Sam68 (Santa Cruz Biotechnology, Inc.) antibody or purified IgGs (negative control) in the presence of protein A magnetic Dynabeads (Life Technologies). Immunoprecipitates were incubated overnight at 4 °C under constant rotation. After stringent washes with high salt buffer (50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% Igepal CA-630 (Sigma I8896), 0.1% SDS, 0.5% sodium deoxycholate), beads were equilibrated with PK buffer (100 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10 mM EDTA). An aliquot (10%) was kept as a control of immunoprecipitation, while the rest was treated with 50 µg proteinase K and incubated for 20 min at 37 °C shaking at 1100 rpm. Then, 7 M urea was added to the PK buffer and incubation was performed for further 20 min at 37 °C and 1100 rpm. The solution was collected and phenol/CHCl3 (Ambion, 9722) was added. After incubation for 5 min at 30 °C shaking at 1100 rpm phases were separated by spinning for 5 min at 13,000 rpm at room temperature. The aqueous layer was transferred into a new tube and precipitated by addition of 0.5 μl glycoblue (Ambion, 9510), 3 M sodium acetate pH 5.5 and 100% ethanol. After mixing, the solution containing retained RNA was precipitated overnight at −20 °C. Purified RNA was reverse transcribed by SuperScript III Reverse Transcriptase (Life Technologies) and the obtained cDNA used for quantitative PCR (qPCR) analysis.
Statistics
A two-tailed Student's t-test was used for statistical comparison of two groups. Two-way analysis of variance (ANOVA) was performed to analyze multiple variables. Pearson's correlation coefficient was used to assess the significance of correlation among IFN-γ analysed by ELISA and IFN-γ analyzed by Luminex; *p < 0.05, **p < 0.01, ***p < 0.001.
References
Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–89.
O’Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 1998;8:275–83.
Anthony RM, Rutitzky LI, Urban JF Jr, Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat Rev Immunol. 2007;7:975–87.
Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41.
Volpe E, Servant N, Zollinger R, Bogiatzi SI, Hupe P, Barillot E, et al. A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nat Immunol. 2008;9:650–7.
Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–82.
Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–9.
Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–81.
Daugaard I, Hansen TB. Biogenesis and function of ago-associated RNAs. Trends Genet. 2017;33:208–19.
Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell . 2011;147:1080–91.
Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–8.
Messina V, Meikar O, Paronetto MP, Calabretta S, Geremia R, Kotaja N, et al. The RNA binding protein SAM68 transiently localizes in the chromatoid body of male germ cells and influences expression of select microRNAs. PLoS One. 2012;7:e39729.
Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86.
Sette C. Post-translational regulation of star proteins and effects on their biological functions. Adv Exp Med Biol. 2010;693:54–66.
Frisone P, Pradella D, Di Matteo A, Belloni E, Ghigna C, Paronetto MP. SAM68: signal transduction and RNA metabolism in human cancer. Biomed Res Int. 2015;2015:528954.
Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74.
Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–49.
Bianchi E, Barbagallo F, Valeri C, Geremia R, Salustri A, De Felici M, et al. Ablation of the Sam68 gene impairs female fertility and gonadotropin-dependent follicle development. Hum Mol Genet. 2010;19:4886–94.
Huot ME, Vogel G, Zabarauskas A, Ngo CT, Coulombe-Huntington J, Majewski J, et al. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol Cell. 2012;46:187–99.
Klein ME, Younts TJ, Castillo PE, Jordan BA. RNA-binding protein Sam68 controls synapse number and local beta-actin mRNA metabolism in dendrites. Proc Natl Acad Sci USA. 2013;110:3125–30.
Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–14.
La Rosa P, Bielli P, Compagnucci C, Cesari E, Volpe E, Farioli Vecchioli S, et al. Sam68 promotes self-renewal and glycolytic metabolism in mouse neural progenitor cells by modulating Aldh1a3 pre-mRNA 3’-end processing. Elife. 2016;5.
Paronetto MP, Cappellari M, Busa R, Pedrotti S, Vitali R, Comstock C, et al. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70:229–39.
Fusaki N, Iwamatsu A, Iwashima M, Fujisawa J. Interaction between Sam68 and Src family tyrosine kinases, Fyn and Lck, in T cell receptor signaling. J Biol Chem. 1997;272:6214–9.
Lang V, Mege D, Semichon M, Gary-Gouy H, Bismuth G. A dual participation of ZAP-70 and scr protein tyrosine kinases is required for TCR-induced tyrosine phosphorylation of Sam68 in Jurkat T cells. Eur J Immunol. 1997;27:3360–7.
Ramakrishnan P, Baltimore D. Sam68 is required for both NF-kappaB activation and apoptosis signaling by the TNF receptor. Mol Cell. 2011;43:167–79.
Fu K, Sun X, Zheng W, Wier EM, Hodgson A, Tran DQ, et al. Sam68 modulates the promoter specificity of NF-kappaB and mediates expression of CD25 in activated T cells. Nat Commun. 2013;4:1909.
Waldmann TA. The structure, function, and expression of interleukin-2 receptors on normal and malignant lymphocytes. Science. 1986;232:727–32.
Bradley LM, Dalton DK, Croft M. A direct role for IFN-gamma in regulation of Th1 cell development. J Immunol. 1996;157:1350–8.
Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell . 2000;100:655–69.
Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–3.
Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol. 2006;177:3721–7.
Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101.
Yang T, Ge B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett. 2018;431:22–30.
Li N, Hebert S, Song J, Kleinman CL, Richard S. Transcriptome profiling in preadipocytes identifies long noncoding RNAs as Sam68 targets. Oncotarget. 2017;8:81994–2005.
Kremer EA, Gaur N, Lee MA, Engmann O, Bohacek J, Mansuy IM. Interplay between TETs and microRNAs in the adult brain for memory formation. Sci Rep. 2018;8:1678.
Paronetto MP, Bernardis I, Volpe E, Bechara E, Sebestyen E, Eyras E, et al. Regulation of FAS exon definition and apoptosis by the Ewing sarcoma protein. Cell Rep. 2014;7:1211–26.
Bielli P, Sette C. Analysis of in vivo interaction between RNA binding proteins and their RNA targets by UV cross-linking and immunoprecipitation (CLIP) method. Bio Protoc. 2017 May 20;7(10).
Matzinger P, Kamala T. Tissue-based class control: the other side of tolerance. Nat Rev Immunol. 2011;11:221–30.
Trinchieri G. Interleukin-10 production by effector T cells: Th1 cells show self control. J Exp Med. 2007;204:239–43.
Zhang H, Song L, Cong H, Tien P. Nuclear protein Sam68 interacts with the enterovirus 71 internal ribosome entry site and positively regulates viral protein translation. J Virol. 2015;89:10031–43.
Lawrence P, Schafer EA, Rieder E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology. 2012;425:40–52.
Henao-Mejia J, Liu Y, Park IW, Zhang J, Sanford J, He JJ. Suppression of HIV-1 Nef translation by Sam68 mutant-induced stress granules and nef mRNA sequestration. Mol Cell. 2009;33:87–96.
Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–78.
Subramanyam D, Blelloch R. From microRNAs to targets: pathway discovery in cell fate transitions. Curr Opin Genet Dev. 2011;21:498–503.
Jeker LT, Bluestone JA. MicroRNA regulation of T-cell differentiation and function. Immunol Rev. 2013;253:65–81.
Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–52.
Adams DO. Molecular interactions in macrophage activation. Immunol Today. 1989;10:33–5.
Bevan MJ. Helping the CD8(+) T-cell response. Nat Rev Immunol. 2004;4:595–602.
Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–6.
Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–9.
Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–42.
van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32.
Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709.
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14.
Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567–76.
Pagliarini V, Pelosi L, Bustamante MB, Nobili A, Berardinelli MG, D’Amelio M, et al. SAM68 is a physiological regulator of SMN2 splicing in spinal muscular atrophy. J Cell Biol. 2015;211:77–90.
Poerio N, Bugli F, Taus F, Santucci MB, Rodolfo C, Cecconi F, et al. Liposomes loaded with bioactive lipids enhance antibacterial innate immunity irrespective of drug resistance. Sci Rep. 2017;7:45120.
Acknowledgements
We thank Vassili Soumelis for helpful suggestions and critical reading of the manuscript. This work was supported by grants from FISM-Fondazione Italiana Sclerosi Multipla- cod2016/R/31 to EV and cod2017/R/24 to CS; Associazione Italiana Ricerca sul Cancro (AIRC) IG 18790 to CS and IG 17278 to MPP. NM was supported by a scholarship from Fondazione Umberto Veronesi.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by Y. Shi
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Volpe, E., Cesari, E., Mercatelli, N. et al. The RNA binding protein Sam68 controls T helper 1 differentiation and anti-mycobacterial response through modulation of miR-29. Cell Death Differ 26, 1169–1180 (2019). https://doi.org/10.1038/s41418-018-0201-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0201-9
This article is cited by
-
Role of Sam68 as an adaptor protein in inflammatory signaling
Cellular and Molecular Life Sciences (2024)
-
Is hydroxychloroquine beneficial for COVID-19 patients?
Cell Death & Disease (2020)
-
BCG vaccination policy and preventive chloroquine usage: do they have an impact on COVID-19 pandemic?
Cell Death & Disease (2020)