Abstract
Androgen receptor (AR) is emerging as a novel prognostic biomarker in triple-negative breast cancer (TNBC), but the underlying mechanisms remain unknown. As accumulating evidence has shown that long non-coding RNAs (lncRNAs) regulate important cancer hallmarks, we hypothesised that AR-regulated lncRNAs might play roles in TNBC progression. Here, we performed experiments with or without DHT treatment in three TNBC cell lines, and we identified an AR negatively induced lncRNA (ARNILA), which correlated with poor progression-free survival (PFS) in TNBC patients and promoted epithelial−mesenchymal transition (EMT), invasion and metastasis in vitro and in vivo. Subsequently, we demonstrated that ARNILA functioned as a competing endogenous RNA (ceRNA) for miR-204 to facilitate expression of its target gene Sox4, which is known to induce EMT and contribute to breast cancer progression, thereby promoting EMT, invasion and metastasis of TNBC. Our findings not only provide new insights into the mechanisms of lncRNA in regulating AR but also suggest ARNILA as an alternative therapeutic target to suppress metastasis of TNBC patients.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC), characterised by a lack of the oestrogen receptor (ER), progesterone receptor (PR) expression and human epidermal growth factor receptor 2 (HER2) amplification, is the most aggressive subtype of tumours associated with poor clinical outcomes [1, 2]. Few effective prognostic markers and targeted approaches are currently available for TNBC. Recently, the oral poly (adenosine diphosphate-ribose) polymerase (PARP) inhibitor olaparib has shown promising antitumour activity in HER2-negative patients with a germline BRCA mutation [3]. BRCA germline mutations have a high prevalence in the TNBC subtype, and thus, olaparib represents the first targeted therapy in this type of cancer. There is an urgent need to develop more therapeutic targets and novel biomarkers to treat TNBC. Androgen receptor (AR), a nuclear receptor showing high structural, functional and topographic similarity to ER and PR, is commonly expressed in breast cancer and has increasingly been shown to be involved in tumour progression, recurrence and metabolic reprogramming [4]. High expression of AR was reported to correlate with prolonged survival of TNBC [5,6,7]. Nevertheless, the underlying mechanisms of AR in regulating tumour progression remain elusive.
Long non-coding RNAs (lncRNAs) are defined as the non-protein-coding transcripts greater than 200 nt in length [8]. LncRNAs were shown to have broad functional roles in regulating important cancer processes, including proliferation [9], apoptosis [10], metastasis [11], and drug resistance [12]. A wide range of models have been proposed to explain the precise mechanisms by which lncRNAs function [13]. These molecules can regulate transcription by serving as guides that recruit transcriptional co-regulators or chromatin-modifying enzymes to a specific DNA region [10] or form ribonucleoprotein complexes by acting as molecular scaffolds to bridge together regulatory proteins [14]. Additionally, lncRNAs may act as decoys that bind microRNAs (miRNAs) or proteins to control the functions of regulatory molecules [15]. Collectively, lncRNAs can function as tumour suppressors or oncogenes by altering these functions to promote or suppress tumour formation, progression and metastasis.
In this study, we aimed to identify the AR-related lncRNAs and investigated the contributions of these lncRNAs to invasion and epithelial−mesenchymal transition (EMT) of TNBC cells and their roles in TNBC metastasis. Here, we identified an AR negatively induced lncRNA (ARNILA) (Ensembl: ENST00000431557) in TNBC and found it was correlated with poor progression-free survival (PFS). Further studies revealed its critical roles in invasion, EMT and metastasis. EMT is the first crucial step in metastasis formation; this process facilitates cancer cell motility and dissemination and promotes dedifferentiation, growth arrest, migration and invasion [16]. We found the ARNILA could sequester miR-204 by functioning as a competing endogenous RNA (ceRNA) to upregulate the miR-204-targeted gene Sox4. Sox4 was recently shown to be abnormally overexpressed in TNBC and correlated with this subtype. Additionally, this gene is considered to play a crucial role in breast cancer progression by orchestrating EMT [17, 18]. We demonstrated that Sox4 is responsible for ARNILA-mediated migration and EMT.
Results
Positive expression of AR is correlated with better prognosis in TNBC
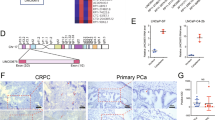
Recently, preclinical and early clinical studies have shown that AR may be a prognostic marker in diagnostics and therapy. We first assessed the relationship between AR and PFS in TNBC in 88 TNBC samples. We found that higher expression of AR was significantly associated with a prolonged PFS (hazard ratio (HR), 0.12; 95% confidence interval (CI), 0.13−0.75; P = 0.011) (Fig. 1a). We confirmed that positive AR expression was correlated with a significant improvement in relapse-free survival (RFS) (HR, 0.45; 95%CI, 0.35–0.59; P < 0.001) in 618 TNBC samples with the online tool Kaplan−Meier Plotter (http://www.kmplot.com/analysis/) [19] (Fig. 1b). Moreover, our previous meta-analysis of nine retrospective studies that included 2463 TNBC patients revealed that higher AR expression was correlated with a prolonged PFS (HR, 0.70; 95%CI, 0.54−0.89; P = 0.005) [20] (Fig. 1c). Moreover, we extracted the expression data for AR from The Cancer Genome Atlas (TCGA) database, which indicated that tumour tissues exhibited significantly decreased AR expression compared with paired normal tissues in 14 TNBC patients (P = 0.001) (Fig. 1d).
AR expression in TNBC and association with survival. a Correlation between AR expression levels and PFS (log-rank test) in 88 TNBC patients. b Correlation between AR expression levels and RFS (log-rank test) in basal-like breast cancer from Kaplan−Meier Plotter (http://www.kmplot.com/analysis/). Patients were stratified into low and high AR expression based on an auto select best cutoff. c Forest plots of hazard ratios for PFS of AR-positive vs AR-negative groups. d AR mRNA levels from TCGA database, including 14 TNBC tumour and paired normal tissues
Identification of AR negatively induced lncRNA ARNILA
Although AR shows potential prognostic value in TNBC, the mechanisms for this correlation remain unknown. Considering the emerging roles of lncRNAs in EMT, invasion and metastasis of breast cancer [21,22,23,24,25], we hypothesised that AR-regulated lncRNAs play a role in TNBC progression. To identify AR-relevant lncRNAs in TNBC, we therefore determined the expression profiles of lncRNAs in three cell lines (MDA-MB-231, Hs578T, MDA-MB-436) treated with or without dihydrotestosterone (DHT). We identified 21 differentially expressed lncRNAs (15 upregulated and 6 downregulated) (fold change ≥2.0, P < 0.05) (Fig. 2a). Further qRT-PCR analysis confirmed the high-throughput profiling results. The lncRNA ENST00000431557 was significantly decreased in DHT-treated cells compared to control cells (Fig. 2b). Accordingly, we chose this uncharacterised lncRNA for further study and named it ARNILA (androgen receptor negatively induced lncRNA). Fluorescence in situ hybridisation (FISH) assays confirmed that ARNILA was primarily located in the cytoplasm of TNBC cells (Fig. 2c) and tissues (Fig. 2d, upper). Notably, cytoplasmic ARNILA expression was significantly altered by DHT, suggesting that ARNILA could act as a ceRNA to absorb miRNAs. A negative correlation was found between AR and ARNILA expression in 88 TNBC cases (P = 0.003) (Fig. 2d). Furthermore, high ARNILA expression was correlated with poor PFS (HR, 2.72; 95%CI, 1.26−5.70; P = 0.012) in TNBC (Fig. 2e).
Identification of the AR negatively induced lncRNA ARNILA. a Heatmap representation of lncRNA microarray data for MDA-MB-231, Hs578T and MDA-MB-436 cells treated with or without DHT (fold change ≥2.0, P < 0.05). b Expression of ARNILA was assayed by qRT-PCR in MDA-MB-231, Hs578T and MDA-MB-436 cells treated with or without DHT. ARNILA levels were normalised to GAPDH. c FISH images showing cellular localisation of ARNILA in MDA-MB-231 and Hs578T cells. d Upper: representative FISH images for ARNILA and AR in paraffin-embedded tissue of TNBC patients. Lower: the correlation of ARNILA and AR expression in the 88 TNBC patients. e Correlation between ARNILA expression levels and PFS (log-rank test) in 88 TNBC patients. f Upper: The primer pair locations within the ANRILA promoter. Lower: ChIP assay of the enrichment of AR on ARNILA promoter in MDA-MB-231 and Hs578T cells. Positive and negative controls are indicated as input and IgG, respectively. The results are presented as the mean ± SD. *P < 0.05, **P < 0.01
It has been reported that AR could serve as a transcriptional repressor by binding to the promoters of target genes [26]. Increased binding of AR to the promoter of ARNILA was observed by chromatin immunoprecipitation (ChIP) assays (Fig. 2f). Two binding sites contain the sequence corresponding to the core motifs of the androgen response element, suggesting that AR may be a transcription factor that downregulates ARNILA. Rapid amplification of cDNA ends (RACE) assays showed that ARNILA is composed of two exons with a full length of 930nt located on chromosome 2 (Supplementary Figure S1).
ARNILA functions as a ceRNA for miR-204 to facilitate Sox4 expression
To test the hypothesis that ARNILA could act as a ceRNA to regulate cancer progression, we first predicted the potential binding sites for 2588 human miRNAs [27] on ARNILA by miRanda [28] and next identified the TNBC survival-associated miRNAs by Kaplan−Meier Plotter [29]. The bioinformatics analysis revealed the top ten survival-associated miRNAs that could bind to ARNILA according to the predicted max binding score (Fig. 3a). From this analysis, miR-204 was shown to bind to ARNILA and also had a positive correlation with a prolonged overall survival (OS) in systemically untreated TNBC patients (HR, 0.31; 95%CI, 0.09−1.00; P = 0.045) (Fig. 3b). Furthermore, Sox4, which is involved in EMT in advanced breast cancer [30], emerged as a validated target of miR-204 [31,32,33,34]. We then performed dual-luciferase reporter assays to confirm the regulatory relationships among ARNILA, miR-204 and Sox4. The luciferase activity of the ARNILA-3′ untranslated region (UTR)-Wt was significantly inhibited by transfection of miR-204 mimics, while ARNILA-3′UTR-Mut (miR-204) did not show a response to miR-204 mimics (P < 0.05) (Fig. 3c). The luciferase activity of Sox4-3’UTR-Wt cells, but not Sox4-3′UTR-Mut (miR-204) cells, was significantly inhibited by transfection with miR-204 mimics (P < 0.05) (Fig. 3d). The luciferase reporter assays demonstrated direct interactions between miR-204 and ARNILA, and miR-204 and Sox4. Notably, the same binding sites shared by ARNILA and Sox4 on miR-204 indicate that ARNILA functions as a ceRNA for miR-204 to facilitate Sox4 expression. Furthermore, we found that miR-204 inhibitor strongly increased ARNILA levels compared with miR-NC, as shown by qRT-PCR (Supplementary Figure S2a). A significant reduction of Sox4 protein in cells transfected with miR-204 mimics was demonstrated by western blot assays (Fig. 3e). Conversely, miR-204 inhibitor transfection resulted in a strong increase in Sox4 protein level (Fig. 3f). ARNILA knockdown decreased Sox4 protein expression, while ARNILA overexpression increased Sox4 protein expression (Supplementary Figure S2b-c, Fig. 3g). Notably, mutations in the miR-204-binding site on ARNILA abolished its regulatory effect on Sox4 (Fig. 3h). Given the regulatory efficiency of ARNILA in Sox4, we hypothesised that ARNILA could also affect other miR-204 targets. Similarly, ARNILA knockdown decreased additional four miR-204 target proteins (BCL2, RAB22A, SIRT1, and FOXA1), while ARNILA overexpression increased the expression of these proteins (Supplementary Figure S3). Although RUNX2 was also shown to be a validated targeted gene of miR-204, its expression was not altered by ARNILA knockdown or overexpression (Supplementary Figure S3). We hypothesised that ectopic expression or knockdown of ARNILA exerted a limited change of miR-204, which is insufficient to repress all target genes at the same time. Additionally, RUNX2 expression could be controlled by other regulatory mechanisms that overwhelmed the effect of the miR-204 alteration. Collectively, these data indicate that ARNILA might function as a molecular sponge of miR-204 to upregulate Sox4 expression.
ARNILA upregulates Sox4 via binding to miR-204. a The top ten putative miRNAs binding to ARNILA sorted according to the max score predicted by miRanda software. The association between miRNA expression and OS in untreated TNBC patients was derived from Kaplan−Meier Plotter (log-rank test). b Correlation between AR expression levels and OS (log-rank test) in TNBC patients from Kaplan−Meier Plotter. Patients were stratified into low and high AR expression based on auto select best cutoff. c Left: putative miR-204 MREs in the 3′UTR of ARNILA generated by miRanda software. The seed sequences are shown in bold and mutant sequences shown in red. Right: luciferase activity of ARNILA-Wt and ARNILA-Mut upon transfection of the indicated miRNA mimics in 293T cells. Data are presented as the ratio of Renilla luciferase activity to firefly luciferase activity. d Left: putative miR-204 MREs in the 3′UTR of Sox4 generated by the TargetScan database. The seed sequences are shown in bold and mutant sequences shown in red. Right: luciferase activity of Sox4-Wt and Sox4-Mut upon transfection of the indicated miRNA mimics in 293T cells. Data are presented as the ratio of Renilla luciferase activity to firefly luciferase activity. e, f Left: western blotting assay of Sox4 expression in MDA-MB-231 and Hs578T cells following miR-204 mimic (e) or inhibitor (f) transfection. Right: quantification of protein levels shown in the left panel. g Upper: western blotting assay of Sox4 expression in MDA-MB-231 and Hs578T cells after transfection with the indicated plasmid. Lower: quantification of protein levels shown in the upper panel. h Upper: western blotting assay of expression of Sox4 in Hs578T cells after transfection with the indicated plasmids. Lower: quantification of protein levels shown in the upper panel. The results are presented as the mean ± SD. *P < 0.05. NS indicates no significant difference
Sox4 is inversely correlated with AR and predicts poor clinical outcome in TNBC
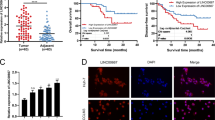
The lncRNA ARNILA was downregulated by AR, indicating that AR is upstream of Sox4 in the pathway. DHT significantly suppressed Sox4 protein level by activating AR in TNBC cells, whereas ARNILA overexpression abolished this suppressive effect (Fig. 4a), indicating that AR negatively regulates Sox4 by inhibiting ARNILA. We analysed RNA-seq data of TCGA database by comparing the AR and Sox4 mRNA levels and found that Sox4 was inversely correlated with AR in 151 TNBC patients (P = 0.028) (Fig. 4b). Moreover, the patients with high Sox4 expression showed a significantly reduced RFS compared with that of patients with low Sox4 expression in TNBC by Kaplan−Meier Plotter [29] (HR, 1.50; 95%CI, 1.16−1.93; P = 0.002) (Fig. 4c). To further examine the correlation between AR and Sox4, we analysed the protein expression in 88 TNBC tissue samples (Fig. 4d). Sox4 expression was negatively correlated with AR expression (P = 0.045) (Fig. 4d). Furthermore, Sox4 was correlated with poor PFS (HR, 2.34; 95%CI, 1.34_6.32; P = 0.008) (Fig. 4e). Patients with different Sox4 and AR levels showed significant differences in PFS (P = 0.009), and notably, patients with high Sox4 and low AR exhibited a substantially shorter PFS than those with low Sox4 and high AR (HR, 0.00; 95%CI, 0.07−0.57; P = 0.003) (Fig. 4f). Taken together, these data serve as additional evidence confirming the regulation of Sox4 by ARNILA. Moreover, the findings indicate that AR has an impact on TNBC clinical outcome by negatively regulating Sox4.
Sox4 is inversely correlated with AR and better survival. a Left: western blotting assay of expression of AR and Sox4 in MDA-MB-231 and Hs578T cells following the indicated treatment. Middle and right: quantification of protein levels shown in the left panel. The results are presented as the mean ± SD. *P < 0.05. b AR and Sox4 expression from TCGA database of 151 TNBC patients. c Correlation between Sox4 expression levels and RFS (log-rank test) in TNBC patients from Kaplan−Meier Plotter. Patients were stratified into low and high Sox4 expression based on auto select best cutoff. d Upper: representative IHC images for AR and Sox4 in paraffin-embedded tissue of TNBC patients. Lower: the correlation of AR and Sox4 expression in the 88 TNBC patients. e Correlation between Sox4 levels and PFS (log-rank test) in 88 TNBC patients. f Correlation between AR-Sox4 combined levels and PFS (log-rank test) in 88 TNBC patients
ARNILA promotes migration, invasion and EMT in vitro
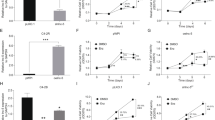
Sox4 has been shown to induce the EMT programme [17, 18], and therefore, we hypothesised that ARNILA might also play a role in EMT. We knocked down ARNILA in MDA-MB-231 cells and overexpressed it in Hs578T cells to further investigate the functions of ARNILA in TNBC progression. ARNILA knockdown significantly decreased the migration of MDA-MB-231 cells, and ARNILA overexpression accelerated the speed of wound closure in Hs578T cells (Fig. 5a). Moreover, silencing ARNILA inhibited invasion, while ectopic expression of ARNILA significantly enhanced invasion as shown by transwell invasion assays (Fig. 5b). MDA-MB-231 cells transfected with a shARNILA plasmid showed significantly increased expression of the epithelial marker E-cadherin, along with a prominent decrease in the mesenchymal marker N-cadherin. Conversely, the gain-of-function study in Hs578T cells revealed that ARNILA overexpression reduced the epithelial marker E-cadherin, along with increased expression of the mesenchymal marker N-cadherin (Fig. 5c). In summary, these data suggest that ARNILA is a positive regulator of migration, invasion and EMT in TNBC cells.
ARNILA promotes migration, invasion and EMT in vitro. a Upper: wound healing assays of MDA-MB-231 and Hs578T cells following the indicated transfections. Lower: quantification of wound closure for cells in the upper panel. b Upper: transwell assays of MDA-MB-231 and Hs578T cells following the indicated transfections. Lower: quantification of migration by cells in the lower panel. c Left: western blotting assay of expression of E-cadherin and N-cadherin in MDA-MB-231 and Hs578T cells following the indicated treatments. Middle and right: quantification of protein levels shown in the left panel. The results are presented as the mean ± SD. *P < 0.05
Sox4 is responsible for ARNILA-mediated migration, invasion and EMT in vitro
We further investigated the mechanisms by which ARNILA promoted malignant processes. Given that ARNILA functions as a ceRNA to upregulate Sox4 expression, we performed functional rescue experiments to determine whether ARNILA promoted tumour progression in a Sox4-dependent manner. Plasmids with shNC (short hairpin RNA of normal control) or shARNILA were transfected into MDA-MB-231 cells that stably ectopically expressed Sox4 or the control, whereas Hs578T cells that stably ectopically expressed ARNILA or the control were transfected with shNC or shSox4 plasmids. Notably, restoration of Sox4 rescued the wound healing rate and invasive capacity in ARNILA-knockdown MDA-MB-231 cells, while Sox4 knockdown significantly abolished ARNILA-mediated migration and invasion in Hs578T cells (Fig. 6a, b). Moreover, ARNILA knockdown failed to suppress the EMT programme when Sox4 was ectopically expressed in MDA-MB-231 cells, while ARNILA overexpression could not restore the EMT process when Sox4 was knocked down in Hs578T cells with regard to the alteration of EMT markers (Fig. 6c). In contrast to ARNILA, miR-204 has a suppressive effect on migration and invasion, as anti-miR-204 significantly enhanced migration and invasion (Supplementary Figure S4). Moreover, decreased miR-204 restored the ARNILA knockdown-mediated repression of migration and invasion (Supplementary Figure S4). Taken together, our findings indicate that Sox4 is responsible for ARNILA-mediated migration, invasion and EMT in TNBC cells.
Sox4 is responsible for ARNILA-mediated migration, invasion and EMT in vitro. a Left: wound healing assays of MDA-MB-231 and Hs578T cells following the indicated transfections. Right: quantification of wound closure for cells in the left panel. b Lower: transwell assays of MDA-MB-231 and Hs578T cells following the indicated transfections. Upper: quantification of migration by cells in the lower panel. c Left: western blotting assays of expression of E-cadherin and N-cadherin in MDA-MB-231 and Hs578T cells following the indicated treatments. Right: quantification of protein levels shown in the left panels. The results are presented as the mean ± SD. *P < 0.05. NS indicates no significant difference
ARNILA promotes breast cancer metastasis in vivo
In addition, we investigated the function of ARNILA in metastasis of breast cancer in vivo by injection of highly aggressive MDA-MB-231 cells that harboured shARNILA or shNC into the tail vein. As shown in Fig. 7a, PET/CT imaging revealed that injection of cells harbouring shNC resulted in lung and liver metastasis in BALB/c mice, whereas lung and liver metastases were significantly reduced in the mice injected with cells harbouring shARNILA. Histological examination confirmed the significantly reduced lung and liver metastases in the shARNILA group compared to the control group (lung, P = 0.013; liver, P = 0.050) (Fig. 7b, c). These data suggest that ARNILA contributes to TNBC metastasis and that silencing of ARNILA inhibits lung and liver metastases in mouse transplant models.
ARNILA promotes metastasis in vivo. a Representative PET/CT images of mice at 1 h after 18F-FDG injection. Images were collected 5 weeks after cell injection. Lung and liver metastases are indicated by white arrows. b Representative images of metastatic nodules in lungs and livers by isolated lung bright-field imaging (top) and haematoxylin and eosin staining (middle and bottom). Lung and liver metastases are indicated by black arrows. c The incidence of lung and liver metastasis in two groups (Fisher’s exact t test). d A schematic diagram of an ARNILA-based signalling circuit in TNBC. In AR-positive TNBC (left), AR significantly represses ARNILA by acting as a transcriptional repressor; hence, ARNILA’s sponge-like function is weak here because of its low expression level, and miR-204 is released to inhibit Sox4. In AR-negative TNBC (right), miR-204 is mostly consumed by ARNILA due to the reduced repression by AR, and Sox4 is consequently expressed at high levels
Discussion
The clinical management of TNBC is challenging because of its relatively aggressive biological behaviour and the lack of effective therapeutic targets. AR has been reported to be expressed in a subset of TNBC patients and implicated in predicting clinical outcome. Emerging evidence has demonstrated that the AR signalling pathway functions in cancer processes. However, the underlying mechanisms by which AR functions in tumour progression as well as in predicting prognosis in TNBC remain unclear. In this study, we identified the lncRNA ARNILA, which was transcriptionally repressed by AR, and showed that it was a promising therapeutic target that was functionally required for invasion and metastasis of TNBC. ARNILA promoted EMT by competitively binding to miR-204, leading to the upregulation of Sox4. In AR-positive tumours, ARNILA was apparently suppressed by AR, resulting in the decreased adsorption of miR-204, thereby releasing miR-204 to inhibit Sox4 expression. Reciprocally, in AR-negative breast cancer, activated ARNILA promoted Sox4 expression by increasing the sequestration of miR-204 (Fig. 7d). It has been reported that Sox4 is not only required but also sufficient for inducing EMT and metastatic spread in breast cancer [18]. Therefore, ARNILA was demonstrated to function in metastasis of breast cancer.
In recent years, the roles of lncRNA in human diseases such as cancer have attracted increased attention. In breast cancer, several lncRNAs have been shown to have prognostic value or even be therapeutic options for patients. Reduced expression of NKILA, an lncRNA induced by nuclear factor kB (NFkB), in response to inflammatory signalling, was linked to metastatic dissemination and poor prognosis in breast cancer [21], suggesting that NKILA serves as a tumour suppressor. In contrast, overexpression of the lncRNA HOTAIR was highly predictive of poor metastasis-free survival or OS in early-stage surgically resected breast cancer. Enforced expression of HOTAIR increased the tumour invasiveness and metastasis of breast cancer cell lines [11]. Knockdown of MALAT1 led to slower tumour growth and reduced migration and metastasis in mouse mammary carcinoma model [35], indicating that HOTAIR and MALAT1 are potential therapeutic targets for breast cancer. Herein, we identified an uncharacterised lncRNA, ARNILA, that was correlated with decreased PFS in TNBC patients. Silencing ARNILA effectively inhibited cancer migration, invasion and metastasis, whereas enforced ARNILA expression promoted tumour progression. Therefore, ARNILA is probably another reliable prognostic biomarker and therapeutic target of TNBC patients.
Several mechanisms illustrating the regulation of cancer cellular process by lncRNA have been proposed, and the cellular location of lncRNA is important to its biological function. Generally, cytoplasmic lncRNAs influence cellular signalling cascades and modulate mRNA stability or translation, whereas nuclear lncRNAs are responsible for RNA processing, transcriptional regulation and chromatin interactions [36]. Increasing evidence has shown that cytoplasmic lncRNAs can act as “RNA sponges” or ceRNAs to sequester miRNAs and thereby reduce their regulatory effect on target mRNAs [37]. For example, lncARSA promoted sunitinib resistance by acting as a ceRNA via competitively binding miR-34/miR-449 to regulate AXL and c-MET in RCC cells [15]. Additionally, HOTAIR could sponge miR-331-3p to facilitate HER2 expression in gastric cancer by functioning as a ceRNA [38]. In our study, we identified the subcellular localisation of ARNILA and found that ARNILA predominately resided in the cytoplasm; thus, ARNILA might function as a ceRNA in TNBC. Subsequent bioinformatics analysis revealed an miRNA response element (MRE) of miR-204 shared by ARNILA and Sox4, and luciferase assays demonstrated that ARNILA and Sox4 both contained functional miR-204 binding sites. A positive regulatory relationship between ARNILA and Sox4 was observed by western blot analyses. Moreover, ARNILA was demonstrated to alter the expression of other miR-204 targets, including BCL2, RAB22A, SIRT1, and FOXA1. These results suggested the molecular sponge-like effect of ARNILA in facilitating expression of Sox4. However, appropriate transcript copy numbers for miRNA and miRNA target site abundance might be required to exert the ceRNA function [39]. We performed the absolute qRT-PCR analysis to quantify the copy numbers of ARNILA and miR-204 in TNBC cells overexpressed ARNILA or the control. Comparable copy numbers of ARNILA and miR-204 were detected per cell in the control group, which is crucial for ceRNA−miRNA interaction. ARNILA overexpression significantly increased the copy numbers of ARNILA and decreased the copy numbers of miR-204 (Supplementary Figure S5), which probably led the miRNA target derepression at the high threshold of added MREs [39]. Further quantitative analyses of ARNILA, miR-204 and transcriptome-wide miR-204 targets might be needed to further verify the ceRNA hypothesis. Notably, in addition to the cytoplasm, ARNILA could also be located in the nucleus of TNBC cells, indicating other possible regulatory mechanisms of ARNILA in cellular biological processes, which require further validation in future work.
In conclusion, our findings first showed that AR negatively regulated the lncRNA ARNILA and provided a comprehensive annotation of this lncRNA in TNBC. We demonstrated that ARNILA was correlated with poor clinical outcome by modulating migration, invasion, EMT and metastasis. Therefore, ARNILA can act not only as a prognostic biomarker but also as a therapeutic target for TNBC patients.
Materials and methods
Tissue samples
Tissue samples were collected from 88 TNBC patients in The First People’s Hospital of Lianyungang and The First Affiliated Hospital of Wenzhou Medical University, China. Inclusion criteria were female, age of 18 years or older, no distant metastasis at the time of diagnosis, and no neoadjuvant chemotherapy.
Cell lines and culture
MDA-MB-231 and Hs578T cells were cultured in RPMI 1640 and DMEM medium (Gibco, USA), respectively, supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin in a 37 °C incubator with 5% CO2. MDA-MB-436 cells were cultured in L-15 medium (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin in a 37 °C incubator without CO2. DHT (Sigma, USA) was dissolved in ethanol, and cells were treated with DHT at 100 nM for 24 h. The same concentration of ethanol was used as the negative control treatment in parallel to DHT treatment.
Plasmids and cell transfection
Plasmids were transfected into cells by Lipofectamine 2000 (Invitrogen, USA) according to the instructions. The plasmids pGpU6/GFP/Neo-ARNILA, pGpU6/GFP/Neo-Sox4, pEX-2 (pGCMV/MCS/IRES/EGFP/Neo)-ARNILA, pEX-2-Sox4, LV5 (LV5-EF1a-GFP/Puro)-Sox4, LV5 (LV5-EF1a-GFP/Puro)-ARNILA and LV3 (pGLV-h1-GFP-Puro)-ARNILA were purchased from GenePharma (Shanghai, China). Sequences were as follows: shARNILA1: 5′-GGTGCAGTTGCTATGGCAATG-3′, shARNILA2: 5′- GAAGAGGCATTCGGTGGATTC-3′, shARNILA3: 5′-GGAGTCAGCTGATGGGTTACA-3′, shARNILA4: 5′-GCAATCGCACAAAGCAATTGT-3′; shSox4: 5′-TGGGCACATCAAGCGACCCAT-3′. The plasmid shARNILA3 was chosen for further study based on the silencing effect.
RNA extraction, reverse transcription, and real-time RT-PCR
Total RNA was extracted by TRIzol reagent (Invitrogen) and reverse-transcribed with a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). Primers were synthesised by Realgene (Nanjing, China) and include the following: forward ARNILA: 5′-AGGAGGCTGCAGGATGTA-3′, reverse ARNILA: 5′-TATGGTTGGAGTCTTCTCCC-3′; forward GAPDH 5′-AGAAGGCTGGGGCTCATTTG-3′, reverse GAPDH 5′-AGGGGCCATCCACAGTCTTC-3′; forward miR-204-5p: 5′-TTCCCTTTGTCATCCTATGCCT-3′, reverse miR-204-5p: 5′- CCGACCGTTATATCGACATCTC-3′; forward U1: 5′-GGGAGATACCATGATCACGAAGGT-3′, reverse U1: 5′-CCACAAATTATGCAGTCGAGTTTCCC-3′. Real-time PCR reactions were conducted with SYBR Premix Ex Taq™ II (TaKaRa). GAPDH was used as the endogenous control.
Microarray analysis
LncRNA expression profiling was performed by an Arraystar Human LncRNA Microarray V3.0 platform (Agilent Technologies). Differentially expressed lncRNAs were identified by P value/false discovery rate filtering.
ChIP assay
ChIP assays were performed by a Pierce Agarose ChIP Kit (Thermo). Sequences of primers 1−10 used for ChIP are listed as follows. Forward (5′-3′): CCACAGTTAGCACAGGGAC, TCCCATTTTTTTTCCCCAATGGAAG, TGCACGGGCTGACTCCACAAA, GGCCATGTGCTTGCCGAC, ACCACTCCAATCACACACAC, GTTTTCTTAAAGAAAATCTTAAAACAAAC, CCCACCACAAGGAGCCCTCGAATTG, ACCCTTTGCGCAGAAGGCAACG, GACAGCTCAGTGGCTGGG, GAGTAGAAAATAAAAGGTCTGACCG; reverse (5′-3′): CAGTGAACCTCACCTATTGAATAAACC, TTACCTCTGTTGCCCTGATTGC, CCAAGTCCACGTCCCTAGAAAG, GTGTATTTCTTTTCTAGTACTTGGTCTG, CAAAAACCCAACACCCTTCCC, ATCAGAACTAATCCAGAGTG, TTCACCAAGGATGGAGAAAGGGAGACAGC, GTGCCAGCACCATTCCTGGTC, TTTGGGTTTTGTTTCTTGCTGTGACTG, ACATGGGGGCAAACTGAGG.
RACE assay
RACE was conducted by using a SMARTer RACE cDNA Amplification Kit (Clontech, Palo Alto, CA). The primers used for the nested-PCR are listed as follows: 5′ RACE-outer: 5′-CGTGTCCGGCATGTAACCCATCAG-3′, 5′ RACE-inner: 5′-GGGAATCCACCGAATGCCTCTTCA-3′; 3′ RACE-outer: 5′-AGATGTTGTCTCTGCAGCACAAGGTAGG-3′, 3′ RACE-inner: 5′-GGTGCAGTTGCTATGGCAATGCCTACAG-3′.
Immunohistochemistry (IHC) and FISH
Tissue sections were deparaffinized and rehydrated, followed by antigen retrieval. For IHC, after primary anti-AR (ab198394, Abcam), anti-Sox4 (ab80261, Abcam) and secondary antibody incubation, the sections were incubated with diaminobenzidine (Sigma) and counterstained with haematoxylin (Sigma). For FISH, the probes of AR and ARNILA were synthesised by GenePharma (Shanghai, China).
Luciferase reporter assay
The wild-type or mutated sequences within the predicted binding sites of the 3′UTR sequence of ARNILA and Sox4 were synthesised, inserted into a luciferase reporter plasmid (GENE, Shanghai, China), and transfected into 293T cells. The luciferase activity was normalised to Renilla luciferase activity after 48 h of transfection.
Western blot
The primary antibodies used include anti-AR (ab108341, Abcam), anti-Sox4 (ab80261, Abcam), anti-E-cadherin (ab40772, Abcam), anti-N-cadherin (ab76011, Abcam), anti-BCL2 (ab32124, Abcam), anti-FOXA1 (ab170933, Abcam), anti-RAB22A (ab137093, Abcam), anti-SIRT1 (ab32441, Abcam), anti-RUNX2 (ab23981, Abcam) and anti-GAPDH (10494-1-AP, Proteintech, USA) antibodies. The secondary antibody was anti-rabbit IgG-HRP (ab6721, Abcam).
Migration and invasion assays
Cells were seeded in six-well plates and grown to 90% confluence, and then, a wound was scratched in the cell monolayer with a 200 μL sterile pipette tip. Cells were cultured in serum-free medium after removing floating cells with PBS. We photographed the edge of a scratch at time 0 and 24 h. Transwell chambers (Corning, USA) were coated with 10 μL of RPMI 1640 or DMEM-diluted Matrigel (BD Biosciences, USA), and cancer cells (2×104) cultured in serum-free medium (200 μL) were added to the upper chamber. The medium (800 μL) containing 10% FBS was added to the lower chamber. The cells in the upper side of the chamber were carefully removed with a cotton swab after 24 h incubation. The migrating cells were fixed with 100% methanol for 30 min and then stained with crystal violet for 10 min.
Tumour xenografts
Animal studies were performed with approval of the Jinling Hospital Animal Care and Use Committee. Female BALB/c nude mice aged 4 weeks were injected with MDA-MB-231 cells (1×107) with stable knockdown of control or ARNILA vectors via tail veins. 18F-FDG PET/CT was performed at 8 weeks after tumour cell injection to monitor the lung and liver metastasis. After PET/CT imaging, all mice were sacrificed to harvest the lung and liver tissues for haematoxylin and eosin staining.
Statistical analysis
Statistical analysis was performed by SPSS (v21), and the figures were plotted by GraphPad Prism software (v6). Student t test and Fisher’s exact t test were used to examine statistical significance of differences between groups. P values less than 0.05 were considered statistically significant, and all analyses were two sided. Data are presented as the mean ± SD.
References
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48.
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–33.
Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33:3225–34.
Ricciardi GR, Adamo B, Ieni A, Licata L, Cardia R, Ferraro G, et al. Androgen receptor (AR), E-cadherin, and Ki-67 as emerging targets and novel prognostic markers in triple-negative breast cancer (TNBC) patients. PLoS ONE. 2015;10:e0128368.
Thike AA, Yong-Zheng Chong L, Cheok PY, Li HH, Wai-Cheong Yip G, Huat Bay B, et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Mod Pathol. 2014;27:352–60.
Tang D, Xu S, Zhang Q, Zhao W. The expression and clinical significance of the androgen receptor and E-cadherin in triple-negative breast cancer. Med Oncol. 2012;29:526–33.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9.
Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–8.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–8.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93.
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–68.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial−mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, et al. SOX4 induces epithelial−mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–608.
Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–83.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Wang Y, Yang F, Wang Y, Zhong F, Guan X. Prognostic role of androgen receptor expression in triple-negative breast cancer. J Clin Oncol. 2015;33:(suppl; abstr 1076).
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81.
Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J, et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454.
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287.
Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Korner C, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;8:1052–64.
Latorre E, Carelli S, Raimondi I, D’Agostino V, Castiglioni I, Zucal C, et al. The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial−mesenchymal transition in breast cancer. Cancer Res. 2016;76:2626–36.
Liu YN, Liu Y, Lee HJ, Hsu YH, Chen JH. Activated androgen receptor downregulates E-cadherin gene expression and promotes tumor metastasis. Mol Cell Biol. 2008;28:7096–108.
Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73.
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363.
Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–46.
Parvani JG, Schiemann WP. Sox4, EMT programs, and the metastatic progression of breast cancers: mastering the masters of EMT. Breast Cancer Res. 2013;15:R72.
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–9.
Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS ONE. 2014;9:e101457.
Yin JJ, Liang B, Zhan XR. MicroRNA-204 inhibits cell proliferation in T-cell acute lymphoblastic leukemia by down-regulating SOX4. Int J Clin Exp Pathol. 2015;8:9189–95.
Wu D, Pan H, Zhou Y, Zhang Z, Qu P, Zhou J, et al. Upregulation of microRNA-204 inhibits cell proliferation, migration and invasion in human renal cell carcinoma cells by downregulating SOX4. Mol Med Rep. 2015;12:7059–64.
Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52.
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92.
Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–76.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81773102, No. 81470357) and a Foundation for Clinical Medicine Science and Technology Special Project of the Jiangsu Province, China (No. BL2014071) (to XG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yang, F., Shen, Y., Zhang, W. et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ 25, 2209–2220 (2018). https://doi.org/10.1038/s41418-018-0123-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0123-6
This article is cited by
-
Metformin and long non-coding RNAs in breast cancer
Journal of Translational Medicine (2023)
-
Identification and validation of telomerase related lncRNAs signature to predict prognosis and tumor immunotherapy response in bladder cancer
Scientific Reports (2023)
-
In Vitro and In Vivo Comparative Analysis of Differentially Expressed Genes and Signaling Pathways in Breast Cancer Cells on Interaction with Mesenchymal Stem Cells
Applied Biochemistry and Biotechnology (2023)
-
Targeted OUM1/PTPRZ1 silencing and synergetic CDT/enhanced chemical therapy toward uveal melanoma based on a dual-modal imaging-guided manganese metal–organic framework nanoparticles
Journal of Nanobiotechnology (2022)
-
Role of androgen receptor signaling pathway-related lncRNAs in the prognosis and immune infiltration of breast cancer
Scientific Reports (2022)