Abstract
Long noncoding RNAs (lncRNAs) have been implicated in the pathogenesis of intracellular pathogens. However, the role and mechanism of the important lncRNAs in Mycobacterium tuberculosis (M.tb) infection remain largely unexplored. Recently, we found that a secreted M.tb Rv1579c (an early secreted target with a molecular weight of 12 kDa, named EST12) protein activates NLRP3-gasdermin D (GSDMD)-mediated pyroptosis and plays a pivotal role in M.tb-induced immunity. In the present study, M.tb and the EST12 protein negatively regulated the expression of a key lncRNA (named lnc-EST12) in mouse macrophages by activating the JAK2-STAT5a signaling pathway. Lnc-EST12, with a size of 1583 bp, is mainly expressed in immune-related organs (liver, lung and spleen). Lnc-EST12 not only reduces the expression of the proinflammatory cytokines IL-1β, IL-6, and CCL5/8 but also suppresses the NLRP3 inflammasome and GSDMD pyroptosis-IL-1β immune pathway through its interaction with the transcription factor far upstream element-binding protein 3 (FUBP3). The KH3 and KH4 domains of FUBP3 are the critical sites for binding to lnc-EST12. Deficiency of mouse lnc-EST12 or FUBP3 in macrophages increased M.tb clearance and inflammation in mouse macrophages or mice. In conclusion, we report a new immunoregulatory mechanism in which mouse lnc-EST12 negatively regulates anti-M.tb innate immunity through FUBP3.
Similar content being viewed by others
Introduction
Mycobacterium tuberculosis (M.tb) is the etiological agent of tuberculosis (TB), and TB remains a severe disease in humans worldwide. According to the World Health Organization Global Tuberculosis Report 2021, ~5.8 million new patients were diagnosed with TB, and ~1.51 million patients with TB died worldwide in 2020; additionally, the COVID-19 pandemic has aggravated the severity of TB [1]. A vaccine based on an attenuated live strain of Mycobacterium bovis, termed Bacillus Calmette-Guerin (BCG), is the only available TB vaccine and has limited protective efficacy in adults [2]. Early-stage disease, extrapulmonary TB, M.tb/human immunodeficiency virus (HIV) coinfection, childhood TB, and multidrug-resistant TB are particularly problematic to diagnose and treat [1, 3]. Therefore, understanding the mechanism of M.tb infection and survival in the host has significant benefits for the prevention and control of TB and the development of new drugs. Macrophages are the primary target host cells of M.tb and play a central role in mycobacterial pathogenesis. Pathogenic mycobacteria have achieved broad evolutionary success through specialization as intracellular pathogens capable of achieving persistent infection by manipulating host macrophages to reside and replicate within them rather than being completely eradicated by innate defense mechanisms [4,5,6].
Regions of deletion (RD) are certain segments missing in the BCG genome relative to M.tb H37Rv or M. bovis [7, 8]. The BCG genome lacks 129 open reading frames from RD1 to RD16, of which RD1-2 regions are the most studied. Recently, studies of the RD1 region (e.g., RD1-encoding ESAT-6 and CFP-10) have revealed many genes that exert significant immunogenic and virulent effects on TB [9]. We recently found that the EST12 (Rv1579c) gene is located in RD3 and is present in both M.tb H37Rv and H37Ra. EST12, a secreted protein, activates NLRP3-gasdermin D (GSDMD)-mediated pyroptosis and plays a pivotal role in M.tb-induced immunity [10].
Long noncoding RNAs (lncRNAs) are a large class of nonprotein-coding transcripts with lengths longer than 200 nucleotides [11] that are involved in many biological processes [12,13,14,15,16,17]. To date, lncRNAs have been revealed to play important roles in regulating gene expression at the epigenetic, transcriptional, and posttranscriptional levels [18,19,20]. For example, lncRNAs participate in immune responses by inhibiting the inflammatory process in macrophages and mice [21], suppressing the activation of abnormal inflammation, and regulating macrophage polarization [22]. LncRNAs exhibit tissue specific, cell-specific or even compartment-specific expression patterns. According to the relative location of lncRNAs to protein-encoding genes in the genome, they have been classified into sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs, intergenic lncRNAs, and enhancer lncRNAs [11, 23]. Although many lncRNAs have been reported to be involved in a variety of diseases, few lncRNAs closely related to specific factors in M.tb have been discovered.
In the present study, we identified a key lncRNA, lnc-EST12, expressed in mouse macrophages that is negatively regulated by M.tb EST12/M.tb. Host lnc-EST12 negatively regulates anti-M.tb innate immunity by suppressing the production of the proinflammatory cytokines IL-1β, IL-6, and CCL5/8 and NLRP3-GSDMD-mediated pyroptosis through the transcription factor far upstream element-binding protein (FUBP).
Results
Identification of lnc-EST12, a key lncRNA that is negatively regulated by M.tb EST12/M.tb in mouse macrophages
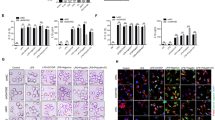
Recently, we found that EST12 (Rv1579c; MW: 12 kDa), a secreted protein of M.tb, activated an NF-κb/AP-1-NLRP3-GSDMD pyroptosis-IL-1β immune pathway in macrophages. We performed a microarray analysis in macrophages to further explore lncRNAs that are involved in M.tb infection. Purified recombinant EST12 protein was used as a stimulant to treat RAW264.7 macrophages for 0, 3, and 24 h (Fig. 1a and Supplementary Fig. 1a). Among the altered lncRNAs, lnc-EST12 (Agilent Mouse lncRNA Microarray Probe No. mmu_8976) was found to be the most downregulated lncRNA after EST12 treatment for 24 h in macrophages (Fig. 1b). In addition, we confirmed that EST12 inhibited the expression of lnc-EST12 in a dose- and time-dependent manner using RT-qPCR (Fig. 1c, d). Fluorescence in situ hybridization (FISH) assays also showed that lnc-EST12 expression decreased in macrophages after treatment with EST12 protein (Fig. 1e, f). We observed that lnc-EST12 mainly accumulated in the cytoplasm, and a small amount was observed in the nucleus (Fig. 1e). Interestingly, lnc-EST12 expression was also decreased in bone marrow-derived macrophages (BMDMs) infected with various mycobacterial strains (bacteria: BMDMs = 10:1, 6 h) (decreased ~70% by H37Rv or H37RvΔEST12::EST12, ~60% by H37RvΔEST12, ~66% by H37Ra strains, ~40% by M. smegmatis and ~50% by M. smegmatis-EST12) (Supplementary Fig. 1b). H37RvΔEST12 and M. smegmatis, which do not encode the EST12 protein, still displayed decreased lnc-EST12 expression, indicating that other factors in these mycobacteria might also cause a decrease in lnc-EST12 expression in addition to EST12. Based on the above data, the EST12 protein of M.tb H37Rv remarkably suppresses lnc-EST12 expression in mouse macrophages.
Mycobacterial EST12 suppresses lnc-EST12 expression in macrophages. a Schematic diagram of the lncRNA screening process in RAW264.7 macrophages after mycobacterial EST12 protein stimulation. b Heatmap of lncRNA expression obtained from microarray data. c, d RT-qPCR analysis of lnc-EST12 expression in RAW264.7 cells treated with different concentrations of EST12 for 12 h (c) or with EST12 (2 μM) for different time points (d). e FISH analysis of RAW264.7 cells treated with EST12 or PBS for 12 h using a Cy3-labeled lnc-EST12 probe (red) and DAPI nuclear staining (blue). f Quantification of the lnc-EST12 fluorescence intensity in each cell in (e). g Dual luciferase reporter assay of the activity of the lnc-EST12 promoter. RAW264.7 cells were cotransfected with the lnc-EST12-luciferase reporter construct (with the lnc-EST12 promoter region −2260 to +80 or its truncations) and the internal control plasmid pRL-TK, followed by EST12 treatment (2 μM, 24 h). The relative luciferase activities are presented as a percentage of values relative to the control group. h EMSA for analysis of STAT5a binding to the lnc-EST12 promoter. The STAT5a complex is indicated by horizontal arrows. i ChIP–qPCR analysis of the binding of STAT5a to the lnc-EST12 promoter region. j Dual luciferase reporter assay of the activity of the lnc-EST12 promoter. RAW264.7 cells were cotransfected with the lnc-EST12-luciferase reporter construct (with the lnc-EST12 promoter region -1000 to +80 or its mutants) and the internal control plasmid pRL-TK, followed by EST12 treatment (2 μM, 24 h). k RT-qPCR analysis of lnc-EST12 expression in RAW264.7 cells transfected with sh-STAT5a or sh-scramble, followed by EST12 treatment, as indicated. l RT-qPCR analysis of lnc-EST12 expression in RAW264.7 cells treated with EST12 (2 μM), gandotinib (JAK2 inhibitor, 10 μM) or STAT5a-IN-1 (STAT5a inhibitor, 10 μM). m WB analysis of the levels of phosphorylated JAK2 and STAT5a in the cytoplasm and nucleus of RAW264.7 cells stimulated with EST12 (2 μM) for different durations. Data in (c, d, g, i–l) are presented as the means ± SD of three independent experiments. **p < 0.01; ***p < 0.001; ****p < 0.0001. ns not significant. Two-tailed unpaired Student’s t test was used to calculate statistical significance (for g, i, j, k)
EST12 negatively regulates lnc-EST12 expression by activating the JAK2-STAT5a signaling pathway
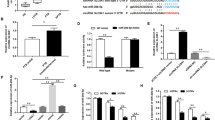
We next analyzed the mechanism underlying the downregulated expression of lnc-EST12 in EST12-stimulated mouse macrophages. We constructed the 2340 bp promoter region of lnc-EST12 and different deletion mutants in the luciferase reporter pGL3 to obtain pGL3-lnc-EST12/deletion-Luc plasmids and transfected them into RAW264.7 cells. As shown in Fig. 1g, the luciferase activity was significantly decreased in the truncations containing the −1000 to −500 bp promoter sequence upon stimulation with EST12 (Fig. 1g). Further bioinformatics analysis of this sequence (−1000 to −500 bp) revealed two independent signal transducer and activator transcription 5a (STAT5a) binding regions (gamma-activated sequence-GAS motif) in LASAGNA-Search 2.0 (uconn.edu).
We performed an EMSA with the corresponding 35-nucleotide DNA probe (Supplementary Table 3) containing the STAT5a transcription factor binding region to determine whether STAT5a bound the lnc-EST12 promoter. Nuclear protein extracts from RAW264.7 cells pretreated with EST12 showed stronger binding to the probe (Fig. 1h, Lane 6 vs. Lane 2) than those from untreated RAW264.7 cells. The binding was abrogated when the extract was pretreated with a 10-fold excess of the unlabeled ‘competitor’ probe (Fig. 1h, Lanes 3 and 7), but the mutant competitor containing substitutions within the STAT5a binding sequence (shown in Supplementary Table 3) did not abolish binding, providing additional evidence for its specificity (Fig. 1h, Lanes 4 and 8). Furthermore, using a chromatin immunoprecipitation (ChIP)-qPCR assay, we observed significantly increased STAT5a binding to the lnc-EST12 promoter in the EST12-treated group compared with the untreated control and the isotype IgG control groups (Fig. 1i). The aforementioned data indicated that STAT5a directly binds to the lnc-EST12 promoter after EST12 treatment.
Additionally, mutation of these STAT5a binding sites completely blocked the downregulation of lnc-EST12-luciferase expression induced by EST12 stimulation (Fig. 1j, Supplementary Fig. 1c), suggesting that EST12 negatively regulated lnc-EST12 expression through a specific interaction between the lnc-EST12 promoter and STAT5a. As expected, STAT5a silencing also abolished the downregulation of lnc-EST12 by EST12 (Fig. 1k and Supplementary Fig. 1d), suggesting that EST12 stimulates STAT5a binding to the lnc-EST12 promoter (Fig. 1h, i) and inhibits lnc-EST12 expression (Fig. 1k). In addition, in the presence of JAK2 or STAT5a inhibitors, the EST12-induced decrease in lnc-EST12 expression was abrogated (Fig. 1l).
Furthermore, we observed an increase in cytoplasmic JAK2 phosphorylation (p-JAK2 at Y1007) and nuclear STAT5a phosphorylation (p-STAT5a at Y694) and a decrease in cytoplasmic p-STAT5a levels by performing nuclear/cytosol separation assays and western blot (WB) analysis (Fig. 1m). However, cytoplasmic and nuclear STAT5b phosphorylation (p-STAT5b) levels showed no difference (Supplementary Fig. 1e), suggesting that EST12 stimulates cytoplasmic JAK2 phosphorylation and the translocation of p-STAT5a from the cytoplasm to the nucleus.
Based on these data, EST12 promotes the activation of the p-JAK2-p-STAT5a pathway and the translocation of p-STAT5a from the cytoplasm to the nucleus. STAT5a binds to the lnc-EST12 promoter to repress lnc-EST12 expression.
Characteristics and distribution of lnc-EST12
By analyzing the genome sequence, we determined that lnc-EST12 represents an intergenic lncRNA located between the Igdcc3 and Parp16 genes and on the antisense strand of mouse chromosome 9 (http://genome.ucsc.edu/cgi-bin/hgBlat) (Fig. 2a). An analysis of the tissue distribution of lnc-EST12 in mice showed that lnc-EST12 is expressed at high levels in the liver, followed by the lung and spleen (Fig. 2b). Lnc-EST12 is expressed at low levels in the kidney and brain and is expressed at very low levels in the heart and muscle (Fig. 2b), suggesting that lnc-EST12 is mainly expressed in immune-related organs (liver, lung and spleen). We also found that EST12 reduced lnc-EST12 expression in both mouse macrophages (including RAW264.7 cells and BMDMs) and DCs (bone marrow-derived dendritic cells, BMDCs) (Fig. 2c). According to the known lncRNA sequence, we determined the full-length sequence of lnc-EST12 to be 1583 bp in size by performing a rapid amplification of cloned cDNA ends (RACE) experiment (Fig. 2d and Supplementary Fig. 2a). Structural prediction analysis showed that full-length lnc-EST12 forms a unique spatial configuration (Supplementary Fig. 2b). A NCBI BLAST search of the human RNA-seq database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the lnc-EST12 sequence as the query displayed no homologous RNA sequence of human origin with significant similarity. Due to the species specificity of most lncRNAs, the homology between mouse lncRNAs and their corresponding human lncRNAs is very low [22, 24, 25].
Characteristics of lnc-EST12 and its predominant distribution in the cytoplasm of macrophages. a schematic diagram of the position of lnc-EST12 in the mouse genome. b Expression and distribution of lnc-EST12 in different organs of mice. RNA was extracted from fresh mouse organs and used for lnc-EST12 detection with RT-qPCR. c RT-qPCR analysis of lnc-EST12 expression in different cells, as indicated, after treatment with EST12 protein. d Agarose gel electrophoresis of PCR products generated by 5′-RACE (left panel) and 3′-RACE (right panel) and schematic diagram of the full-length lnc-EST12 with 5′-RACE and 3′-RACE sequences (lower panel). e RT-qPCR analysis of the effect of lnc-EST12 overexpression or knockdown on adjacent gene expression (Igdcc3 and Parp16) in RAW264.7 cells. f FISH analysis of the cellular distribution of lnc-EST12 in RAW264.7 cells with a lnc-EST12 probe (Cy3, red) and DAPI (nuclear staining, blue). g The cellular distribution of lnc-EST12 in RAW264.7 cells was detected using RT-qPCR. Data are presented as the means ± SD of three independent experiments. ns not significant. Two-tailed unpaired Student’s t test was used to calculate the statistical significance of the data shown in (c) (vs. 0 h) and (e) (sh-lnc-EST12 vs. pSilencer-U6, pcDNA3.1-lnc-EST12 vs. pcDNA3.1)
In addition, the overexpression or knockdown of lnc-EST12 had no effect on the expression of its neighboring genes Igdcc3 and Parp16 (Fig. 2e). FISH assays showed that lnc-EST12 was primarily located in the cytoplasm of macrophages (Fig. 2f). Furthermore, we used a nuclear/cytosol separation assay and found that ~96% of lnc-EST12 was distributed in the cytoplasm of mouse macrophages (Fig. 2g). Thus, mouse lnc-EST12 is mainly expressed in immune-related organs (liver, spleen and lung), macrophages and DCs.
Lnc-EST12 overexpression suppresses inflammatory cytokine expression, while lnc-EST12 KO exerts the opposite effects
We constructed lnc-EST12 KO mice by deleting the 3 kb genomic locus containing lnc-EST12 to further investigate the function of lnc-EST12 (Supplementary Fig. 3a). We confirmed the deletion of lnc-EST12 from the genomic DNA of lnc-EST12 KO mice by Sanger sequencing and genomic PCR (Supplementary Fig. 3b, c). We performed unbiased transcriptome profiling using RNA-seq in wild-type (WT) and lnc-EST12 KO BMDMs stimulated with EST12 proteins for 0, 6 and 24 h to assess the effect of lnc-EST12 deficiency on gene expression in macrophages (Fig. 3a). Gene ontology (GO) enrichment analysis showed that cytokine activity, chemokine activity, cytokine receptors, and chemokine receptors (CCRs) were significantly overrepresented in the differentially expressed genes between lnc-EST12 KO and WT BMDMs (Supplementary Fig. 3d). We further confirmed that lnc-EST12 knockout induced the expression of the inflammatory cytokines IL-1β, IL-6 and CCL5/8 in lnc-EST12 KO BMDMs treated with EST12 using RT-qPCR and ELISA (Fig. 3b, c, Supplementary Fig. 3g, h). The levels of IL-1β and IL-6 were increased 1.5- to 3-fold in lnc-EST12-KO BMDMs compared to WT BMDMs at both the mRNA (Fig. 3b) and protein levels (Fig. 3c) at 24 h after EST12 stimulation. Overexpression of lnc-EST12 in lnc-EST12 KO BMDMs by lentivirus infection (LV-lnc-EST12) inhibited the expression of the IL-1β and IL-6 mRNAs and proteins (Fig. 3d, e, Supplementary Fig. 3e). Similar results were observed for CCL5 and CCL8 (Supplementary Fig. 3i, j). As Gm40527 partly overlaps with lnc-EST12, we further determined the possible impact of Gm40527 on inflammatory cytokine expression. We reconstructed the recombinant lentivirus LV-shGm40527 by knocking down a specific fragment of Gm40527 with no overlap with lnc-EST12 (Supplementary Fig. 3a, shGm40527 knockdown position is indicated by black triangle). Our results showed that knockdown of Gm40527 using LV-shGm40527 significantly decreased Gm40527 mRNA expression but did not affect the expression of lnc-EST12 and cytokines (IL-1β and IL-6) and chemokines (CCL5/8), as determined using RT-qPCR (Supplementary Fig. 3a, f).
Lnc-EST12 overexpression suppresses inflammatory cytokine expression, while lnc-EST12 KO exerts the opposite effects. a Heatmap of differentially expressed genes in lnc-EST12 KO BMDMs compared to WT BMDMs at 6 and 24 h after EST12 treatment. b IL-1β and IL-6 mRNA levels in WT/lnc-EST12 KO BMDMs treated with EST12 (2 μM) for 6 and 24 h. c IL-1β and IL-6 protein levels in WT/lnc-EST12 KO BMDMs treated with EST12 (2 μM) for 6 and 24 h, as analyzed using ELISA. d IL-1β and IL-6 mRNA levels in WT/lnc-EST12 KO BMDMs infected with LV-Ctl (control) or LV-lnc-EST12, followed by EST12 treatment (2 μM, 24 h). e IL-1β and IL-6 protein levels in WT/lnc-EST12 KO BMDMs infected with LV-Ctl or LV-lnc-EST12, followed by EST12 treatment (2 μM, 24 h). Data are presented as the means ± SD of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns not significant. LV lentivirus. Ctl control. lnc KO lnc-EST12 KO. Two-tailed unpaired Student’s t test was used to calculate the statistical significance of the data shown in (b, c) (vs. WT), and one-way ANOVA followed by Sidak’s multiple comparisons test was used for the data shown in (d, e)
Based on the data presented above, lnc-EST12 suppresses inflammatory cytokine expression in mouse macrophages, while lnc-EST12 KO exerts the opposite effects.
Lnc-EST12 binds the KH3 and KH4 domains of FUBP3
Next, we explored the host proteins interacting with lnc-EST12. Biotin-labeled lnc-EST12 was used for the pulldown of RAW264.7-cell lysates, followed by SDS–PAGE analysis (Fig. 4a). Differentially abundant protein bands were subjected to mass spectrometry (MS) analysis (Supplementary Fig. 4a). The specific proteins were verified by lnc-EST12 RNA pulldown and WB analysis, and FUBP3 was confirmed to be the specific protein for binding lnc-EST12 (Fig. 4b). We confirmed the interaction of lnc-EST12 with FUBP3 using RIP (RNA Binding Protein Immunoprecipitation) and confocal colocalization analysis (Fig. 4c, Supplementary Fig. 4b), and more colocalization between lnc-EST12 and FUBP3 was observed in the control group than in the EST12 group. Decreased expression of both cytoplasmic and nuclear lnc-EST12 was observed after EST12 treatment (Supplementary Fig. 4b), which indicated that decreased expression of lnc-EST12 might reduce the interaction between lnc-EST12 and FUBP3. We also used biotin-labeled full-length lnc-EST12 and different truncated lnc-EST12 constructs, as indicated, for RNA pulldown of RAW264.7-cell lysates. Only full-length lnc-EST12 bound to FUBP3, and each truncated lnc-EST12 fragment lost its binding ability (Fig. 4d).
Lnc-EST12 promotes FUBP3 binding to the promoters of proinflammatory cytokines and then suppresses cytokine expression. a SDS–PAGE analysis of proteins purified from the in vitro binding assay using biotinylated lnc-EST12 or antisense control RNA and RAW264.7-cell extracts. Differentially abundant protein bands were subjected to mass spectrometry analysis. b WB confirmed the lnc-EST12 and FUBP3 interaction in vitro. RAW264.7-cell lysates were incubated with biotin-labeled lnc-EST12 and detected using anti-FUBP3, anti-FUBP2, and anti-HNRNPD antibodies. c RIP-qPCR analysis was performed to determine the binding of lnc-EST12 to FUBP3 in RAW264.7 cells. d Schematic of lnc-EST12 deletion mutants used in RNA–protein binding assays. The RNA–protein binding assay was performed using biotinylated full-length or deletion mutants of lnc-EST12 and the lysate from RAW264.7 cells. The proteins pulled down by biotinylated lnc-EST12 were captured using streptavidin beads and subjected to WB with an antibody against FUBP3. e Schematic of HA-FUBP3 deletion mutants used in RNA–protein binding assays. f FUBP3 binds lnc-EST12 through the KH3 and KH4 domains. The RNA–protein binding assay was performed using biotinylated lnc-EST12 and overexpressed HA-FUBP3 and mutants in RAW264.7 cells. The biotinylated lnc-EST12 pulldown proteins were captured using streptavidin beads and subjected to immunoblotting against HA. g ChIP–qPCR assay of the binding of the FUBP3 protein to the promoter region of cytokines. ChIP-purified DNA was analyzed using qPCR with primers targeting the promoter site of the indicated genes. h ChIP–qPCR analysis of FUBP3 binding to the promoter region of cytokines in BMDMs treated with or without EST12 for 24 h. i ChIP–qPCR analysis of FUBP3 binding to the promoter region of cytokines in BMDMs expressing ectopic lnc-EST12 or LV-Ctl (control) that were treated with or without EST12 for 24 h. j, k The mRNA (j) and protein (k) levels of IL-1β and IL-6 in BMDMs infected with LV-Ctl, LV-lnc-EST12, LV-lnc-EST12 with LV-sh-scramble or LV-sh-FUBP3, followed by stimulation with EST12 (2 μM) for 24 h. LV, lentivirus. Ctl, control. Data are presented as the means ± SD of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns not significant. Two-tailed unpaired Student’s t test was used to calculate the statistical significance of the data shown in (c) (vs. IgG), g (vs. IgG), h (vs. Control), and (i) (vs. LV-Ctl), and one-way ANOVA followed by Sidak’s multiple comparisons test was used to calculate the statistical significance of the data shown in (j, k)
The FUBP3 protein contains four KH (K homology) repeats (from KH1 to KH4), which are considered the key domains for FUBP3 binding to RNA or DNA [26]. Therefore, FUBP3 with different KH domain mutations, Mut A (with KH1 to KH4 mutants), Mut B (KH1 mutant), Mut C (KH2 mutant), Mut D (KH3 mutant) and Mut E (KH4 mutant), were constructed (Supplementary Fig. 4c, d). RAW264.7 cells were transfected with pcDNA3.1 expression plasmids carrying the full-length FUBP3 sequence or its various KH site-specific mutants with HA tag for 24 h, and then cell lysates were pulled down with lnc-EST12 RNA, followed by WB analysis with an anti-HA tag monoclonal antibody (mAb). The full-length FUBP3 protein and its various KH mutant constructs were expressed at high levels with a molecular weight of 63 kDa, and we observed that lnc-EST12 only bound FUBP3 and FUBP3-Mut B/C but no longer bound FUBP3-Mut A, D and E (Fig. 4e, f). These data suggest that the KH3 and KH4 domains of FUBP3 are the critical binding sites for lnc-EST12.
The lnc-EST12-FUBP3 interaction promotes FUBP3 binding to the promoters of proinflammatory cytokines and then suppresses proinflammatory cytokine expression
ChIP was performed, followed by RT-qPCR analysis, to determine whether FUBP3 functions as a transcription factor to regulate proinflammatory cytokine expression by binding to the promoter regions of cytokines. The results showed that the FUBP3 protein indeed bound to the promoter regions of IL-1β, IL-6, CCL5, and CCL8 (Fig. 4g). Furthermore, the binding of FUBP3 to these promoter regions decreased when BDMDs were stimulated with the EST12 protein (Fig. 4h) and increased after lnc-EST12 overexpression in both resting and EST12-treated BMDMs (Fig. 4i). We also constructed a lentivirus expression vector by knocking down FUBP3 and confirmed the ~70% knockdown efficiency of FUBP3 protein expression by Lv-sh-FUBP3 in BMDMs using WB analysis (Supplementary Fig. 4e). Notably, lnc-EST12 suppressed the expression of IL-1β, IL-6, and CCL5/8 mRNA and protein in BMDMs stimulated with EST12, as evidenced by the RT-qPCR and ELISA results; however, after FUBP3 knockdown, the expression levels of IL-1β, IL-6 and CCL5/8 were increased (Fig. 4j, k and Supplementary Fig. 4f, g), suggesting that lnc-EST12 suppresses proinflammatory cytokine expression through FUBP3.
Based on the findings described above, our data revealed that lnc-EST12 enhanced the binding of FUBP3 to chromatin at the promoters of proinflammatory cytokines (IL-1β, IL-6, etc.), thus suppressing inflammatory cytokine expression. We postulate that lnc-EST12 binding to FUBP3 promotes the translocation of FUBP3 from the cytoplasm to the nucleus, and lnc-EST12 per se does not enter the nucleus (Supplementary Fig. 4b), similar to the p-JAK-p-STAT5 interaction promoting p-STAT5 translocation into the nucleus, but p-JAK does not enter the nucleus.
Lnc-EST12 suppresses EST12-induced NLRP3 inflammasome assembly and GSDMD-mediated pyroptosis via FUBP3
Previously, we found that the EST12 protein promoted NLRP3-GSDMD-mediated pyroptosis in macrophages [10]. Here, the expression levels of the constitutive proteins NLRP3, ASC and Caspase-1 were all increased in lnc-EST12 KO BMDMs compared to WT BMDMs (Fig. 5a), suggesting that lnc-EST12 suppresses the assembly of the NLRP3 inflammasome. The level of the N-terminal fragment of GSDMD (indicating GSDMD cleavage) was increased in lnc-EST12 KO BMDMs stimulated with the EST12 protein compared to WT BMDMs (Fig. 5b). In contrast, the cleavage and activation of GSDMD were inhibited after the overexpression of lnc-EST12 in lnc-EST12 KO BMDMs by lentivirus infection (LV-lnc-EST12) (Fig. 5c). WT, lnc-EST12 KO and GSDMD-KO BMDMs infected with LV-Ctl (control) or LV-lnc-EST12 and then treated with the EST12 protein were analyzed for cytotoxicity using the LDH assay. We found that lnc-EST12 knockout further enhanced the BMDM cytotoxicity and pyroptosis induced by EST12, and rescue of lnc-EST12 KO BMDMs with LV-lnc-EST12 alleviated this effect (Fig. 5d). GSDMD-KO attenuated lnc-EST12-mediated inhibition of BMDM pyroptosis (Fig. 5d). Therefore, lnc-EST12 suppresses the EST12-induced assembly of NLRP3 inflammasomes, as well as GSDMD-mediated pyroptosis of mouse macrophages.
Lnc-EST12 inhibits EST12-induced NLRP3 inflammasome assembly and pyroptosis via FUBP3. a WT and lnc-EST12 KO BMDMs were treated with the EST12 protein (2 μM) and subjected to IP with an anti-NLRP3 antibody and IB with antibodies against NLRP3, caspase-1 and ASC. b WT and lnc-EST12 KO BMDMs treated with the EST12 protein (2 μM) were analyzed for GSDMD activation using WB. c Lnc-EST12 KO BMDMs infected with LV-Ctl or LV-lnc-EST12 for 24 h and then treated with EST12 protein (2 μM) were analyzed for GSDMD activation using WB. d Effects of lnc-EST12 knockout and overexpression on the cytotoxicity of BMDMs stimulated with EST12. WT BMDMs and GSDMD-KO BMDMs were prepared with isolated WT and GSDMD-KO mouse bone marrow cells by incubating them with 10% FBS plus M-CSF for 7 days. WT, lnc-EST12 KO and GSDMD-KO BMDMs infected with LV-Ctl (control) or LV-lnc-EST12 for 24 h and treated with the EST12 protein (2 μM) were analyzed for cytotoxicity using the LDH assay. e Effects of FUBP3 knockdown on GSDMD activation following EST12 stimulation. BMDMs were infected with LV-Ctl or LV-shFUBP3 and stimulated with the EST12 protein (2 μM) for the indicated time, and then GSDMD activation was detected using WB. f Effects of FUBP3 knockdown on macrophage cytotoxicity induced by EST12. BMDMs infected with LV-Ctl, LV-lnc-EST12, or LV-lnc-EST12 with LV-sh-scramble or LV-sh-FUBP3 for 24 h and treated with the EST12 protein (2 μM) were analyzed for cytotoxicity using an LDH assay. LV lentivirus. Ctl control. lnc KO lnc-EST12 KO. Data are presented as the means ± SD of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. One-way ANOVA followed by Sidak’s multiple comparisons test was used to calculate the statistical significance of the data shown in (d, f)
Next, we determined whether FUBP3 was involved in the suppressive effect of lnc-EST12 on GSDMD pyroptosis in mouse macrophages. The cleavage of GSDMD was enhanced in FUBP3-silenced BMDMs stimulated with the EST12 protein compared to WT BMDMs (Fig. 5e). Overexpression of lnc-EST12 inhibited mouse macrophage pyroptosis induced by EST12, while FUBP3 knockdown in BMDMs reversed this effect (Fig. 5f). These results indicate that lnc-EST12 suppresses EST12-induced pyroptosis in a FUBP3-dependent manner.
Based on these results, the lnc-EST12 and FUBP3 interaction plays an important role in lnc-EST12-mediated inhibition of proinflammatory cytokine expression and pyroptosis.
Deficiency of lnc-EST12 or FUBP3 in macrophages increases both M.tb clearance and inflammation
Next, we examined whether lnc-EST12 KO macrophages and mice would show increased levels of inflammatory cytokines and decreased mycobacterial survival in mouse macrophages and in vivo. Both WT and Lnc-EST12 KO BMDMs were infected with M.tb H37Rv for 6 h. Extracellular bacteria were removed by washes with PBS, and intracellular bacteria were counted. Knockout of lnc-EST12 reduced mycobacterial CFUs and survival in mouse macrophages (Fig. 6a). Overexpression of lnc-EST12 increased the mycobacterial CFUs in a FUBP3-dependent manner, since FUBP3 silencing in BMDMs reversed this effect (Fig. 6b). WT and lnc-EST12 KO BMDMs were infected with WT H37Rv, H37RvΔEST12, and H37RvΔEST12::EST12. As expected, we observed increased levels of the inflammatory cytokines IL-1β and IL-6 and decreased mycobacterial CFUs in lnc-EST12 KO BMDMs compared with WT BMDMs (Supplementary Fig. 5e–g), suggesting that deficiency of lnc-EST12 in BMDMs increases both M.tb clearance and inflammation.
Lnc-EST12 knockout in macrophages increased M.tb clearance and inflammation in mice. a Bacterial burden in WT and lnc-EST12 KO BMDMs infected with M.tb H37Rv at an MOI of 10:1 (bacteria: cells = 10:1) for 6 h. Extracellular bacteria were removed by washes with PBS, and intracellular bacteria were counted. b Effects of FUBP3 knockdown on M.tb survival in macrophages. BMDMs were infected with LV-Ctl, LV-lnc-EST12, LV-lnc-EST12 with LV-sh-scramble or LV-sh-FUBP3 for 24 h and then infected with M.tb H37Ra at an MOI of 10:1 (bacteria: cells = 10:1) for 6 h. c–f IL-1β and IL-6 levels in the serum (c), spleen (d), liver (e), and lung (f) of WT and lnc-EST12 KO mice i.n. infected with H37Ra (5 × 104 CFUs/mouse) for 30 days were analyzed using ELISA. Data are presented as the means ± SD (n = 4–6 mice per group). g Bacterial burden in the lung and spleen from WT and lnc-EST12 KO mice infected with H37Ra. Data are presented as the means ± SD (n = 4–6 mice per group). h, i Acid-fast staining (h) and HE staining (i) of lung tissues from WT or lnc-EST12 KO mice infected with H37Ra or uninfected controls. LV lentivirus. Ctl control. lnc KO lnc-EST12 KO. Data are presented as the means ± SD of three independent experiments. *p < 0.05; **p < 0.01. ns not significant. Two-tailed unpaired Student’s t test was used to calculate the statistical significance of the data shown in (a–g) (vs. WT)
Furthermore, M.tb H37Ra, a slow-growing mycobacterium, was administered to mice through an intranasal infection at a dose of 106 CFUs/mouse [27,28,29,30]. Thirty days after infection, we observed significantly increased serum IL-1β, IL-6, CCL5, and CCL8 levels in lnc-EST12 KO mice (Fig. 6c and Supplementary Fig. 5a). In addition, similar results for the expression of these cytokines were observed in the spleen, liver and lung organs of the lnc-EST12 KO mice (Fig. 6d–f and Supplementary Fig. 5b–d). Through the bacterial colony counting experiment, we observed decreased colony numbers of H37Ra in the lung and spleen organs of lnc-EST12 KO mice (Fig. 6g). Acid-fast staining of lung tissues from H37Ra-infected mice also showed decreased numbers of acid-fast positive bacilli in lnc-EST12 KO mice compared with WT mice (Fig. 6h). We did not find classic TB lesions in the lungs of H37Ra-infected mice (Fig. 6i), but we could observe obvious inflammatory responses in these mouse infection models. The histopathological analysis of alveolar tissue from the lnc-EST12 KO mice showed increased inflammatory lymphocyte infiltration and smaller intact alveolar spaces compared with the WT mice (Fig. 6i). Therefore, overexpression of lnc-EST12 increases the survival of M.tb, while deficiency of lnc-EST12 or FUBP3 in mouse macrophages increases M.tb clearance and inflammation.
Lnc-EST12 promotes mycobacterial growth and suppresses IL-1β and IL-6 production after adoptive transfer in animal experiments
In an adoptive transfer experiment, clodronate liposomes eliminated both macrophages and DCs [31,32,33]. We found that ~88% of F4/80+ macrophages were depleted in the mouse lungs after injection of clodronate liposomes for 3 days (the macrophage percentage in the lungs decreased from 11.7% in the liposome control group to 1.36% in the clodronate depletion group), and ~80% of F4/80+ macrophages were restored in the mouse lung after injection of clodronate liposomes plus adoptive transfer of C57BL/6 mouse BMDMs (increased from 1.36% in the clodronate depletion group to 9.43% in the clodronate depletion plus adoptive transfer group) by flow cytometry (FCM) analysis (Supplementary Fig. 5i). We subsequently adoptively transferred WT or lnc-EST12 KO BMDMs into clodronate liposome-treated WT C57BL/6 mice to examine lnc-EST12 expression in lung F4/80+ macrophages by RT-qPCR. We found much less lnc-EST12 expression in the group with adoptive transfer of lnc-EST12 KO BMDMs than in the group with adoptive transfer of WT BMDMs (Supplementary Fig. 5h). Then, the mice were intravenously (i.v.) infected with M. smegmatis (M.s, a fast-growing mycobacterium) and M.s-EST12 (M.s carrying pMV261-EST12 that can express EST12 protein) for 3 days (Fig. 7a). EST12 was confirmed to be expressed in M. s-EST12 but not in WT M.s by WB (Supplementary Fig. 5j). M.s-EST12 significantly increased IL-1β and IL-6 production (Fig. 7b, c) and decreased bacterial CFUs in the lung, liver and spleen organs (Fig. 7d) in the group with adoptive transfer of lnc-EST12 KO BMDMs compared to the group with adoptive transfer of WT BMDMs. These adoptive transfer experiments strongly suggest that lnc-EST12 promotes mycobacterial growth by suppressing IL-1β and IL-6 production in vivo.
Adoptive transfer of lnc-EST12 KO macrophages enhances inflammatory cytokine production and bacterial clearance. a Procedure used for the WT and lnc-EST12 KO BMDM adoptive transfer experiment. b–d IL-6 (b) and IL-1β (c) levels and bacterial burden (d) in the liver, spleen, and lung of mice adoptively transferred with WT or lnc-EST12 KO BMDMs and i.v. infected with M.s or M.s-EST12 (M.s expressing EST12) (5 × 104 CFUs/mouse) for 5 days. Data are presented as the means ± SD (n = 4–6 mice per group) from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns, not significant. Two-tailed unpaired Student’s t test was used to calculate the statistical significance of differences shown in (b–d) (vs. WT)
Discussion
LncRNAs are widely involved in various physiological activities. LncRNA-Mirt2 has been reported to inhibit the excessive activation of LPS-induced inflammation and regulate inflammatory damage in the body to achieve a reasonable level [22]. LncRNA-Neat1 increases the activation of caspase-1, thereby promoting the production of IL-1β and the pyroptosis of macrophages [34]. LncRNAs also play important roles in the infection process of M.tb [35, 36]. LncRNA-CD244, which is highly expressed in CD8+ T cells of patients with active TB, inhibits the production of IFN-γ and TNF-α in CD8+ T cells and inhibits host clearance of M.tb [37]. Although several lncRNAs related to TB have been discovered in recent years, the role and mechanism of the important lncRNAs related to the key factors of M.tb during infection remain largely unexplored. Therefore, a particularly important goal is to discover a lncRNA related to an M.tb key factor/protein and to explore its function and mechanism in vitro and in vivo.
Previously, we found that EST12, a secreted protein of M.tb, activated an NF-κb/AP-1-NLRP3-GSDMD pyroptosis-IL-1β immune pathway in macrophages [10]. In this study, we identified a key lncRNA, lnc-EST12, which was negatively regulated by the M.tb EST12 protein through the activation of the p-JAK2-p-STAT5a signaling pathway in mouse macrophages. We also documented that EST12-stimulated STAT5a binding to the lnc-EST12 promoter and then repressed lnc-EST12 expression. Similarly, other reports also showed that STAT5a binds and represses target gene expression [38]. Lnc-EST12 is more likely to act as a general responder to mycobacterial infection, since the host immune responses are activated following the decreased expression of lnc-EST12, and its expression is downregulated by either ETS12-expressing or ETS12-nonexpressing mycobacterial strains (Supplementary Fig. 1b). EST12 promotes the activation of the p-JAK2-p-STAT5a pathway and the translocation of p-STAT5a from the cytoplasm to the nucleus. STAT5a binds to the lnc-EST12 promoter to repress lnc-EST12 expression (Supplementary Fig. 6). Importantly, lnc-EST12 suppressed antimycobacterial innate immunity through its interaction with FUBP3. Notably, lnc-EST12 binding to FUBP3 promoted the binding of the transcription factor FUBP3 to the promoter regions of cytokines such as IL-1β and IL-6, thus suppressing the expression of these cytokines. Lnc-EST12 also suppressed EST12-induced NLRP3 inflammasome assembly and GSDMD-mediated pyroptosis via FUBP3. These results indicate that lnc-EST12, which is negatively regulated by mycobacterial EST12, represses the innate inflammatory immune response and pyroptosis through its interaction with FUBP3 (Supplementary Fig. 6).
The expression levels of various cytokines and chemokines play important roles in M.tb infection [39]. Among them, M.tb infection activates the NLRP3 inflammatory pathway in macrophages, promoting the secretion of IL-1β and IL-18 [40, 41]. IL-1β directly kills M.tb in macrophages, upregulates TNF-α expression and promotes cell apoptosis. IL-6 exerts a dual immunomodulatory effect on M.tb infection [42, 43]. Generally, IL-6 is presumed to induce the proliferation and differentiation of B cells and T cells, activate macrophages, and enhance the killing effect of NK cells, thereby resisting M.tb infection. CCL5 contributes to early, protective, M.tb-specific immunity, primarily by recruiting early antigen-specific IFN-γ-producing T cells to M.tb-infected lungs [44, 45]. CCL8 is expressed at high levels in patients with TB. Its mechanism of action is mainly to recruit lymphocytes and other immune cells to the inflammatory site to exert an antibacterial effect [46,47,48]. Consistently, we found that M.tb EST12 increased IL-6, IL-1β and CCL5/8 expression in mouse macrophages and promoted M.tb clearance, while lnc-EST12 or FUBP3 suppressed IL-6, IL-1β, and CCL5/8 expression and promoted M.tb survival in mice (Figs. 4j, k, 6).
Macrophages eliminate intracellular Mtb through various mechanisms, including the production of nitric oxide (NO) and cytokines and the activation of autophagy and pyroptosis [49,50,51]. At the same time, in the long-term struggle with the host, M.tb has also successfully evolved many strategies to ensure its survival. Pyroptosis is a type of proinflammatory programmed cell death [52, 53]. Studies have shown that a variety of bacteria and viruses induce pyroptosis, thereby regulating their survival and replication in the host [54,55,56]. Pyroptosis destroys the living environment of M.tb in macrophages and subsequently kills M.tb and inhibits its replication [57, 58]. In addition, M.tb avoids clearance by evading pyroptosis in macrophages [59]. Both the virulent strain M.tb H37Rv and the attenuated strain H37Ra activate NLRP3 inflammasomes in mouse macrophages [41, 60, 61]. The secreted ESAT-6 protein of M.tb activates the NLRP3 inflammasome of the retinal pigment epithelium (RPE) [40]. Previous research in our laboratory also proved that the H37Rv-secreted protein EST12 activated the NLRP3 inflammasome and pyroptosis of macrophages [10]. Here, lnc-EST12, which is negatively regulated by EST12, suppressed the expression of inflammatory cytokines (IL-6 and IL-1β) and chemokines (CCL5 and CCL8) in mouse macrophages and inhibited the activation of the NLRP3 inflammatory pathway by inhibiting the assembly of inflammasomes.
FUBPs are a family of single-stranded DNA-binding proteins that include FUBP1, FUBP2, and FUBP3 [62]. The central structural region in the middle of FUBPs contains four KH domains, which are important sites for FUBP binding [26, 63]. Studies have found that FUBPs bind to RNAs and proteins and have all the characteristics of general transcription factors [64,65,66]. FUBP3 has been implicated in cell proliferation, differentiation, migration, and carcinogenesis. In addition, FUBP3 binds to the 5’UTR of EV71 and subsequently promotes virus replication [67]. Although FUBP3 plays an important role in many diseases, its function in TB has not yet been reported. In the present study, we first found that lnc-EST12 bound to FUBP3, not FUBP1/2, and the interaction of lnc-EST12 and FUBP3 depended on the complete sequence of lnc-EST12 and the KH3 and KH4 domains of FUBP3. We also found that the lnc-EST12-FUBP3 interaction promoted FUBP3 binding to the promoters of proinflammatory cytokines (IL-1β, IL-6, CCL5, and CCL8) (Fig. 4i) and then suppressed the expression of these proinflammatory cytokines (Fig. 4j, k). Decreased expression of both cytoplasmic and nuclear lnc-EST12 after EST12 treatment might reduce the interaction between lnc-EST12 and FUBP3 (not due to lnc-EST12 translocation into the nucleus after binding with FUBP3), thus causing increased expression of inflammatory cytokines. Overall, lnc-EST12 binds to the FUBP3 protein and thus suppresses the expression of inflammatory cytokines and NLRP3 inflammasome-GSDMD-mediated pyroptosis in mouse macrophages, which further promotes M.tb survival.
The proinflammatory pyroptosis pathway has been widely targeted by therapeutics. The FDA-approved drug anakinra exerts its effect by inhibiting IL-1β [68]. Because of its role in inflammation, GSDMD has become a hot target for the development of therapeutics. Some studies have identified small-molecule GSDMD inhibitors. For example, the drug DSF used to treat chronic alcoholism was found to be an inhibitor of GSDMD-mediated pore formation [69]. The therapeutic drug DMF for multiple sclerosis was found to prevent the interaction between GSDMD and caspase, limiting the processing and oligomerization of GSDMD and suppressing pyroptosis [70]. Although inhibiting GSDMD may become an effective treatment strategy, interfering with the antibacterial activity of GSDMD may also increase the sensitivity to infection, and the innate immune response is complex. Similarly, our discovery of the interaction of lnc-EST12 and FUBP3, which suppress pyroptosis, might also have potential use for disease therapeutics (such as for the treatment of multiple sclerosis). Certainly, each potential treatment strategy must be rigorously verified.
In the adoptive transfer experiment, the clodronate liposomes eliminated both macrophages and DCs. Therefore, both macrophages and DCs may be involved in adoptive transfer experiments. Lnc-EST12 expression was decreased in BMDMs and BMDCs stimulated by EST12 protein (Fig. 2c), suggesting that mouse lnc-EST12 might be expressed in both macrophages and DCs. In the future, more studies are needed to determine whether lnc-EST12 functions in other immune cells (such as DCs, T cells and B cells) and plays a similar role.
In addition, we conducted detailed analyses of human/mouse sequential homology of lnc-EST12 by analyzing GenBank databases for potential transcripts but could not identify a clear human sequential homolog for this lncRNA. Recently, a large number of studies on mouse lncRNAs have been reported and published [22, 71,72,73,74,75,76], and no human/mouse homologies of these lncRNAs have been found in these studies [22, 71,72,73,74,75,76]. For example, lncRNA MUNC regulates the transcription of different promyogenic factors in mice [71]; a novel mouse lncRNA Discn fine-tunes replication protein A (RPA) availability to promote genomic stability [72]; a mouse long noncoding RNA-lncFAO regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation [73]; and a mouse macrophage-specific lncRNA-MAARS regulates apoptosis and atherosclerosis by tethering HuR in the nucleus [74]. LncRNAs exhibit low sequence conservation, and most mouse lncRNAs tend not to have sequences homologous to those in humans [77,78,79,80,81]; however, the mouse lncRNA research platform still provides a useful reference for research on human-related diseases [22, 71,72,73,74,75,76]. Furthermore, although there have been reports about human lncRNAs with no homologous sequence to that of mouse lncRNAs, there remains a possibility for the existence of similar structures or functions between mouse and human lncRNAs [82]. It was reported that human, mouse and zebrafish lncRNA Cyrano, which were identified according to synteny loci (located on the same chromosome and with the same downstream/upstream genes), did not share sequence homology with each other in whole sequence alignments; however, human or mouse lncRNA orthologs could phenotypically rescue zebrafish lncRNA loss of function, which is crucial for embryonic development [78]. Thus, there is a possibility for the existence of lncRNAs between mice and humans with similar structures or functions but with no homologous sequences [78, 82]. Therefore, whether lnc-EST12 functions in humans needs further investigation, and the potential human homolog of lnc-EST12 with a similar structure or function remains to be explored in future studies. Our discovery of lnc-EST12 provides potential benefits for further understanding the immune mechanism of human TB.
In conclusion, lnc-EST12 inhibits the activation of inflammation and pyroptosis in mouse macrophages. Deficiency of lnc-EST12 or FUBP3 decreases mycobacterial survival and increases inflammation in mouse macrophages or mice. Here, we report a new immunoregulatory mechanism in which host lnc-EST12 negatively regulates anti-M.tb innate immunity through FUBP3 (Supplementary Fig. 6). The discovery of this lncRNA further improves our understanding of the intracellular struggle and survival mechanism after M.tb infection and may provide insights into the development of potential anti-tuberculosis treatments that target the interaction between host lnc-EST12 and FUBP3.
Materials and methods
Cell culture and stimulation
The mouse macrophage line RAW264.7 was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, China) supplemented with 10% heat-inactivated FBS (fetal bovine serum; Gibco, China) in a humidified incubator with 5% CO2 at 37 °C. BMDMs were differentiated from isolated mouse bone marrow cells by incubating them with 10% FBS plus macrophage colony-stimulating factor (M-CSF; 40 ng/mL; Peprotech, China) for 7 days. BMDCs were differentiated from isolated mouse bone marrow cells by culturing them for 7 days in complete RPMI 1640 (Gibco, China) containing granulocyte M-CSF (GM-CSF; 20 ng/mL; Peprotech, China) and IL-4 (20 ng/mL; Peprotech, China). A total of 2 × 106 cells were seeded in 60 mm cell culture plates, and after an overnight culture, the cells were treated with ultrapure EST12 proteins at a 2 μM concentration for different periods.
Bacterial culture
M.tb H37Rv [strain American Type Culture Collection (ATCC) 27294] and H37Ra [strain American Type Culture Collection (ATCC) 25177] were maintained on Lowenstein-Jensen medium. M. smegmatis (strain ATCC 19420), Escherichia coli DH5α (strain ATCC 25922), and E. coli BL-21 (stain ATCC BAA-1025) were propagated from laboratory stocks (School of Medicine, Wuhan University, Wuhan, China). E. coli DH5α and BL-21 (DE3) were grown in flasks using LB medium. The mycobacterial strains were grown in Middlebrook 7H9 broth (BD Biosciences, USA) supplemented with 10% oleic acid–albumin–dextrose–catalase (BD Biosciences, USA) and 0.05% Tween 80 (Sigma–Aldrich, USA) or on Middlebrook 7H10 agar (BD Biosciences, USA) supplemented with 10% OADC [83].
Agilent mouse lncRNA microarray assay
The Agilent Mouse lncRNA Microarray (4x180K, Design ID: 049801) was analyzed by Shanghai OE Biotech Co. Ltd. of China. Sample labeling, microarray hybridization and washing were performed according to the manufacturer’s standard protocols. Briefly, total RNA was extracted from RAW264.7 cells in the control group and EST12 treatment group with TRIzol (Invitrogen, USA), transcribed to double-strand cDNAs, synthesized into cRNAs and labeled with cyanine-3-CTP. The labeled cRNAs were hybridized to the microarray. After washing, the arrays were scanned using an Agilent Scanner G2505C (Agilent Technologies).
Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used to analyze array images and obtain raw data. GeneSpring software (GX V11.5, Agilent Technologies) was employed to perform the basic analysis of the raw data. Differentially expressed lncRNAs were then identified based on the fold change. The threshold set for upregulated and downregulated genes was a fold change ≥2.0. Then, GO and KEGG analyses were conducted to determine the functions of these differentially expressed genes.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, USA). One microgram of RNA was used to synthesize cDNAs with the RT Master Mix (Toyobo, Japan). Real-time PCR was performed using SYBR Green real-time PCR mix (Toyobo, Japan) with specific primers (Supplementary Tables 1, 2). The RT-qPCRs were cycled using an ABI StepOnePlus instrument (Applied Biosystems, USA) under standard cycling conditions. Target gene expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative RNA levels were calculated using the comparative cycle threshold (CT) method (2−ΔΔCT method), where CT indicates the amplification cycle number at which the fluorescence generated within a reaction increases above a defined threshold fluorescence, and ΔΔCT = experimental sample (CTtarget gene − CTGAPDH) − control sample (CTtarget gene − CTGAPDH).
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of IL-6, IL-1β, CCL5, and CCL8 were determined using ELISA kits according to the manufacturer’s instructions (4A Biotech, China). Briefly, BMDMs were stimulated with EST12 for 0, 6, and 24 h. Cell debris was removed by centrifugation, and the cell supernatant was collected for ELISA. For the detection of cytokines in animal serum and immune tissues, mice were infected with H37Ra for 30 days and then sacrificed to obtain serum and liver, spleen, and lung tissues. The tissues were ground and centrifuged to obtain supernatants for ELISA.
Electrophoretic mobility shift assay (EMSA)
The EMSA was performed using chemiluminescent EMSA kits according to the manufacturer’s instructions (Beyotime, China). Briefly, synthetic oligonucleotides containing the STAT5a transcription factor binding region were used as probes for EMSA and are listed in Supplementary Table 3. Double-stranded oligonucleotides were generated by annealing the synthetic oligonucleotides with their respective complementary sequences. Complementary oligonucleotides of equal quantity (10 μM each) were annealed in a thermocycler (Techgene, UK) at the following temperatures: 88 °C for 2 min, 65 °C for 10 min, 37 °C for 10 min and 25 °C for 5 min. A nonlabeled probe competitor and its mutant competitor (100 μM) were used (shown in Supplementary Table 3. Nuclear proteins were extracted from RAW264.7 cells and incubated with biotin-labeled DNA probes (10 μM) and/or competitor or mutant competitor (100 μM). Then, the mixtures were separated on native PAGE gels, transferred to nitrocellulose membranes, incubated with HRP-labeled streptavidin, and developed using a UVP bioimaging system (UVP, USA).
Rapid amplification of cloned cDNA ends (RACE)
Total RNA was extracted from RAW264.7 cells using TRIzol reagent (Invitrogen, USA), and 5′- or 3′-RACE was performed with a SMARTer RACE 5′/3′ Kit (Takara, Japan) according to the manufacturer’s instructions. The following primers used for 5′- or 3′-RACE were designed based on the known sequence information: 5′ specific primer-1-CTTCCCTGGGACCTTCAGCCCTAGCCCTCA; 5′ specific primer-2-CGAAACATTGGCTTCCCTGGGACCTTCAGC; 3′ specific primer-1-CTGCTTTAAGTGACCTACACAGCCAAGCGT; and 3′ specific primer-2-TGTTTCCTCTGGTGGACCCCCTTTTCCAGC.
Sample preparation and RNA-seq
WT and lnc-EST12 KO BMDMs were stimulated with EST12 (2 μM) for 0, 6, or 24 h. The supernatant was discarded, and 500 μL of TRIzol (Invitrogen, USA) was added. The samples were then transferred to a centrifuge tube and sent to Novogene Company (Beijing, China) for sequencing and analysis. Briefly, mRNA was purified from total RNA using poly-T oligo-conjugated magnetic beads. Fragmentation was performed in the presence of divalent cations in First Strand Synthesis Reaction Buffer (5X). The cDNA templates were synthesized using random hexamer primers and M-MuLV Reverse Transcriptase. Adaptors with hairpin loop structures were ligated to the cDNAs. The cDNA fragments 370–420 bp in length were preferentially purified with an AMPure XP system (Beckman Coulter, Beverly, USA). Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed using the Agilent Bioanalyzer 2100 system.
Raw data (raw reads) in fastq format were first processed using in-house Perl scripts. Reads were aligned to the reference genome using HISAT2 v2.0.5. FeatureCounts v1.5.0-p3 was used to count the read numbers mapped to each gene. Differential expression between two conditions/groups (two biological replicates per condition) was analyzed using the DESeq2 R package (1.20.0). GO enrichment analysis of differentially expressed genes was implemented using the clusterProfiler R package, in which gene length bias was corrected.
Single RNA fluorescent in situ hybridization (FISH)
The red fluorescent Cy3-labeled probe (Ribo-lncRNA FISH Probe Mix) against lnc-EST12 was designed by RiboBio Company (Guangzhou, China) and was detected using a Fluorescent In Situ Hybridization Kit (RiboBio, China) according to the manufacturer’s instructions. Briefly, RAW264.7 cells grown on cover slips in 24-well plates with the indicated treatment were fixed with 4% (v/v) paraformaldehyde for 10 min at room temperature and then washed three times with cold PBS. The cells were permeabilized in PBS containing 0.5% Triton X-100 for 5 min at 4 °C and then blocked with prehybridization buffer for 30 min at 37 °C. Cells were then incubated with a probe in hybridization buffer (2.5 μL, 20 μM probe in 250 μL hybridization buffer) overnight at 37 °C in the dark. After hybridization, the cells were washed in the dark with washing buffer (4 × SSC/2 × SSC/1 × SSC) and then stained with DAPI for 10 min. The cells were washed three times with PBS and then imaged with a ZEISS confocal microscope under an oil objective.
LDH release assay
The LDH release assay was performed using the CytoTox 96 NonRadioactive Cytotoxicity assay kit (Promega, USA) according to the manufacturer’s protocol. BMDMs were seeded in 12-well plates (2 × 106 cells per well), infected with lentiviruses (multiplicity of infection (MOI) = 10:1) for 24 h, and then stimulated with EST12 (20 μg/mL) for 12 h. Cell supernatants were collected to quantify the amount of LDH released from cells. The percentage of specific killing was calculated as follows: specific killing% = (LDH released from the experimental group − LDH released spontaneously)/(total LDH released − LDH released spontaneously) × 100%. All experiments were performed in triplicate.
Animal infection model
GSDMD-KO mice were kindly provided by Professor Feng Shao (National Institute of Biological Sciences, Beijing) [10, 84]. Lnc-EST12 KO mice were generated by deleting the lnc-EST12 genomic locus (3 kb) with the CRISPR–Cas9 strategy. The sgRNAs designed for CRISPR binding were as follows: sgRNA-1, 5′-GAAGAATACGGAGACCCTTCTGG and sgRNA-2, 5′-AGATGCTCGCCGTGCAATCCAGG. M.tb H37Ra is a slow-growing mycobacterium, so the infection procedure for its mouse model usually lasts for 30 days. While M. smegmatis is a fast-growing mycobacterium, the infection procedure for mouse models is usually 3-4 days. All WT and lnc-EST12 KO mice (female) were infected with H37Ra (5 × 104 CFUs of M.tb/50 μL diluted with PBS) by intranasal administration on Day 0. On Day 30, the mice were euthanized. Serum cytokine production was assessed using ELISA. The sections of lung tissues and spleen tissues were fixed with 4% paraformaldehyde, and pathological sections were examined with hematoxylin and eosin (HE) staining.
All mice were maintained in microisolator cages, fed a standard laboratory diet and water, and housed in the animal colony at the animal center of Wuhan University. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication no. 86-23, revised 1985). All animal protocols were approved by the Institutional Animal Care and Use Committee of the Institute of Model Animals of Wuhan University (nos. S01317101C, S01317012S, and 2018022).
Generation of recombinant lentivirus
The lentiviral vector plasmids (SBI, USA) containing the lnc-EST12 or shFUBP3 sequence were cotransfected into 293T cells with the helper plasmid pMD2. G (Addgene, 12259) and pSPAX (Addgene, 12260). After 60 h, the cell supernatant was collected and centrifuged at 12,000 × g to remove cell debris. The supernatant was transferred to an ultracentrifuge tube, and 2 mL of a 20% sucrose solution was gently added to the bottom and centrifuged at 25,000 × g for 2 h. The supernatant was discarded, and 100 μL of ice-cold PBS was added to resuspend the pellet. A lentivirus containing only green fluorescence protein (GFP) was used as a negative control (LV-Ctl).
RNA pulldown and mass spectrometry (MS)
Biotin-labeled lncRNAs were transcribed using the Biotin RNA Labeling Mix and T7 RNA polymerase in vitro (RiboBio, China) and purified using the RNeasy Mini Kit (QIAGEN, Germany) with on-column digestion of DNA. Biotinylated lnc-EST12, antisense lnc-EST12, was incubated with RAW264.7-cell lysates (containing RNase inhibitor) overnight at 4 °C. The interacting complexes were purified with streptavidin beads for 3 h at room temperature and visualized by performing Coomassie Brilliant Blue staining for mass spectrometry or immunoblotting using a specific antibody against FUBP3. For mass spectrometry, a specific band present in the experimental lane was extracted (the corresponding region in the control lane was also extracted) for further analysis.
Chromatin immunoprecipitation (ChIP)-qPCR
BMDMs were infected with LV-Ctl or LV-lnc-EST12 for 24 h and then treated with or without the EST12 protein for 12 h (20 μg/mL). The growth media was removed, and the cells were rinsed three times with cold PBS. Then, the cells were treated with formaldehyde to a final concentration of 1% at room temperature for 10 min. Glycine was added to the cells to a final concentration of 0.125 M to stop the crosslinking reaction. Cells were harvested into cold PBS by scraping and transferred to a 1.5 mL microcentrifuge tube. After centrifugation at 1000 × g for 5 min at 4 °C, the formaldehyde-crosslinked cells were collected and resuspended in 1 mL of nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS, and 1 mM PMSF). Chromatin was sheared to an average size of 100-500 bp by sonication and then centrifuged (10 min, 10,000 × g, 4 °C). Sixty microliters of supernatant was diluted 10-fold with 540 μL of ChIP dilution buffer (1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, and 16.7 mM Tris-HCl, pH 8.0) and then incubated with anti-FUBP3 (Abcam, UK, ab181025) or anti-rabbit IgG (Millipore, Germany, 12–370) overnight at 4 °C with rotation. Then, 50 μL of Protein A/G magnetic beads (MCE, USA, HY-K0202) was added to each sample, and the incubation continued for 5 h at 4 °C on a rotating platform. The beads were pelleted and then washed sequentially with low-salt buffer (150 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.1% SDS, 0.5% Triton X-100, and 2 mM EDTA), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, and 500 mM NaCl), and LiCl buffer (0.25 M LiCl, 1% sodium deoxycholate, 10 mM Tris-HCl pH 8.0, 1% NP-40, and 1 mM EDTA), followed by two washes with TE buffer (1 mM EDTA and 10 mM Tris-HCl, pH 8.0). Chromatin was eluted from the beads by two incubations with 100 μL of elution buffer (100 mM NaHCO3 and 1% SDS), the Na+ concentration was adjusted to 300 mM with 5 M NaCl, and the crosslinks were reversed by an overnight incubation in a 65 °C water bath. The samples were then incubated with 0.1 mg/mL RNase A for 1 h at 37 °C followed by 1 mg/mL proteinase K for 2 h at 55 °C. DNA was purified by phenol extraction and ethanol precipitation and then used for qPCR.
Hematoxylin–eosin (H&E) and Ziehl–Neelsen staining
H&E staining was performed according to the instructions provided by the manufacturer of the reagent (Solarbio, China). Briefly, mouse tissues were immobilized, embedded in paraffin, and then sectioned. Paraffin slides were dewaxed and hydrated through two dewaxing steps with xylene for 5 min each, soaked in anhydrous ethanol, 95, 80, and 70% ethanol for 2 min each, and finally soaked in distilled water for 2 min. The dewaxed sections were stained with hematoxylin solution for 10 min, washed with tap water and soaked in differentiation solution for 30 s. The sections were then transferred to warm water at 50 °C for 5 min, stained with eosin solution for 1 min and rinsed with tap water. Sections were dehydrated with a graded series of alcohol solutions and cleared in xylene.
Ziehl–Neelsen staining was performed according to the instructions provided by the manufacturer of the reagent (Baso, China). Briefly, mouse tissues were fixed, embedded in paraffin, and then sectioned. Paraffin sections were dewaxed and hydrated through two dewaxing steps with xylene for 5 min each, soaked in anhydrous ethanol, 95, 80, and 70% ethanol for 2 min each, and finally soaked in distilled water for 2 min. The dewaxed sections were stained with Carbolfuchsin staining solution for 10 min and then washed with tap water for 30 s. Acid alcohol solution was added from the outer edge of the upper end of the tissue to cover the glass slide, discolored for 1 min, and then washed with tap water for 30 s. Sections were then stained with a methylene blue solution for 30 s. After washing with tap water, the sections were allowed to dry before microscopic examination.
The mounted sections were then examined and photographed using a Leica Aperio VERSA 8 (Leica, Germany). The staining intensity was analyzed using ImageScope version 12 software (Leica, Germany).
Macrophage adoptive transfer animal experiment
WT mice were i.v. injected with 200 μL of clodronate liposomes (Liposoma, USA) 2 days in advance according to the manufacturer’s instructions. On Day 0, all the mice were i.v. injected with WT or lnc-EST12 KO BMDMs (106/mice). Two days later, the mice were i.n. infected with M.s or M.s-EST12 (105 CFUs/mouse). On the 5th day, the mice were sacrificed, and the serum, spleen, liver, and lung tissues were collected and processed for the next step.
Statistical analysis
GraphPad Prism software (GraphPad Software Inc., La Jolla, CA) was used for statistical analyses. Significant differences between groups were analyzed using two-tailed unpaired Student’s t test or one-way ANOVA followed by Sidak’s multiple comparisons test. Data are presented as the means ± standard deviations (SD). A p value < 0.05 was considered significant.
References
World Health Organization. Global tuberculosis report 2020. Geneva: WHO Press; 2021.
Mangtani P, Abubakar I, Ariti C, Beynon R, Pimpin L, Fine PE, et al. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis. 2014;58:470–80.
Lawn SD, Churchyard G. Epidemiology of HIV-associated tuberculosis. Curr Opin HIV AIDS. 2009;4:325–33.
Eum SY, Kong JH, Hong MS, Lee YJ, Kim JH, Hwang SH, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–8.
Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annu Rev Pathol. 2012;7:353–84.
Repasy T, Lee J, Marino S, Martinez N, Kirschner DE, Hendricks G, et al. Intracellular bacillary burden reflects a burst size for Mycobacterium tuberculosis in vivo. PLoS Pathog. 2013;9:e1003190.
Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M-bovis. J Bacteriol. 1996;178:1274–82.
Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–3.
Ganguly N, Siddiqui I, Sharma P. Role of M-tuberculosis RD-1 region encoded secretory proteins in protective response and virulence. Tuberculosis. 2008;88:510–7.
Qu Z, Zhou J, Zhou Y, Xie Y, Jiang Y, Wu J, et al. Mycobacterial EST12 activates a RACK1-NLRP3-gasdermin D pyroptosis-IL-1beta immune pathway. Sci Adv. 2020;66:eaba4733.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69.
Chen J, Ao LF, Yang J. Long non-coding RNAs in diseases related to inflammation and immunity. Ann Transl Med. 2019;7:494.
Fathizadeh H, Hayat SMG, Dao S, Ganbarov K, Tanomand A, Asgharzadeh M, et al. Long non-coding RNA molecules in tuberculosis. Int J Biol Macromol. 2020;2020:340–6.
Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–6.
Hu GQ, Tang QS, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–8.
Wang P, Xu J, Wang Y, Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358:1051–5.
Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747–57.
Carpenter S, Atianand M, Aiello D, Ricci E, Gandhi P, Hall LL, et al. LincRNA-Cox2 is a long noncoding RNA induced by TLRs that mediates both activation and repression of immune response genes. Cytokine. 2013;63:251.
Walther K, Schulte LN. The role of lncRNAs in innate immunity and inflammation. RNA Biol. 2021;18:587–603.
Zhang Q, Chao TC, Patil VS, Qin Y, Tiwari SK, Chiou J, et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019;38:e100041.
Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–85.
Du M, Yuan L, Tan X, Huang D, Wang X, Zheng Z, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. 2017;8:2049.
Ma LN, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–34.
Nitsche A, Stadler PF. Evolutionary clues in lncRNAs. Wiley Interdiscip Rev RNA. 2017;8:1.
Scacalossi KR, van Solingen C, Moore KJ. Long non-coding RNAs regulating macrophage functions in homeostasis and disease. Vasc Pharmacol. 2019;114:122–30.
Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–6.
Wang J, Thorson L, Stokes RW, Santosuosso M, Huygen K, Zganiacz A, et al. Single mucosal, but not parenteral, immunization with recombinant adenoviral-based vaccine provides potent protection from pulmonary tuberculosis. J Immunol. 2004;173:6357–65.
Pepponi I, Stylianou E, van Dolleweerd C, Diogo GR, Paul MJ, Drake PMW, et al. Immune-complex mimics as a molecular platform for adjuvant-free vaccine delivery. PLoS ONE. 2013;8:e60855.
Choi JA, Cho SN, Lee J, Son SH, Nguyen DT, Lee SA, et al. Lipocalin 2 regulates expression of MHC class I molecules in Mycobacterium tuberculosis-infected dendritic cells via ROS production. Cell Biosci. 2021;11:175.
Cheng AC, Yang KY, Chen NJ, Hsu TL, Jou R, Hsieh SL, et al. CLEC9A modulates macrophage-mediated neutrophil recruitment in response to heat-killed Mycobacterium tuberculosis H37Ra. PLoS ONE. 2017;12:e0186780.
Bennett CL, Clausen BE. DC ablation in mice: promises, pitfalls, and challenges. Trends Immunol. 2007;28:525–31.
Ward NL, Loyd CM, Wolfram JA, Diaconu D, Michaels CM, McCormick TS. Depletion of antigen-presenting cells by clodronate liposomes reverses the psoriatic skin phenotype in KC-Tie2 mice. Br J Dermatol. 2011;164:750–8.
Zhang Y, Shlomchik WD, Joe G, Louboutin JP, Zhu J, Rivera A, et al. APCs in the liver and spleen recruit activated allogeneic CD8+ T cells to elicit hepatic graftversus-host disease. J Immunol. 2002;169:7111–8.
Zhang PF, Cao LM, Zhou RB, Yang XL, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10:1495.
Li M, Cui J, Niu W, Huang J, Feng T, Sun B, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun. 2019;509:803–9.
Sun WW, Lou H, Cao J, Wang P, Sha W, Sun Q. LncRNA MEG3 control Mycobacterium Tuberculosis infection via controlled MiR-145-5p expression and modulation of macrophages proliferation. Microb Pathog. 2020;149:104550.
Wang Y, Zhong HL, Xie XD, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8(+) T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci USA. 2015;112:E3883–92.
Zhou J, Guo C, Wu H, Li B, Zhou LL, Liang AB, et al. Dnmt3a is downregulated by Stat5a and mediates G0/G1 arrest by suppressing the miR-17-5p/Cdkn1a axis in Jak2(V617F) cells. Bmc Cancer. 2021;21:1213.
Cooper AM, Mayer-Barber KD, Sher A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol. 2011;4:252–60.
Basu S, Fowler BJ, Kerur N, Arnvig KB, Rao NA. NLRP3 inflammasome activation by mycobacterial ESAT-6 and dsRNA in intraocular tuberculosis. Micro Pathog. 2018;114:219–24.
Dorhoi A, Nouailles G, Jorg S, Hagens K, Heinemann E, Pradl L, et al. Activation of the NLRP3 inflammasome by Mycobacterium tuberculosis is uncoupled from susceptibility to active tuberculosis. Eur J Immunol. 2012;42:374–84.
Nagabhushanam V, Solache A, Ting LM, Escaron CJ, Zhang JY, Ernst JD. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J Immuno. 2003;171:4750–7.
Cheekatla SS, Tripathi D, Venkatasubramanian S, Nathella PK, Paidipally P, Ishibashi M, et al. NK-CD11c+Cell Crosstalk in Diabetes Enhances IL-6-Mediated Inflammation during Mycobacterium tuberculosis Infection. Plos Pathogens. 2016;12:e1005972.
Sanchez-Castanon M, Baquero IC, Sanchez-Velasco P, Farinas MC, Ausin F, Leyva-Cobian F, et al. Polymorphisms in CCL5 promoter are associated with pulmonary tuberculosis in northern Spain. I Int J Tuberc Lung D. 2009;13:480–5.
Vesosky B, Rottinghaus EK, Stromberg P, Turner J, Beamer G. CCL5 participates in early protection against Mycobacterium tuberculosis. J Leukoc Biol. 2010;87:1153–65.
Millington KA, Innes JA, Hackforth S, Hinks TSC, Deeks JJ, Dosanjh DPS, et al. Dynamic relationship between IFN-gamma and IL-2 profile of Mycobacterium tuberculosis-specific T cells and antigen load. J Immunol. 2007;178:5217–26.
Ruhwald M, Bodmer T, Maier C, Jepsen M, Haaland MB, Eugen-Olsen J, et al. Evaluating the potential of IP-10 and MCP-2 as biomarkers for the diagnosis of tuberculosis. Eur Respir J. 2008;32:1607–15.
Dulek DE, Newcomb DC, Goleniewska K, Cephus J, Zhou WS, Reiss S, et al. Allergic airway inflammation decreases lung bacterial burden following acute klebsiella pneumoniae infection in a neutrophil-and CCL8-dependent manner. Infect Immun. 2014;82:3723–39.
Braverman J, Stanley SA. Nitric oxide modulates macrophage responses to Mycobacterium tuberculosis infection through activation of HIF-1alpha and repression of NF-kappaB. J Immunol. 2017;199:1805–16.
Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell Mol Immunol. 2017;14:963–75.
Chai QY, Wang L, Liu CH, Ge BX. New insights into the evasion of host innate immunity by Mycobacterium tuberculosis. Cell Mol Immunol. 2020;17:901–13.
Beckwith KS, Beckwith MS, Ullmann S, Saetra RS, Kim H, Marstad A, et al. Plasma membrane damage causes NLRP3 activation and pyroptosis during Mycobacterium tuberculosis infection. Nat Commun. 2020;11:2270.
He WT, Wan HQ, Hu LC, Chen PD, Wang X, Huang Z, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1 beta secretion. Cell Res. 2015;25:1285–98.
Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–14.
Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007;9:2562–70.
Liu X, Lieberman J. A Mechanistic understanding of pyroptosis: the fiery death triggered by invasive infection. Adv Immunol. 2017;135:81–117.
Fogel N. Tuberculosis: a disease without boundaries. Tuberculosis. 2015;95:527–31.
Lerner TR, Borel S, Gutierrez MG, Borel S. The innate immune response in human tuberculosis. Cell Microbiol. 2015;17:1277–85.
Korb VC, Chuturgoon AA, Moodley D. Mycobacterium tuberculosis: manipulator of protective immunity. Int J Mol Sci. 2016;17:131.
Zhang Q, Jiang X, He W, Wei K, Sun J, Qin X, et al. MCL plays an anti-inflammatory role in Mycobacterium tuberculosis-induced immune response by inhibiting NFkappaB and NLRP3 inflammasome activation. Mediators Inflamm. 2017;2017:2432904.
Lee HM, Kang J, Lee SJ, Jo EK. Microglial activation of the NLRP3 inflammasome by the priming signals derived from macrophages infected with mycobacteria. Glia. 2013;61:441–52.
DavisSmyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–87.
Chung HJ, Liu JH, Dundr M, Nie ZQ, Sanford S, Levens D. FBPs are calibrated molecular tools to adjust gene expression. Mol Cell Biol. 2006;26:6584–97.
Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip Rev RNA. 2010;1:230–9.
He LS, Liu JH, Collins I, Sanford S, O’Connell B, Benham CJ, et al. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–44.
Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. Bmc Cancer. 2008;8:369.
Huang HI, Chang YY, Lin JY, Kuo RL, Liu HP, Shih SR, et al. Interactome analysis of the EV71 5 ‘ untranslated region in differentiated neuronal cells SH-SY5Y and regulatory role of FBP3 in viral replication. Proteomics. 2016;16:2351–62.
Kyriazopoulou E, Poulakou G, Milionis H, Metallidis S, Adamis G, Tsiakos K, et al. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: a double-blind, randomized controlled phase 3 trial. Nat Med. 2021;27:1752–60.
Ou AT, Zhang JX, Fang YF, Wang R, Tang XP, Zhao PF, et al. Disulfiram-loaded lactoferrin nanoparticles for treating inflammatory diseases. Acta Pharmacol Sin. 2021;42:1913–20.
Humphries F, Shmuel-Galia L, Ketelut-Carneiro N, Li S, Wang B, Nemmara VV, et al. Succination inactivates gasdermin D and blocks pyroptosis. Science. 2020;369:1633–7.
Przanowska RK, Weidmann CA, Saha S, Cichewicz MA, Jensen KN, Przanowski P, et al. Distinct MUNC lncRNA structural domains regulate transcription of different promyogenic factors. Cell Rep. 2022;38:110361.
Wang L, Li JZ, Zhou H, Zhang WD, Gao J, Zheng P. A novel lncRNA Discn fine-tunes replication protein A (RPA) availability to promote genomic stability. Nat Commun. 2021;12:5572.
Nakayama Y, Fujiu K, Yuki R, Oishi Y, Morioka MS, Isagawa T, et al. A long non-coding RNA regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation. Proc Natl Acad Sci USA. 2020;117:14365–75.
Simion V, Zhou HY, Haemmig S, Pierce JB, Mendes S, Tesmenitsky Y, et al. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat Commun. 2020;11:6135.
Hao KL, Lei W, Wu HC, Wu J, Yang ZZ, Yan SP, et al. LncRNA-Safe contributes to cardiac fibrosis through Safe-Sfrp2-HuR complex in mouse myocardial infarction. Theranostics. 2019;9:7282–97.
Li P, Ruan XB, Yang L, Kiesewetter K, Zhao Y, Luo HT, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–67.
Ma Y, Jiang CF, Li P, Cao H. In vivo functional analysis of nonconserved human lncRNAs using a humanized mouse model. Methods Mol Biol. 2021;2254:339–47.
Ransohoff JD, Wei YN, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–57.
Salih MM, Carpenter S. What sequencing technologies can teach us about innate immunity. Immunol Rev. 2022;305:9–28.
Ramilowski JA, Yip CW, Agrawal S, Chang JC, Ciani Y. Kulakovskiy IV, et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020;30:1060–72.
Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–55.
Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–3.
Yuan C, Qu ZL, Tang XL, Liu Q, Luo W, Huang C, et al. Mycobacterium tuberculosis Mannose-Capped Lipoarabinomannan Induces IL-10-Producing B Cells and Hinders CD4(+)Th1 Immunity. iScience. 2019;11:13–30.
Shi JJ, Zhao Y, Wang K, Shi XY, Wang Y, Huang HW, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5.
Acknowledgements
We acknowledge the staff at the Animal Biosafety Level 3 Laboratory (ABSL-III) of the Wuhan University School of Medicine (NO. 18021B) and the Second Military Medical University of Shanghai (No. 18002) for their help with the M.tb infection experiment. This project was supported by grants from the National Grand Program on Key Infectious Disease of China (2017ZX10201301-006 and 2012ZX10003002-015), the National Key R&D Program of China (2018YFA0507603), the National Natural Science Foundation of China (91740120, 22077097 and 21721005), the National Outstanding Youth Foundation of China (81025008), Hubei Province’s Outstanding Medical Academic Leader Program (523-276003), Hubei Province Key R&D Program (2020BCB020), Research and Innovation Team Project of Hubei Provincial Health Commission (WJ2021C002), the Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC 2018002), and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
XLZ designed and supervised the research. QY conducted the experiments, analyzed the data, and wrote the paper. DX, ZQ, JW, YZ, YW, and HX assisted in the experimental execution. YX and XLZ revised the paper. All authors contributed to and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Yao, Q., Xie, Y., Xu, D. et al. Lnc-EST12, which is negatively regulated by mycobacterial EST12, suppresses antimycobacterial innate immunity through its interaction with FUBP3. Cell Mol Immunol 19, 883–897 (2022). https://doi.org/10.1038/s41423-022-00878-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-022-00878-x